Abstract
The primary macrophage growth factor, colony-stimulating factor-1 (CSF-1), is homodimeric and exists in 3 biologically active isoforms: a membrane-spanning, cell-surface glycoprotein (csCSF-1) and secreted glycoprotein (sgCSF-1) and proteoglycan (spCSF-1) isoforms. To investigate the in vivo role of the chondroitin sulfate glycosaminoglycan (GAG) chain of spCSF-1, we created mice that exclusively express, in a normal tissue-specific and developmental manner, either the secreted precursor of spCSF-1 or the corresponding precursor in which the GAG addition site was mutated. The reproductive, hematopoietic tooth eruption and tissue macrophage defects of CSF-1-deficient, osteopetrotic Csf1op/Csf1op mice were corrected by transgenic expression of the precursors of either sgCSF-1 or spCSF-1. Furthermore, in contrast to the transgene encoding csCSF-1, both failed to completely correct growth retardation, suggesting a role for csCSF-1 in the regulation of body weight. However, spCSF-1, in contrast to sgCSF-1, completely resolved the osteopetrotic phenotype. Furthermore, in transgenic lines expressing different concentrations of sgCSF-1 or spCSF-1, spCSF-1 more efficiently corrected Csf1op/Csf1op defects of tooth eruption, eyelid opening, macrophage morphology, and B-cell deficiency than sgCSF-1. These results indicate an important role of the CSF-1 chondroitin sulfate proteoglycan in in vivo signaling by secreted CSF-1. (Blood. 2006;107:786-795)
Introduction
The effects of the primary regulator of macrophage survival, proliferation, and differentiation, colony-stimulating factor-1 (CSF-1), also known as macrophage colony-stimulating factor, are mediated by the CSF-1 receptor (CSF-1R), encoded by the c-fms proto-oncogene.1-4 There are 3 biologically active, homodimeric isoforms of CSF-1: a secreted glycoprotein (sgCSF-1),5-7 a secreted proteoglycan (spCSF-1),8,9 and a membrane-spanning, cell-surface glycoprotein (csCSF-1)10-13 (reviewed in Pixley and Stanley14 ). Circulating CSF-1 (spCSF-1 and sgCSF-1) is thought to be synthesized primarily by endothelial cells and selectively maintains certain macrophage populations, including Kupffer cells.15,16 Local synthesis of csCSF-1 is sufficient for the regulation of several other tissue macrophage populations.17 However, it is likely that the many different tissue cell types that synthesize CSF-1 synthesize all 3 CSF-1 isoforms and that the secreted isoforms also regulate locally (reviewed in Pixley and Stanley14 ).
The secreted and cell-surface CSF-1 isoforms are respectively derived from full-length and truncated homodimeric membrane-spanning precursors. The amino-terminal 150 amino acids of both types of precursor are identical and when linked by an interchain disulfide bond, as in the full-length proteins, are sufficient for in vitro biologic activity. The remainder of the precursor sequences determines how the isoforms are processed and expressed. Differential proteolysis from the full-length precursor in the secretory vesicle yields the secreted isoforms: the secreted glycoprotein by N-terminal cleavage and the secreted proteoglycan, containing a longer polypeptide chain on which a single 18-kDa chondroitin sulfate glycosaminoglycan (GAG) chain per monomeric subunit is attached, by C-terminal cleavage. Both spCSF-1 and sgCSF-1 accumulate rapidly in the extracellular milieu after fusion of the secretory vesicle with the plasma membrane. In contrast, csCSF-1 is derived from the truncated precursor encoded by an alternatively spliced mRNA in which the regions encoding the proteolytic cleavage and GAG addition sites are spliced out. Following fusion of the vesicle with the plasma membrane, this uncleaved precursor is expressed stably on the cell surface as a membrane-spanning glycoprotein. All 3 isoforms are glycosylated with N- and O-linked carbohydrates (reviewed in Pixley and Stanley14 ).
Mice homozygous for the mutation osteopetrotic (Csf1op) bear an inactivating mutation in the CSF-1 gene and are CSF-1 deficient.18-20 These Csf1op/Csf1op mice are osteoclast deficient, toothless, and have a low growth rate and defects in fertility and neural development21 (reviewed in Pixley and Stanley14 ; Pollard and Stanley22 ; and Cohen et al23 ). They are also deficient in tissue macrophages.16,20,24 The restoration of circulating CSF-1 in Csf1op/Csf1op mice by daily subcutaneous injection of CSF-1 selectively corrects some,16,25-27 but not all,16 of the osteopetrotic defects. However, essentially complete correction was observed by expression of a transgene (TgC) encoding the full-length CSF-1 precursor driven by a 3.13-kb Csf1 promoter and the first intron that conferred normal tissue-specific and developmental expression.28 These results suggested that CSF-1 regulates CSF-1R-expressing cells both locally and humorally. This conclusion was confirmed by transgenic expression of csCSF-1 using the same driver (TgCS transgene). In contrast to either CSF-1 injection or TgC expression, Csf1op/Csf1op mice expressing TgCS were devoid of circulating CSF-1. In contrast to TgC expression, TgCS expression only partially rescued the Csf1op/Csf1op phenotype.17
These studies with the TgCS transgenic mice emphasized the importance of the secreted isoforms, sgCSF-1 and spCSF-1. To directly investigate the contribution of these isoforms and to examine the role of the chondroitin sulfate GAG chain in vivo, we created mice that exclusively express, in a normal tissue-specific and developmental pattern, the secreted precursors of either sgCSF-1 or spCSF-1. We report significant differences in the phenotypes of these mice that indicate an important in vivo role for the CSF-1 proteoglycan.
Materials and methods
Mouse generation, maintenance, and genotyping
Mutant and transgenic mice were maintained on the FVB/NJ background as described previously.17 The plasmid bearing the full-length CSF-1 cDNA driven by CSF-1 promoter and the first intron28 was used in the construction of the secreted proteoglycan precursor (TgSPP) and the secreted glycoprotein precursor (TgSGP) transgenes. For the TgSPP construct, the BamHI restriction site in the cDNA just before the transmembrane domain (TM) was mutated by partial digestion with BamHI and blunting by Klenow polymerase to generate an insertion of 4 nucleotides with a resultant frameshift and a premature termination codon (S454I-D455L-P456-STOP) just prior to the TM. For the TgSGP construct, the BamHI restriction site in intron 1 was eliminated by partial BamHI digestion and blunting and the resultant plasmid digested with BamHI and HindIII (partial) to excise the BamHI-HindIII fragment within the coding region that contains the GAG addition site. The excised fragment was replaced with the corresponding polymerase chain reaction (PCR)-amplified fragment mutated (S276L-G277A) at the GAG addition site (SGXG/A). This plasmid was then digested with BamHI and blunted to introduce the S454I-D455L-P456-STOP mutation into the final TgSGP construct, as described for TgSPP. The production of transgenic mice and genotyping were carried out as described for the full-length (TgN(FLCsf1)Ers)28 and cell-surface (TgN(CSCsf1)Ers)17 transgenic mice. The sppCSF-1 and sgpCSF-1 transgenes were designated as TgN(SPPCsf1)Ers (here abbreviated to TgSPP) and TgN(SGPCsf1)Ers (here abbreviated to TgSGP), according to the Mouse Nomenclature Rules and Guidelines.67 To determine transgene homozygosity by real-time quantitative PCR, mouse DNA was PCR-amplified using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and primer pairs that amplified a 54-nucleotide sequence at the exon 4-exon 5 junction of the transgene and a 68-nucleotide sequence of the mouse connective tissue growth factor gene as internal standard.
X-ray analysis, CSF-1 RIA, immunoblotting, histochemistry, immunohistochemistry, and flow cytometry
Radiographs and CSF-1 radioimmunoassays29,30 were carried out as described previously.17 To identify the spCSF-1, sgCSF-1, and csCSF-1 isoforms expressed in cells, cloned, immortalized skin fibroblast lines31 were prepared from different mouse lines, and the conditioned medium (CM) was harvested at 6 to 8 days of confluent culture in 1% fetal bovine serum (Invitrogen, Carlsbad, CA) in alpha MEM (Sigma, St Louis, MO) for analysis of the secreted CSF-1. Cell lysates were prepared8 for analysis of csCSF-1. CM was concentrated approximately 30-fold by ultrafiltration and the concentrated CM (∼ 0.7 μg of CSF-1) precleared (16 hours, 2 mg/mL) with anti-rat IgG-coupled beads (Pierce, Rockford, IL) and incubated (16 hours, 1 mg/mL) with rat anti-mouse CSF-1 monoclonal antibody YYG10629 -coupled beads, prior to elution with sodium dodecyl sulfate (SDS) sample buffer without beta-mercaptoethanol. The eluted proteins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) in a 4% to 20% acrylamide gradient in the absence of reducing agent, transferred (14 hours at 30 V, then 6 hours at 70 V) to a PVDF membrane using 25 mM Tris-Glycine, 20% methanol without SDS, and Western blotted with a rabbit anti-murine polyclonal CSF-1 antibody (a gift from Cetus, Emeryville, CA). The histochemical staining for tartrate-resistant acid phosphatase (TRAP) in femoral sections was carried out as described.32-34 For eye histology, heads were fixed (48 hours) in 10% buffered formalin decalcified (48 hours) using Immunocal (Decal, Congers, NY), the brain was removed, and the skull was cut midsagittally and then coronally at an angle of 60° anterior to the midline. The fragment containing the eye was then embedded and serially sectioned. F4/80+ cells in tissue sections of at least 2 mice of a particular genotype at each age were stained and quantitated in a blinded fashion as described previously.16 Frozen 5-μm sections of spleen were stained for marginal metallophilic macrophages using the rat monoclonal antibody MOMA-1.35 For light microscopy, a Zeiss Axiophot (Figures 3A, 4) or Zeiss Axioskop 2 plus (Figure 5A-C) was used, imaging with 35 mm Kodak 64T film (Eastman Kodak, Rochester, NY) and Axiovision software, respectively. Objectives used were Plan-Neofluar 2.5 ×/0.075 numeric aperture (NA) (Figures 4 first and third rows and 5A top row), 10 ×/0.3 NA (Figure 5A middle and bottom rows), 25 ×/0.8 (Figures 3A, 4 second and fourth rows, 5B), and Plan-Apochromat 63 ×/1.4 NA (oil) (Figure 5C). Flow cytometric analysis of csCSF-1 on primary skin fibroblasts and for hematologic parameters in bone marrow, spleen, and blood were carried out as described previously.17
Bone histomorphometric analysis
Femora from 2-month-old mice were fixed, decalcified, embedded, and sectioned as described previously.4,32 For static bone parameters, frontal sections (5 μm thick) through distal femurs were stained with hematoxylin and eosin. Trabecular tissue in a 1.2-mm wide × 2.2-mm long area located just proximal to the distal growth plate cartilage and excluding trabeculae connected to the cortex was measured at an objective magnification of × 20 using a semiautomatic image analysis system (Osteomeasure; Osteometrics, Atlanta, GA).36,37 The parameters examined are listed in Figure 6A. The parameters and formulas used for their calculations are based on the recommendation of the American Society for Bone and Mineral Research Nomenclature Committee.38
Bone microcomputed tomography and bending test
The 2-month-old femurs from male mice were analyzed using a compact cone-beam type tomography or desktop micro-CT (micro-CT40; Scanco Medical, Bassersdorf, Switzerland). For the measurement, the femurs were placed in a cylindrical sample holder of plexiglass filled with buffered saline in order to preserve the sample for the duration of the measurement. For distal femurs, 600 microtomographic slices with a slice increment of 6 μm were acquired (starting at a point just distal to growth plate). The same femurs were mechanically tested ex vivo in 4-point bending. All tests were done at room temperature and at a rate of 3 mm/min using an Instron-5543 testing system (Canton, MA). The load-displacement plot from each mechanical test was analyzed for structural strength properties.
Statistical analysis
Data were expressed as means plus or minus standard deviations. Student t test was used to test significance. Differences were considered statistically significant for comparisons of data sets yielding P values less than or equal to .05.
Results
Csf1op/Csf1op; TgSPP/+ and Csf1op/Csf1op; TgSGP/+ mice express circulating CSF-1 but not cell-surface CSF-1 and differ in the timing of incisor eruption
TgN(SPPCsf1)Ers (TgSPP) and TgN(SGPCsf1)Ers (TgSGP) transgene expression were driven by a fragment of the mouse Csf1 gene containing 3.13 kb of the promoter and the entire first intron that was previously shown to drive essentially wild-type CSF-1 expression in mice.28 TgSGP differs from TgSPP in possessing a blocking mutation (S276L-G277A) in the Ser-Gly-X-Gly/Ala consensus for GAG addition39 (Figure 1A). Four of 13 TgSPP (TgSPP2, TgSPP7, TgSPP10, and TgSPP13) and 4 of 6 TgSGP (TgSGP2, TgSGP4, TgSGP5, and TgSGP6) founders generated and maintained on the FVB/NJ inbred background exhibited germ-line transmission, normal fertility, and transmission on the Csf1op/Csf1op background (Table 1). A detailed characterization of mice expressing each of these transgenes on the Csf1op/Csf1op background revealed similar phenotypes within each transgene type, although for the Csf1op/Csf1op; TgSGP/+ mice the phenotype also depended on the transgene expression level (Table 1).
General phenotypes of mice of the Csf1op/Csf1op; TgSPP/+ and Csf1op/Csf1op; TgSGP/+ lines
. | Transgenic correction of Csf1op/Csf1op phenotype at 2 mo* . | . | . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | F4/80+ cell densities . | . | . | . | ||||||||
| Founder . | OP . | FL . | EPL . | TPC . | IE, d . | L . | S . | K . | BM . | ||||||||
| wt | 4 | 4 | 4 | 4 | 7-8 | 4 | 4 | 4 | 4 | ||||||||
| Csf1op/Csf1op | 0 | 0 | 0 | 0 | ∞ | 0 | 0 | 0 | 0 | ||||||||
| TgSPP2 | 4 | 4 | 4 | 4 | 7-8 | 3 | 4 | 4 | 4 | ||||||||
| TgSPP7 | 4 | 4 | 4 | 4 | 7-8 | 2 | 3 | 2 | 4 | ||||||||
| TgSPP8† | 4 | 4 | 4 | 4 | 7-8 | 4 | 3 | 3 | 3 | ||||||||
| TgSPP10 | 4 | 4 | 4 | 4 | 7-8 | 4 | 3 | 4 | 3 | ||||||||
| TgSPP11‡ | 4 | 4 | 4 | 4 | 7-8 | 4 | 4 | 3 | 4 | ||||||||
| TgSPP13 | 4 | 4 | 4 | 4 | 7-8 | 3 | 3 | 3 | 3 | ||||||||
| TgSGP2 | 4 | 3.5 | 4 | 4 | 7-8 | 3 | 3 | 4 | 4 | ||||||||
| TgSGP4 | 1 | 2 | 2 | 7 | 12-13 | 4 | 3 | 2 | 3 | ||||||||
| TgSGP5 | 3 | 4 | 3 | 4 | 7-8 | 4 | 3.5 | 3 | 3.5 | ||||||||
| TgSGP6 | 3 | 4 | 4 | 6 | 9-10 | 2 | 2 | 3 | 3 | ||||||||
. | Transgenic correction of Csf1op/Csf1op phenotype at 2 mo* . | . | . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | F4/80+ cell densities . | . | . | . | ||||||||
| Founder . | OP . | FL . | EPL . | TPC . | IE, d . | L . | S . | K . | BM . | ||||||||
| wt | 4 | 4 | 4 | 4 | 7-8 | 4 | 4 | 4 | 4 | ||||||||
| Csf1op/Csf1op | 0 | 0 | 0 | 0 | ∞ | 0 | 0 | 0 | 0 | ||||||||
| TgSPP2 | 4 | 4 | 4 | 4 | 7-8 | 3 | 4 | 4 | 4 | ||||||||
| TgSPP7 | 4 | 4 | 4 | 4 | 7-8 | 2 | 3 | 2 | 4 | ||||||||
| TgSPP8† | 4 | 4 | 4 | 4 | 7-8 | 4 | 3 | 3 | 3 | ||||||||
| TgSPP10 | 4 | 4 | 4 | 4 | 7-8 | 4 | 3 | 4 | 3 | ||||||||
| TgSPP11‡ | 4 | 4 | 4 | 4 | 7-8 | 4 | 4 | 3 | 4 | ||||||||
| TgSPP13 | 4 | 4 | 4 | 4 | 7-8 | 3 | 3 | 3 | 3 | ||||||||
| TgSGP2 | 4 | 3.5 | 4 | 4 | 7-8 | 3 | 3 | 4 | 4 | ||||||||
| TgSGP4 | 1 | 2 | 2 | 7 | 12-13 | 4 | 3 | 2 | 3 | ||||||||
| TgSGP5 | 3 | 4 | 3 | 4 | 7-8 | 4 | 3.5 | 3 | 3.5 | ||||||||
| TgSGP6 | 3 | 4 | 4 | 6 | 9-10 | 2 | 2 | 3 | 3 | ||||||||
OP indicates osteopetrosis; FL, femur length; EPL, epiphyseal plate length; TPC, TRAP-positive cells; IE, incisor eruption; L, liver; S, spleen; K, kidney; and BM, bone marrow.
From X-ray measurements, histochemistry, and immunohistochemistry. Correction assessed using an arbitrary scale (for each trait, wt = 4 and Csf1op/Csf1op = 0). Of a total of 19 founder mice (13 TgSPP and 6 TgSGP), 6 transgenes (TgSPP1, TgSPP4, TgSPP5, TgSPP6, TgSPP9, TgSGP1) were not transmissible and 3 (TgSPP3, TgSPP12, TgSGP3) were not transmissible on the Csf1op/Csf1op background.
Non-Mendelian transmission on the Csf1op/Csf1op background.
Lower rate of fertility.
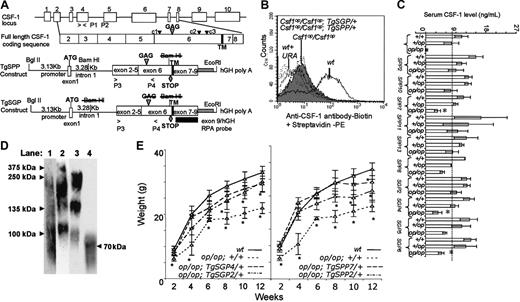
Transgene construction, CSF-1 expression, and growth curves for transgenic mice. (A) Genomic organization of the mouse CSF-1 gene and transgenic constructs. Top diagram: CSF-1 gene showing intron-exon boundaries and the full-length coding sequence with positions of the glycosaminoglycan addition site (large arrowhead), the 3 major putative cleavage sites (small arrowhead) that generate sgCSF-1 (c1) and spCSF-1 (c2 or c3), and the transmembrane domain (TM). Bottom diagram: TgSPP and TgSGP transgenes, prepared by cloning the exons 2 to 9 fragment of the cDNA encoding a full-length CSF-1 downstream of the 3.13-kb CSF-1 promoter and first intron fragment. An additional hGH polyA signal fragment was added at the 3′ end of the cDNA. The exons 2 to 9 fragments used for both TgSGP and TgSPP contained a stop codon at amino acid 456 (diamond) to ensure that they were secreted. The fragment used for TgSGP also contained a mutation (S276L-G277A) in the unique glycosaminoglycan (chondroitin sulfate) addition site (SGXG/A). P1 and P2 indicate the oligonucleotide primer pairs used to detect the Csf1op mutation in exon 4; P3 and P4, the primer pairs for amplifying a region between exons 2 to 4 to distinguish the transgene from the wild-type gene; and the exon 9 RPA probe was used to detect exon 9-containing mRNA expression. Relevant restriction sites are also indicated. Struck-out text indicates mutations of sites. (B) Fluorescence-activated cell sorter (FACS) analysis of cell-surface expression of CSF-1 in skin fibroblasts from transgenic lines. Expression profiles for wt, Csf1op/Csf1op; +/+ (filled), wt + URA (wt with unrelated antibody), Csf1op/Csf1op; TgSPP2/+, Csf1op/Csf1op; TgSPP7/+, Csf1op/Csf1op; TgSGP2/+, and Csf1op/Csf1op; TgSGP4/+ are shown. (C) Circulating CSF-1 concentrations in TgSPP and TgSGP mouse serum measured with a radioimmunoassay (RIA) that detects only biologically active CSF-1 and both spCSF-1 and sgCSF-1 equivalently (± SD, n > 5 for each line). +/+ indicates wild type; +/op, Csf1op heterozygotes; and op/op, Csf1op homozygotes. *P < .01 (compared with Csf1op/+). (D) Nonreducing SDS-PAGE and anti-CSF-1 Western blot of immunoaffinity purified CSF-1 from conditioned media (lane 1, wt; lane 2, Csf1op/Csf1op; TgSPP/+; lane 3, Csf1op/Csf1op; TgSGP/+) and cell lysates (lane 4, Csf1op/Csf1op; TgCS/+) from immortalized skin fibroblasts. (E) Growth curves. Left panel: wt, Csf1op/Csf1op; TgSGP2/+, Csf1op/Csf1op; TgSGP4/+, Csf1op/Csf1op; +/+. Right panel: wt, Csf1op/Csf1op; TgSPP2/+, Csf1op/Csf1op; TgSPP7/+. *Significantly different from wt; P ≤ .05; n ≥ 6 mice at each time point.
Transgene construction, CSF-1 expression, and growth curves for transgenic mice. (A) Genomic organization of the mouse CSF-1 gene and transgenic constructs. Top diagram: CSF-1 gene showing intron-exon boundaries and the full-length coding sequence with positions of the glycosaminoglycan addition site (large arrowhead), the 3 major putative cleavage sites (small arrowhead) that generate sgCSF-1 (c1) and spCSF-1 (c2 or c3), and the transmembrane domain (TM). Bottom diagram: TgSPP and TgSGP transgenes, prepared by cloning the exons 2 to 9 fragment of the cDNA encoding a full-length CSF-1 downstream of the 3.13-kb CSF-1 promoter and first intron fragment. An additional hGH polyA signal fragment was added at the 3′ end of the cDNA. The exons 2 to 9 fragments used for both TgSGP and TgSPP contained a stop codon at amino acid 456 (diamond) to ensure that they were secreted. The fragment used for TgSGP also contained a mutation (S276L-G277A) in the unique glycosaminoglycan (chondroitin sulfate) addition site (SGXG/A). P1 and P2 indicate the oligonucleotide primer pairs used to detect the Csf1op mutation in exon 4; P3 and P4, the primer pairs for amplifying a region between exons 2 to 4 to distinguish the transgene from the wild-type gene; and the exon 9 RPA probe was used to detect exon 9-containing mRNA expression. Relevant restriction sites are also indicated. Struck-out text indicates mutations of sites. (B) Fluorescence-activated cell sorter (FACS) analysis of cell-surface expression of CSF-1 in skin fibroblasts from transgenic lines. Expression profiles for wt, Csf1op/Csf1op; +/+ (filled), wt + URA (wt with unrelated antibody), Csf1op/Csf1op; TgSPP2/+, Csf1op/Csf1op; TgSPP7/+, Csf1op/Csf1op; TgSGP2/+, and Csf1op/Csf1op; TgSGP4/+ are shown. (C) Circulating CSF-1 concentrations in TgSPP and TgSGP mouse serum measured with a radioimmunoassay (RIA) that detects only biologically active CSF-1 and both spCSF-1 and sgCSF-1 equivalently (± SD, n > 5 for each line). +/+ indicates wild type; +/op, Csf1op heterozygotes; and op/op, Csf1op homozygotes. *P < .01 (compared with Csf1op/+). (D) Nonreducing SDS-PAGE and anti-CSF-1 Western blot of immunoaffinity purified CSF-1 from conditioned media (lane 1, wt; lane 2, Csf1op/Csf1op; TgSPP/+; lane 3, Csf1op/Csf1op; TgSGP/+) and cell lysates (lane 4, Csf1op/Csf1op; TgCS/+) from immortalized skin fibroblasts. (E) Growth curves. Left panel: wt, Csf1op/Csf1op; TgSGP2/+, Csf1op/Csf1op; TgSGP4/+, Csf1op/Csf1op; +/+. Right panel: wt, Csf1op/Csf1op; TgSPP2/+, Csf1op/Csf1op; TgSPP7/+. *Significantly different from wt; P ≤ .05; n ≥ 6 mice at each time point.
On this background, TgSPP2, TgSPP10, and TgSPP13 expressed concentrations of circulating CSF-1 approximating Csf1op/+ heterozygote levels, whereas TgSPP7 expressed half this concentration. TgSGP2 expressed concentrations approximating heterozygote levels, whereas TgSGP4 expressed at half this concentration and TgSGP5 and TgSGP6 at concentrations intermediate between those of TgSGP2 and TgSGP4 (Figure 1C). As expected, the level of circulating CSF-1 was increased when each transgene was expressed on the Csf1op/+ background and was even more increased on the +/+ background (Figure 1C). Furthermore, compared with the csCSF-1 expression observed on skin fibroblasts from wild-type (wt) mice, skin fibroblasts from Csf1op/Csf1op; Tg/+ mice expressing TgSPP or TgSGP failed to express CSF-1 on their surface (Figure 1B). These studies indicate that both TgSPP and TgSGP transgenes encode proteins that are exclusively secreted and can reconstitute wild-type levels of circulating CSF-1.
When the circulating CSF-1 concentration in Csf1op/Csf1op mice expressing TgSPP or TgSGP approximated wt homozygote concentrations, the gross mutant phenotype of the Csf1op/Csf1op mouse was corrected. In contrast, when expressed at approximately half this concentration, TgSPP corrected more efficiently than TgSGP. This was reflected in lower tissue macrophage densities, a delay in the timing of tooth eruption, and a failure to correct the skeletal abnormalities in the Csf1op/Csf1op; TgSGP/+ mice (Table 1). Because the lower expressing transgenic lines expressing serum CSF-1 at half the Csf1op/+ concentration revealed phenotypic differences between TgSPP and TgSGP mice, we focused our attention on these lines (Csf1op/Csf1op; TgSPP7/+ and Csf1op/Csf1op; TgSGP4/+), as well as those expressing serum CSF-1 at higher concentrations (Csf1op/Csf1op; TgSPP2/+ and Csf1op/Csf1op; TgSGP2/+).
Circulating CSF-1 in wt mice is comprised of approximately equal concentrations of sgCSF-1 and spCSF-1.40 Csf1op/Csf1op; TgSPP7/+ and Csf1op/Csf1op; TgSGP4/+ mice respectively express circulating spCSF-1 and sgCSF-1 at the concentrations found in wt heterozygotes, whereas Csf1op/Csf1op; TgSPP2/+ and Csf1op/Csf1op; TgSGP2/+ respectively express circulating spCSF-1 and sgCSF-1 at the concentrations found in wt homozygous mice. To check the expression of secreted and csCSF-1 isoforms, they were immunoaffinity-purified from immortalized skin fibroblast CM and cell lysate, respectively, and examined by nonreducing SDS-PAGE and Western blotting with anti-CSF-1 antibody (Figure 1D). As expected,8 fibroblasts from wt mice (Figure 1D, lane 1) secreted the 375-kDa homomeric and 250-kDa heteromeric proteoglycans, 135-kDa and 100-kDa glycoproteins, and some lower Mr glycoproteins. Csf1op/Csf1op; TgSPP/+ fibroblasts secreted the 375-kDa and the 250-kDa proteoglycan forms and a 100-kDa glycoprotein form (Figure 1D, lane 2). In contrast, the Csf1op/Csf1op; TgSGP/+ fibroblasts failed to secrete CSF-1 with the characteristic 375-kDa and 250-kDa Mrs of the proteoglycans and instead secreted 260-kDa, 135-kDa, and 90-kDa glycoproteins (Figure 1D, lane 3) (in contrast to wt mice, where sgCSF-1 is smaller due to its generation by proteolytic cleavage on the amino-terminal side of the GAG addition site of the proteoglycanylated precursor, longer, less cleaved forms of the sgCSF-1 are secreted by Csf1op/Csf1op; TgSGP/+ fibroblasts because the GAG addition site is mutated and thus uncleaved precursors contribute to the sgCSF-1 pool). Consistent with the expression of spCSF-1 in Csf1op/Csf1op; TgSPP/+, but not Csf1op/Csf1op; TgSGP/+ fibroblasts, 3 times more 35SO42- was incorporated into secreted CSF-1 from the former than from the latter (data not shown, some incorporation of 35SO42- also occurs in the glycoprotein8 ). In summary, TgSPP predominantly encodes spCSF-1 with a small amount of sgCSF-1 due to cleavage at site c1 (Figure 1A), whereas TgSGP encodes glycoprotein exclusively. As expected, Csf1op/Csf1op; TgCS/+ fibroblasts, which fail to secrete CSF-1,17 expressed a 70-kDa csCSF-1 (Figure 1D, lane 4).
Reduced growth rates but normal male and female reproductive function of mice expressing spCSF-1 or sgCSF-1
Important aspects of the Csf1op/Csf1op phenotype are the markedly retarded growth rate21 and female and male reproductive defects (Pollard and Stanley22 ; and Cohen et al23 ). We have previously shown that both full-length CSF-1 and csCSF-1 transgenes fully correct the growth retardation defects of Csf1op/Csf1op mice. In contrast, the growth of Csf1op/Csf1op; TgSGP/+ and Csf1op/Csf1op; TgSPP/+ mice was intermediate between the growth curves of the wild-type and Csf1op/Csf1op, although the results for Csf1op/Csf1op; TgSPP7/+ were not significantly different at several time points (Figure 1D). In view of the normal femur lengths of the mice exclusively expressing TgSPP7, TgSPP2, and TgSGP2, this result, together with the wild-type growth of Csf1op/Csf1op; TgCS/+ mice,17 suggests an important role for csCSF-1 in maintaining a normal growth rate.
CSF-1 plays an important role in the regulation of ovulation, the estrous cycle, and lactation in females and of libido in males.23 In contrast to Csf1op/Csf1op mice, both Csf1op/Csf1op; TgSPP/+ and Csf1op/Csf1op; TgSGP/+ lines were able to nurture their progeny normally. To test the reproductive function of these mice, we carried out reciprocal matings between the Csf1op/Csf1op; Tg/+ mice and Csf1op/+; +/+ mice (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article). The results of these crosses indicate that the litter sizes of all reciprocal crosses were similar and that the percentages of genotypes were as expected from Mendelian segregation of the transgene and the lower survival of Csf1op/Csf1op mice. These results are consistent with normal reproductive function of mice expressing sgCSF-1 or spCSF-1.
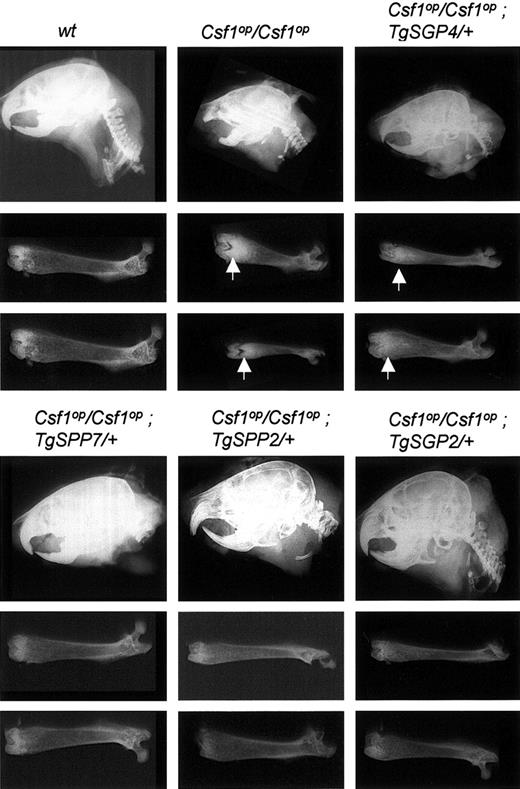
More efficient correction of the osteopetrosis of CSF-1 deficiency by TgSPP than by TgSGP
Csf1op/Csf1op mice exhibit osteopetrosis due to a paucity of osteoclasts. The osteopetrosis is marked by increased bone density in X-radiograms of bones, which in femurs is associated with a highly trabecularized distal metaphysis (Figure 2). The failure of these osteoclast-deficient mice to remodel bone results in skeletal abnormalities, including a smaller overall skeletal size, shortening and thickening of the long bones, shortening of the facial bones, and a “doming” of the skull.21 Mice exclusively expressing csCSF-1 have a residual osteopetrosis, indicating a role for the secreted CSF-1 isoforms in the generation and regulation of functional osteoclasts.17 To determine the relative roles of sgCSF-1 and spCSF-1 in osteoclastogenesis and function, we initially examined X-radiograms of mice exclusively expressing the sgCSF-1 or spCSF-1 precursors (Figure 2). Mice exclusively expressing lower than wt levels of circulating sgCSF-1 (Csf1op/Csf1op; TgSGP4/+, Csf1op/Csf1op; TgSGP6/+) had radio-opaque femurs at 2 months of age (Figure 2; Table 1). In contrast, femurs from spCSF-1 mice (Csf1op/Csf1op; TgSPP7/+, Csf1op/Csf1op; TgSPP8/+) expressing half wt levels of circulating CSF-1 were indistinguishable from those of wt mice (Figure 2; Table 1). Femurs from Csf1op/Csf1op; TgSGP2/+ or Csf1op/Csf1op; TgSPP2/+ mice that express wt levels of circulating CSF-1 were also indistinguishable from wild type (Figure 2), and in Csf1op/Csf1op; TgSGP4/TgSGP4 mice there was a partial reduction in the osteopetrosis and volume of trabecular bone compared with Csf1op/Csf1op; TgSGP4/+ mice (Figure 3A-B). These results demonstrate that spCSF-1 is significantly more efficient at correcting the Csf1op/Csf1op osteopetrosis than sgCSF-1, but correction can be observed at higher concentrations of sgCSF-1.
As shown previously, at 1 week of age Csf1op/Csf1op mice, in contrast to wt mice, lack osteoclasts and possess large tracts of trabecular bone (Figure 4). By 2 months of age, however, osteoclasts are apparent and the delayed resorption of trabecular bone has been initiated.25 At both 1 week and 2 months of age, sections of femurs from Csf1op/Csf1op mice heterozygous for TgSPP7 and TgSPP2 stained with hematoxylin and eosin and for tartrate-resistant acid phosphatase (TRAP) were indistinguishable from wt femurs, except that, at 1 week of age, Csf1op/Csf1op; TgSPP2/+ mice possessed more trabecular bone in the distal metaphyseal region (Figure 4). Furthermore, histomorphometry of femoral sections of these mice at 2 months of age similarly revealed no significant difference in percent bone volume to total volume, thickness, spacing, or number of trabeculae from the same parameters in wt femurs (Figure 3B). In contrast, femurs from the Csf1op/Csf1op; TgSGP4/+ mice were deficient in osteoclasts and histologically indistinguishable from Csf1op/Csf1op femurs at 1 week (Figure 4A), but showed intense TRAP positivity compared with wt or Csf1op/Csf1op sections by 2 months of age (Figure 4B). Despite intense TRAP staining in these femurs, the distal metaphyseal femoral regions were profoundly trabecularized (Figure 4B), suggesting that these mice might be osteosclerotic. Of interest, the Csf1op/Csf1op; TgSGP2/+ mice, despite their higher level of expression of sgCSF-1, failed to completely resorb the trabecular bone at both 2 days (data not shown) and at 2 months of age (Figure 4B). While the histomorphometric parameters of these mice did not detect these abnormalities (Figure 3B), those of Csf1op/Csf1op; TgSGP4/+ and Csf1op/Csf1op; TgSGP4/TgSGP4 mice were intermediate between the wt and Csf1op/Csf1op parameters (Figure 3B). Thus, the histologic data confirm that spCSF-1 more efficiently corrects the osteopetrotic defects than sgCSF-1.
Comparison of the skeletal development of the Csf1op/Csf1opmice expressing varying levels of TgSGP and TgSPP CSF-1. Radiograms of the head and femurs of 2-month-old mice. Each femur is from a different mouse. Vertical arrows indicate regions with increased radiopacity compared with wt mice.
Comparison of the skeletal development of the Csf1op/Csf1opmice expressing varying levels of TgSGP and TgSPP CSF-1. Radiograms of the head and femurs of 2-month-old mice. Each femur is from a different mouse. Vertical arrows indicate regions with increased radiopacity compared with wt mice.
Dosage effect of TgSGP. (A) Comparison of 2-month-old Csf1op/Csf1op; TgSGP4/+ and Csf1op/Csf1op; TgSGP4/TgSGP4 femurs. Top panel: x-ray. Bottom panel: histology of the bone sections. Low-power photomicrographs of comparable fields of 5-μm sections of distal femoral metaphyses stained with H&E. Bar, 50 μm. (B) Histomorphometric analyses of femurs from 2-month-old mice (average from femurs from 2 mice). Dose-dependent correction of skeletal parameters in mice expressing TgSGP but not TgSPP. Also note correction by TgCS. BV/TV indicates bone volume/total volume; Tb.Sp, trabecular spacing; and Tb.N, trabecular number.
Dosage effect of TgSGP. (A) Comparison of 2-month-old Csf1op/Csf1op; TgSGP4/+ and Csf1op/Csf1op; TgSGP4/TgSGP4 femurs. Top panel: x-ray. Bottom panel: histology of the bone sections. Low-power photomicrographs of comparable fields of 5-μm sections of distal femoral metaphyses stained with H&E. Bar, 50 μm. (B) Histomorphometric analyses of femurs from 2-month-old mice (average from femurs from 2 mice). Dose-dependent correction of skeletal parameters in mice expressing TgSGP but not TgSPP. Also note correction by TgCS. BV/TV indicates bone volume/total volume; Tb.Sp, trabecular spacing; and Tb.N, trabecular number.
To further characterize the bone phenotypes, we carried out micro-CT and a 4-point bending test of 2-month femurs. Consistent with the developmental analysis (Figure 4), micro-CT revealed significant abnormalities in bone volume, number of trabeculae, trabecular spacing, trabecular thickness, second moment of area/inertia around A/P and M/L axes, and width across the M/L axis for Csf1op/Csf1op; TgSGP4/+ mice (Table 2). These abnormalities were shared with Csf1op/Csf1op mice, which, in addition, possessed decreased cortical thickness. Abnormalities in some of these and other parameters were also observed for Csf1op/Csf1op; TgSGP2/+ and Csf1op/Csf1op; TgSPP2/+ femurs, but not for Csf1op/Csf1op; TgSPP7/+ femurs. Femurs from mice exclusively expressing csCSF-1 (Csf1op/Csf1op; TgCS5/+), which we have previously shown are partially osteopetrotic,17 exhibited abnormalities in most parameters. Consistent with the micro-CT data, the bending test analysis indicated that Csf1op/Csf1op, Csf1op/Csf1op; TgSGP4/+, and Csf1op/Csf1op; TgCS5/+ femurs exhibited increased stiffness and ultimate load compared with wt femurs (Table 3). Csf1op/Csf1op; TgSGP4/+, Csf1op/Csf1op; TgSGP2/+, and Csf1op/Csf1op; TgSPP2/+ were more plastic than wt femurs, exhibiting increased ultimate stress without an increase in yield stress, whereas Csf1op/Csf1op; TgCS5/+ were less plastic than wt femurs, exhibiting increases in both parameters. Accurate determination of the femur lengths revealed that mice exclusively expressing sgCSF-1 or csCSF-1, but none of the other transgenic mice, had femurs shorter than wt mice. In summary, the histologic, histomorphometric, micro-CT, and biomechanical analyses demonstrate that mice either exclusively expressing sgCSF-1, or spCSF-1 at concentrations higher than the circulating CSF-1 concentration in normal homozygous wt mice, exhibit abnormal bone development, whereas mice expressing spCSF-1 at half the circulating concentration of normal heterozygote mice exhibit normal bone development.
Micro-CT of femurs from 2-month-old male Csf1op/Csf1op; TgSPP/+ and Csf1op/Csf1op; TgSGP/+ mice
. | wt . | Csf1op/Csf1op . | Csf1op/Csf1op; TgSGP4/+ . | Csf1op/Csf1op; TgSGP2/+ . | Csf1op/Csf1op; TgSPP7/+ . | Csf1op/Csf1op; TgSPP2/+ . | Csf1op/Csf1op; TgCS5/+ . |
|---|---|---|---|---|---|---|---|
| 3D-BV/TV, % | 0.22 ± 0.02 | 0.80 ± 0.023* | 0.55 ± 0.01* | 0.24 ± 0.01 | 0.24 ± 0.026 | 0.26 ± 0.022* | 0.29 ± 0.052* |
| 3D-Tb.N, 1/mm | 6.6 ± 0.19 | 13.6 ± 1.78* | 11.8 ± 0.79* | 7.3 ± 0.23* | 6.5 ± 0.24 | 7.9 ± 0.35* | 7.7 ± 0.5* |
| Tb. Th, mm | 0.04 ± 0.002 | 0.11 ± 0.007* | 0.08 ± 0.004* | 0.04 ± 0.002 | 0.04 ± 0.002 | 0.04 ± 0.003 | 0.04 ± 0.005 |
| Tb.Sp, mm | 0.16 ± 0.004 | 0.11 ± 0.05† | 0.10 ± 0.013† | 0.14 ± 0.005† | 0.16 ± 0.007 | 0.13 ± 0.007† | 0.13 ± 0.01† |
| Imax, mm4 | 0.23 ± 0.011 | 0.21 ± 0.045 | 0.29 ± 0.028* | 0.21 ± 0.016 | 0.25 ± 0.017 | 0.24 ± 0.018 | 0.40 ± 0.03* |
| Imin, mm4 | 0.12 ± 0.007 | 0.21 ± 0.05* | 0.14 ± 0.011* | 0.11 ± 0.008† | 0.13 ± 0.005 | 0.11 ± .006† | 0.16 ± 0.012* |
| Area, mm2 | 0.85 ± 0.037 | 0.63 ± 0.049† | 0.85 ± 0.09 | 0.87 ± 0.048 | 0.85 ± 0.031 | 0.88 ± 0.028 | 1.17 ± 0.127* |
| Av.Cor.Th., mm | 0.22 ± 0.013 | 0.17 ± 0.01† | 0.21 ± 0.012 | 0.24 ± 0.016 | 0.21 ± 0.011 | 0.23 ± 0.01 | 0.13 ± 0.011† |
| Width (A/P), mm | 1.20 ± 0.026 | 1.25 ± 0.034‡ | 1.23 ± 0.029 | 1.13 ± 0.038† | 1.24 ± 0.013 | 1.15 ± 0.028‡ | 1.23 ± 0.021 |
| Width (M/L), mm | 1.68 ± 0.05 | 1.74 ± 0.124 | 1.89 ± 0.045* | 1.67 ± 0.031 | 1.71 ± 0.024 | 1.74 ± 0.066 | 1.91 ± 0.053* |
. | wt . | Csf1op/Csf1op . | Csf1op/Csf1op; TgSGP4/+ . | Csf1op/Csf1op; TgSGP2/+ . | Csf1op/Csf1op; TgSPP7/+ . | Csf1op/Csf1op; TgSPP2/+ . | Csf1op/Csf1op; TgCS5/+ . |
|---|---|---|---|---|---|---|---|
| 3D-BV/TV, % | 0.22 ± 0.02 | 0.80 ± 0.023* | 0.55 ± 0.01* | 0.24 ± 0.01 | 0.24 ± 0.026 | 0.26 ± 0.022* | 0.29 ± 0.052* |
| 3D-Tb.N, 1/mm | 6.6 ± 0.19 | 13.6 ± 1.78* | 11.8 ± 0.79* | 7.3 ± 0.23* | 6.5 ± 0.24 | 7.9 ± 0.35* | 7.7 ± 0.5* |
| Tb. Th, mm | 0.04 ± 0.002 | 0.11 ± 0.007* | 0.08 ± 0.004* | 0.04 ± 0.002 | 0.04 ± 0.002 | 0.04 ± 0.003 | 0.04 ± 0.005 |
| Tb.Sp, mm | 0.16 ± 0.004 | 0.11 ± 0.05† | 0.10 ± 0.013† | 0.14 ± 0.005† | 0.16 ± 0.007 | 0.13 ± 0.007† | 0.13 ± 0.01† |
| Imax, mm4 | 0.23 ± 0.011 | 0.21 ± 0.045 | 0.29 ± 0.028* | 0.21 ± 0.016 | 0.25 ± 0.017 | 0.24 ± 0.018 | 0.40 ± 0.03* |
| Imin, mm4 | 0.12 ± 0.007 | 0.21 ± 0.05* | 0.14 ± 0.011* | 0.11 ± 0.008† | 0.13 ± 0.005 | 0.11 ± .006† | 0.16 ± 0.012* |
| Area, mm2 | 0.85 ± 0.037 | 0.63 ± 0.049† | 0.85 ± 0.09 | 0.87 ± 0.048 | 0.85 ± 0.031 | 0.88 ± 0.028 | 1.17 ± 0.127* |
| Av.Cor.Th., mm | 0.22 ± 0.013 | 0.17 ± 0.01† | 0.21 ± 0.012 | 0.24 ± 0.016 | 0.21 ± 0.011 | 0.23 ± 0.01 | 0.13 ± 0.011† |
| Width (A/P), mm | 1.20 ± 0.026 | 1.25 ± 0.034‡ | 1.23 ± 0.029 | 1.13 ± 0.038† | 1.24 ± 0.013 | 1.15 ± 0.028‡ | 1.23 ± 0.021 |
| Width (M/L), mm | 1.68 ± 0.05 | 1.74 ± 0.124 | 1.89 ± 0.045* | 1.67 ± 0.031 | 1.71 ± 0.024 | 1.74 ± 0.066 | 1.91 ± 0.053* |
Data (mean ± SD) from 4 femurs.
3D-BV/TV, % indicates percentage of bone volume to total volume; 3D-Tb.N, 1/mm, number of trabeculae; Tb.Th, mm, trabecular thickness; Tb.Sp, mm, trabecular spacing; A/P indicates anterior/posterior; M/L, medio/lateral; Imax, second moment of area/inertia around A/P axis; Imin, second moment of area/inertia around M/L axis; area, mid-shaft cross-sectional area; and Av.Cor.Th., average cortical thickness.
P < .05; higher than wt.
P < .05; lower than wt.
P .08; trend but not significantly different from wt.
Biomechanical properties of femurs of 2-month-old male Csf1op/Csf1op; TgSPP/+ and Csf1op/Csf1op; TgSGP/+ mice
. | wt . | Csf1op/Csf1op . | Csf1op/Csf1op; TgSGP4/+ . | Csf1op/Csf1op; TgSGP2/+ . | Csf1op/Csf1op; TgSPP7/+ . | Csf1op/Csf1op; TgSPP2/+ . | Csf1op/Csf1op; TgCS5/+ . |
|---|---|---|---|---|---|---|---|
| Ultimate load, N | 15.2 ± 0.84 | 17 ± 4.14 | 26.4 ± 5.49* | 16.3 ± 1.8 | 15.4 ± 3.5 | 18.5 ± 2.45* | 26.2 ± 2.07* |
| Yield load, N | 11.3 ± 3.84 | 10.9 ± 7.17 | 20.1 ± 7.54 | 10.5 ± 4.32 | 10.5 ± 2.51 | 13.5 ± 3.5 | 23.4 ± 1.08* |
| Stiffness, N/mm | 30.1 ± 5.66 | 47.9 ± 6.99* | 59.5 ± 11.64* | 29.9 ± 6.63 | 36.1 ± 8.63 | 38.3 ± 6.23 | 64.3 ± 7.06* |
| Flexural modulus, N/mm2 | 2352 ± 433.3 | 1778 ± 496.2 | 2714 ± 1239 | 2764 ± 670.3 | 2756 ± 659.7 | 3151 ± 88.5† | 3884 ± 411* |
| Length, mm | 13.8 ± 0.13 | 11.1 ± 0.32‡ | 11.71 ± 0.16‡ | 13.3 ± 0.29‡ | 13.8 ± 0.15 | 13.7 ± 0.12 | 13.5 ± 0.12‡ |
| Ultimate stress, N/mm2 | 99 ± 3.61 | 92 ± 26.24 | 125.8 ± 26.85† | 118.8 ± 4.61† | 102.0 ± 22.35 | 125 ± 17.94† | 135.9 ± 16.3* |
| Yield stress, N/mm2 | 73.6 ± 21.94 | 60.2 ± 45.52 | 97.8 ± 38.2 | 76.5 ± 32.66 | 69.8 ± 18.03 | 98.5 ± 16.9 | 121.6 ± 14.3† |
. | wt . | Csf1op/Csf1op . | Csf1op/Csf1op; TgSGP4/+ . | Csf1op/Csf1op; TgSGP2/+ . | Csf1op/Csf1op; TgSPP7/+ . | Csf1op/Csf1op; TgSPP2/+ . | Csf1op/Csf1op; TgCS5/+ . |
|---|---|---|---|---|---|---|---|
| Ultimate load, N | 15.2 ± 0.84 | 17 ± 4.14 | 26.4 ± 5.49* | 16.3 ± 1.8 | 15.4 ± 3.5 | 18.5 ± 2.45* | 26.2 ± 2.07* |
| Yield load, N | 11.3 ± 3.84 | 10.9 ± 7.17 | 20.1 ± 7.54 | 10.5 ± 4.32 | 10.5 ± 2.51 | 13.5 ± 3.5 | 23.4 ± 1.08* |
| Stiffness, N/mm | 30.1 ± 5.66 | 47.9 ± 6.99* | 59.5 ± 11.64* | 29.9 ± 6.63 | 36.1 ± 8.63 | 38.3 ± 6.23 | 64.3 ± 7.06* |
| Flexural modulus, N/mm2 | 2352 ± 433.3 | 1778 ± 496.2 | 2714 ± 1239 | 2764 ± 670.3 | 2756 ± 659.7 | 3151 ± 88.5† | 3884 ± 411* |
| Length, mm | 13.8 ± 0.13 | 11.1 ± 0.32‡ | 11.71 ± 0.16‡ | 13.3 ± 0.29‡ | 13.8 ± 0.15 | 13.7 ± 0.12 | 13.5 ± 0.12‡ |
| Ultimate stress, N/mm2 | 99 ± 3.61 | 92 ± 26.24 | 125.8 ± 26.85† | 118.8 ± 4.61† | 102.0 ± 22.35 | 125 ± 17.94† | 135.9 ± 16.3* |
| Yield stress, N/mm2 | 73.6 ± 21.94 | 60.2 ± 45.52 | 97.8 ± 38.2 | 76.5 ± 32.66 | 69.8 ± 18.03 | 98.5 ± 16.9 | 121.6 ± 14.3† |
Data (mean ± SD) were collected from 4 femurs.
N indicates Newton.
P < .05; higher than wt.
P < .09; trend, but not significantly different from wt.
P < .05; lower than wt.
Histology of bone marrow of Csf1op/Csf1opmice expressing varying amounts of TgSPP and TgSGP. Low-power photomicrographs (× 25) above high-power photomicrographs (× 400) (boxed areas in low-power images) of midsagittal 5-μm sections of the distal femoral metaphyses of (A) 1-week-old mice stained for TRAP and counterstained with hematoxylin and (B) 2-month-old mice (top panels stained with H&E; bottom panels, serial sections stained for TRAP and counterstained with hematoxylin). * Osteoclasts; TR, trabecular region; and EP, epiphyseal plate.
Histology of bone marrow of Csf1op/Csf1opmice expressing varying amounts of TgSPP and TgSGP. Low-power photomicrographs (× 25) above high-power photomicrographs (× 400) (boxed areas in low-power images) of midsagittal 5-μm sections of the distal femoral metaphyses of (A) 1-week-old mice stained for TRAP and counterstained with hematoxylin and (B) 2-month-old mice (top panels stained with H&E; bottom panels, serial sections stained for TRAP and counterstained with hematoxylin). * Osteoclasts; TR, trabecular region; and EP, epiphyseal plate.
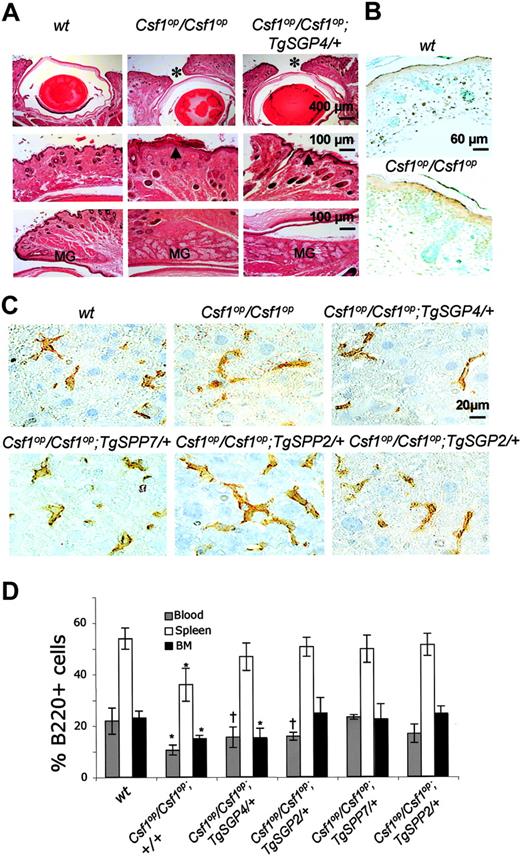
TgSGP dose-dependent correction of incisor eruption and eyelid opening in the Csf1op/Csf1op; Tg/+ mice
Incisors erupt at 7 to 8 days of age in FVB/NJ mice but fail to erupt in Csf1op/Csf1op mice on the same background. Of interest, Csf1op/Csf1op; TgSGP4/+ and Csf1op/Csf1op; TgSGP6/+ mice, expressing approximately half the Csf1op/+ level of circulating CSF-1, exhibited delayed incisor eruption (Table 1), whereas Csf1op/Csf1op; TgSGP5/+ and Csf1op/Csf1op; TgSGP2/+ mice, expressing high circulating CSF-1 concentrations, did not. This dosage-dependent delay in incisor eruption was exhibited only by mice exclusively expressing the secreted CSF-1 glycoprotein, as the timing of tooth eruption in Csf1op/Csf1op; TgSPP7/+ and Csf1op/Csf1op; TgSPP8/+ mice, expressing levels of circulating CSF-1 equivalent to Csf1op/Csf1op; TgSGP4/+ mice, was normal.
An unreported phenotype of Csf1op/Csf1op mice is incomplete eyelid opening. This phenotype has a 100% penetrance in Csf1op/Csf1op mice on the FVB/NJ background. Csf1op/Csf1op; TgSGP4/+ mice also possessed this trait with 100% penetrance, but the phenotype was less severe (Figure 5A). It was not observed in mice expressing sgCSF-1 at higher levels or in mice expressing spCSF-1. Histologic studies of 6-week-old mice revealed a chronic active blepharitis with hyperkeratosis of the eyelids in affected mice (Figure 5A). In addition to the abnormally thickened epidermis of the eyelid epithelia, meibomian gland hyperplasia also contributed to the eyelid pathology. Postnatal apoptosis of the eyelid epithelium has been shown to be a crucial factor in eyelid opening.41 Supporting this, eyelids from 2-day-old Csf1op/Csf1op mice, compared with wt mice, failed to accumulate apoptotic cells (Figure 5B), indicating that CSF-1 plays a role in apoptotic cell generation.
Transgenic expression of secreted CSF-1 in Csf1op/Csf1op mice restores tissue macrophage densities in a dose-dependent manner but with a less dendritic macrophage morphology in response to sgCSF-1
In contrast to the bone marrow macrophage and osteoclast deficiencies of Csf1op/Csf1op mice, which spontaneously recover with age, the deficiencies of most other tissue macrophages remain.16,42,43 Normal tissue-specific and developmental expression of full-length CSF-1 (TgC) in Csf1op/Csf1op mice restored wild-type tissue macrophage densities,28 whereas expression of csCSF-1 (TgCS),17 or restoration of circulating CSF-1 by CSF-1 injection,16 failed to restore normal macrophage densities in many tissues, suggesting important roles for the spCSF-1 and sgCSF-1 in tissue macrophage regulation. This was confirmed in an analysis of several tissue macrophage populations with the macrophage marker F4/80+,44 where macrophage density showed a strong dependence on secreted CSF-1 concentration, but with little difference between mice exclusively expressing sgCSF-1 and those expressing spCSF-1 (Table 4). Of interest, liver macrophages from mice expressing spCSF-1 were significantly more spread than in mice expressing sgCSF-1 at both 2 days (data not shown) and 3 months of age (Figure 5C). In contrast to the complete restoration of F4/80+ macrophages in the Csf1op/Csf1op; TgSGP2/+ and Csf1op/Csf1op; TgSPP2/+ tissues, in neither of these lines were MOMA-1-expressing marginal metallophillic macrophages restored to wild-type levels (Table 4). Together with earlier studies,16,17 these data indicate that secreted spCSF-1 and sgCSF-1 play important roles in maintaining tissue macrophage densities in a dose-dependent manner.
Macrophage densities in the tissues of Csf1op/Csf1op; TgSGP/+ and Csf1op/Csf1op; TgSPP/+ mice
. | Liver cells/mm2 . | . | . | . | . | . | . | |
|---|---|---|---|---|---|---|---|---|
| Genotype . | 2-d-old mouse . | 3-mo-old mouse . | Dermis cells/mm . | Kidney cells/mm2 . | Bladder cells/mm2 . | Tendon cells/mm2 . | MMM cells/mm2 . | |
| wt | 744 ± 71 | 413 ± 10 | 245 ± 12 | 160 ± 17 | 161 ± 29 | + + + + | + + + + | |
| Csf1op/Csf1op | 526 ± 47* | 121 ± 18* | 157 ± 25* | 44 ± 11* | 42 ± 4* | − | − | |
| Csf1op/Csf1op; TgSGP4/+ | 457 ± 31* | 248 ± 5* | 246 ± 27 | 82 ± 9* | 154 ± 20 | + + | + + | |
| Csf1op/Csf1op; TgSGP2/+ | 1251 ± 75† | 464 ± 28 | 326 ± 53† | 198 ± 13 | 184 ± 29 | + + + + | + + + | |
| Csf1op/Csf1op; TgSPP7/+ | 902 ± 79 | 309 ± 38* | 229 ± 16 | 62 ± 14* | 189 ± 23 | + + + | + + | |
| Csf1op/Csf1op; TgSPP2/+ | 1129 ± 72† | 498 ± 75 | 297 ± 11† | 163 ± 20 | 201 ± 19 | + + + + | + + + | |
. | Liver cells/mm2 . | . | . | . | . | . | . | |
|---|---|---|---|---|---|---|---|---|
| Genotype . | 2-d-old mouse . | 3-mo-old mouse . | Dermis cells/mm . | Kidney cells/mm2 . | Bladder cells/mm2 . | Tendon cells/mm2 . | MMM cells/mm2 . | |
| wt | 744 ± 71 | 413 ± 10 | 245 ± 12 | 160 ± 17 | 161 ± 29 | + + + + | + + + + | |
| Csf1op/Csf1op | 526 ± 47* | 121 ± 18* | 157 ± 25* | 44 ± 11* | 42 ± 4* | − | − | |
| Csf1op/Csf1op; TgSGP4/+ | 457 ± 31* | 248 ± 5* | 246 ± 27 | 82 ± 9* | 154 ± 20 | + + | + + | |
| Csf1op/Csf1op; TgSGP2/+ | 1251 ± 75† | 464 ± 28 | 326 ± 53† | 198 ± 13 | 184 ± 29 | + + + + | + + + | |
| Csf1op/Csf1op; TgSPP7/+ | 902 ± 79 | 309 ± 38* | 229 ± 16 | 62 ± 14* | 189 ± 23 | + + + | + + | |
| Csf1op/Csf1op; TgSPP2/+ | 1129 ± 72† | 498 ± 75 | 297 ± 11† | 163 ± 20 | 201 ± 19 | + + + + | + + + | |
Mice used for dermis samples were 2 days old; those for kidney samples, 2 months old; those for bladder samples, 3 months old; tendon, 2 days old; and MMM, 6 weeks old. The antibody for all samples except MMM was F4/80; for MMM, it was MOMA-1.
Data (mean ± SD) are taken from 12 low-power fields of tissues from 2 mice of each genotype. Data are mean ± SD.
MMM indicates marginal metallophyllic macrophages; +, the degree of correction of MMM density in spleen and F4/80+ cell density in tendons of different transgenic lines in comparison with the wt; −, no correction.
P < .05; count lower than wt.
P < .05; count higher than wt.
Eyelid opening, macrophage morphology, and B-cell distribution in Csf1op/Csf1op; TgSGP/+ and Csf1op/Csf1op; TgSPP/+ mice. (A) Incomplete eyelid opening in Csf1op/Csf1op and Csf1op/Csf1op; TgSGP/+ mice. H&E staining of eyelids from 6-week-old mice. Top panels: narrow gap between the eyelids marked by an asterisk in Csf1op/Csf1op and Csf1op/Csf1op; TgSGP4/+ mice. Middle panels: chronic active blepharitis with hyperkeratosis marked by arrowhead. Bottom panels: meibomian gland (MG) hyperplasia of the eyelids of these same mice compared with the eyelids of wt mice. (B) Lack of apoptosis in the eyelids of 2-day-old Csf1op/Csf1op mice compared with wt control mice. Brown dots indicate TUNEL-positive cells. (C) Reduced dendritic morphology of F4/80-stained macrophages in 3-month-old Csf1op/Csf1op; TgSGP/+ liver. (D) B220+ cells as a percentage of total white blood cells in blood, spleen, and bone marrow of mice expressing TgSPP and TgSGP on Csf1op/Csf1op background. Data are given as mean ± SD. *P < .05 and †P < .08, for comparison with wt.
Eyelid opening, macrophage morphology, and B-cell distribution in Csf1op/Csf1op; TgSGP/+ and Csf1op/Csf1op; TgSPP/+ mice. (A) Incomplete eyelid opening in Csf1op/Csf1op and Csf1op/Csf1op; TgSGP/+ mice. H&E staining of eyelids from 6-week-old mice. Top panels: narrow gap between the eyelids marked by an asterisk in Csf1op/Csf1op and Csf1op/Csf1op; TgSGP4/+ mice. Middle panels: chronic active blepharitis with hyperkeratosis marked by arrowhead. Bottom panels: meibomian gland (MG) hyperplasia of the eyelids of these same mice compared with the eyelids of wt mice. (B) Lack of apoptosis in the eyelids of 2-day-old Csf1op/Csf1op mice compared with wt control mice. Brown dots indicate TUNEL-positive cells. (C) Reduced dendritic morphology of F4/80-stained macrophages in 3-month-old Csf1op/Csf1op; TgSGP/+ liver. (D) B220+ cells as a percentage of total white blood cells in blood, spleen, and bone marrow of mice expressing TgSPP and TgSGP on Csf1op/Csf1op background. Data are given as mean ± SD. *P < .05 and †P < .08, for comparison with wt.
Mice exclusively expressing sgCSF-1 tend to have lower B-cell numbers
Apart from their macrophage deficiencies, young Csf1op/Csf1op mice have a compensatory extramedullary splenic hematopoiesis secondary to their reduced bone marrow cellularity, as well as a decrease in B lymphocytes.28 To examine the effects of TgSGP and TgSPP on the hematologic abnormalities of Csf1op/Csf1op mice, we analyzed total cellularity and CD45.1, B220, CD3, Gr1, and Mac1 lineage marker expression in peripheral blood, bone marrow, and spleens of 2-month-old wt, Csf1op/Csf1op, Csf1op/Csf1op; TgSGP/+, and Csf1op/Csf1op; TgSPP/+ mice. Csf1op/Csf1op; TgSGP4/+, Csf1op/Csf1op; TgSGP2/+, Csf1op/Csf1op; TgSPP7/+, and Csf1op/Csf1op; TgSPP2/+ mice exhibited no significant differences from wt mice in all these parameters, except for B-cell numbers estimated using the B-cell marker, B220 (Figure 5D). In Csf1op/Csf1op; TgSGP4/+ mice, bone marrow B220+ cell concentrations were significantly lower than in wt mice, and their concentrations in the blood of Csf1op/Csf1op; TgSGP4/+ and Csf1op/Csf1op; TgSGP2/+ mice were also lower (P < .08). In contrast, the B-cell concentrations were indistinguishable from wt in Csf1op/Csf1op; TgSPP/+ mice.
Discussion
The present study, together with previous studies of Csf1op/Csf1op mice either expressing the full-length CSF-1 precursor encoding all 3 CSF-1 isoforms,28 or exclusively expressing csCSF-1,17 indicates that the different CSF-1 isoforms regulate differentially within tissues. While the exclusive expression of csCSF-1 in CSF-1-null (Csf1op/Csf1op) mice was able to normalize several aspects of development, it failed to correct many others, suggesting an important role of the secreted CSF-1 isoforms.17 We here confirm this observation, demonstrating that at wt levels of expression, the secreted CSF-1s rescue Csf1op/Csf1op defects more efficiently than csCSF-1. Furthermore, comparing secreted CSF-1 precursors that differ only in a single 18 000-kDa chondroitin sulfate chain, we show that the Csf1op/Csf1op defects are more efficiently and substantially rescued in mice expressing spCSF-1 than in mice exclusively expressing sgCSF-1. Incisor eruption, skeletal abnormalities, eyelid opening, the dendritic macrophage morphology, and the reduction in B-cell number of Csf1op/Csf1op mice were more efficiently rescued in Csf1op/Csf1op; TgSPP/+ mice than in Csf1op/Csf1op; TgSGP/+ mice. Indeed, the spCSF-1 precursor was not only better than the sgCSF-1 precursor but was almost as effective as the full-length CSF-1 precursor in rescuing Csf1op/Csf1op defects. Thus, our studies indicate an important role for the CSF-1 chondroitin sulfate chain in CSF-1 localization and/or signaling and emphasize the different, but overlapping, actions of the CSF-1 isoforms.
One aspect of the CSF-1-deficient phenotype was not corrected by either of the secreted CSF-1s. The failure of spCSF-1 or sgCSF-1, compared with csCSF-1,17 to completely correct the growth retardation of Csf1op/Csf1op mice, despite the complete rescue of skeletal size in Csf1op/Csf1op; TgSPP/+ mice, suggests that the csCSF-1 is required for normal adult body weight. The mechanism underlying this effect of csCSF-1 could be a CSF-1-dependent, macrophage-mediated increase in adipocyte size,45,46 since adipocytes express CSF-145 and are in close contact with macrophages in adipose tissue.46
We have previously shown that fibroblasts from Csf1op/Csf1op; TgC/+ mice expressing the full-length CSF-1 precursor display 30% of the cell surface CSF-1 of Csf1op/+ fibroblasts,28 due either to the membrane-spanning, cell-surface isoform or to association of sgCSF-1 or spCSF-1 with the extracellular matrix. Using the same method, we here demonstrate that neither sgCSF-1 nor spCSF-1 are expressed on the surface of fibroblasts. Thus, while the bulk of cell-surface CSF-1 is derived from the CSF-1 precursor encoded by a truncated mRNA in which the exon 6 sequence encoding the fragment containing the GAG addition site and all the proteolytic cleavage sites used to release the secreted isoforms has been spliced out, a small proportion of cell-surface sgCSF-1 and/or spCSF-1 can result from expression of the uncleaved, full-length precursor.
Stem cell factor (SCF) and flt ligand (FL) belong to the same growth factor family as CSF-1, act via similar class III receptor tyrosine kinases, and possess similarly generated cell-surface and secreted isoforms (reviewed in Broudy47 and Lyman and Jacobsen48 ). Cell-surface SCF has been shown to stimulate more prolonged signaling than secreted SCF by delaying internalization of the SCF receptor,49 and the cell-surface and secreted SCF isoforms have been shown to possess distinct roles in the migration of lateral neural crest cells50 and in erythropoiesis.51 Similarly, previous studies,17,28 in conjunction with the present study, indicate that cell-surface and secreted CSF-1s have shared and specialized roles. However, neither SCF nor FL possess GAG addition sites.
The present study clearly shows that the increased isoform diversity due to GAG addition has important consequences for CSF-1 function. Three distinct mechanisms by which spCSF-1 might exert its differential effects are differential stability or activity, selective localization at specific tissue sites, or differential signal transduction. In vitro experiments with the secreted human CSF-1 proteoglycan suggest that the chondroitin sulfate GAG chain and the C-terminal protein sequence containing it have negative effects on the CSF-1-stimulated proliferation but may also increase stability.52 Relevant to selective localization, spCSF-1 has been shown to differentially bind to collagen V,53 low-density lipoprotein,54 and bone55 and to be extractable from the bone matrix.56 These localization mechanisms could contribute to the differential effect of spCSF-1 that we have observed in the TgSPP mice.
There now exist several examples of modulation of growth factor signal transduction by proteoglycans. The presence of cell-surface heparan sulfate proteoglycan (HSPG) is necessary for the optimum binding of fibroblast growth factor to its cell-surface receptor and its mitogenic activity.57,58 Furthermore, cytokine signaling is modulated by the interaction of extracellular galectin-3 with the N-glycans present in the cytokine receptors.59 Whether spCSF-1 functions differentially through interactions of its chondroitin sulfate chain with ECM or cell-surface molecules remains to be investigated.
Not all interactions that have been attributed to spCSF-1 are necessarily mediated by the GAG component. spCSF-1 has a longer core protein than sgCSF-1, and, independent of the GAG chain, the core protein component specific to human spCSF-1 (approximately, amino acids 220-444) has been shown to bind basic fibroblast growth factor and neutralize its mitogenic activity in vitro.60 The in vivo role of this core component of spCSF-1 remains to be investigated. As TgSGP expresses this protein core, it represents an appropriate control for analysis of the effects of the chondroitin sulfate component of spCSF-1. However, one cannot regard TgSGP mice as mice exclusively expressing the sgCSF-1 found in wt mice, the vast majority of which does not contain amino acids 220 to 444.
The present study points to a critical role of the chondroitin sulfate moiety of spCSF-1 in regulation by CSF-1 in normal mice. How this GAG modulates the physiologic and developmental processes regulated by spCSF-1 is of obvious interest. Furthermore, the presence of CSF-1 has been shown to be deleterious in several mouse models of chronic disease, including atherosclerosis,3,61 obesity,46 collagen-induced arthritis,62 and renal inflammation,63 as well as the enhancement of tumor cell progression and metastasis.64-66 With the development of mice expressing the different CSF-1 isoforms, the roles played by the csCSF-1, spCSF-1, and sgCSF-1 in these diseases and in early hematopoiesis can be studied.
Prepublished online as Blood First Edition Paper, October 6, 2005; DOI 10.1182/blood-2005-05-1822.
Supported by National Institutes of Health grants CA32551 (E.R.S.), the Albert Einstein College of Medicine Cancer Center grant 5P30-CA13330, an American Society of Hematology Fellow Scholar Award (X.-M.D.), and a Leukemia and Lymphoma Society Special Fellow Award (X.-M.D.).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr G. R. Ryan for assembly of the transgenic constructs. We thank Xiao-Hua Zong and Ranu Basu for technical assistance and members of the AECOM transgenic, FACS, histopathology, and analytical imaging facilities for assistance in different aspects of this work. We thank Dr G. Kraal for the MOMA-1 antibody and Dr D. A. Hume for the F4/80 antibody.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal