Abstract
We assessed primary cutaneous large B-cell lymphoma, leg type (PCLBCL, leg type; n = 13), and primary cutaneous follicle center lymphoma (PCFCL; n = 19) for somatic hypermutation (SHM) of BCL6, and aberrant SHM of MYC, RhoH/TTF, and PAX5. We demonstrate SHM of BCL6 in 8 PCLBCLs (62%), leg type, and 7 PCFCL patients (37%), and aberrant SHM in PAX5, RhoH/TTF, and/or MYC in 7 PCLBCLs (54%), leg type, and 10 PCFCL patients (53%). The majority of mutations consisted of single base-pair substitutions (n = 54) with rare deletions/insertions (n = 4), and displayed molecular features typical of the SHM process. Quantitative real-time PCR and immunohistochemical stainings for activation-induced cytidine deaminase, which is indispensable for SHM, demonstrated significantly higher expression in PCLBCL, leg type. Our results suggest that (aberrant) SHM may contribute to the pathogenesis of PCLBCL, leg type, and PCFCL and is not restricted to diffuse large B-cell lymphomas with an aggressive clinical behavior.
Introduction
The recently published World Health Organization-European Organization for Research and Treatment of Cancer classification for primary cutaneous lymphomas distinguishes 2 main groups of primary cutaneous large B-cell lymphomas (PCLBCLs): primary cutaneous follicle center lymphomas (PCFCLs) with an indolent behavior (5-year survival > 95%) and PCLBCLs, leg type, with an intermediate prognosis (5-year survival = 50%).1
Recent investigations identified (aberrant) somatic hypermutation (SHM) of proto-oncogenes as a novel mechanism of genetic lesion in diffuse large B-cell lymphomas (DLBCLs).2 Normally, the SHM process introduces point mutations in the variable region of immunoglobulin genes (IgV) of germinal center (GC) B cells, allowing affinity maturation of the humoral immune response.3-5 Remarkably, the SHM process in normal GC B cells is not limited to the IgV sequences but can also target, albeit at a much lower rate, BCL6, CD95, and the B-cell receptor accessory proteins CD79b and CD79a. 6-10
In more than half of the patients with DLBCL, AIDS-associated DLBCL, and Burkitt lymphomas, aberrant activity of the SHM process affects regulatory as well as coding sequences from multiple gene loci, including proto-oncogenes PIM1, PAX5, RhoH/TTF, and MYC.2,11 In contrast, mutations of PIM1, PAX5, RhoH/TTF, and MYC do not occur at a significant level in normal GC B cells.2 Expression of AID (activation-induced cytidine deaminase) has been identified as an absolute requirement for SHM.12,13 Recent studies demonstrated expression of AID mRNA and protein in Burkitt lymphomas and systemic DLBCL.14-19
These observations resulted in the concept that (aberrant) SHM plays a role in lymphomagenesis of aggressive B-cell lymphomas by mutating multiple proto-oncogenes.
We investigated the occurrence of (aberrant) SHM of BCL6, PAX5, MYC, and RhoH/TTF in PCFCL and PCLBCL, leg type. In addition, the expression of AID mRNA and protein was studied in both entities.
Study design
Patient material and RNA and DNA extraction
Frozen pretreatment skin biopsies of 19 patients with PCFCL and 17 patients with PCLBCL, leg type, with the histology of a DLBCL and containing 60% or more neoplastic cells were included in this study. Frozen lymph node samples from 2 patients with follicular hyperplasia were included as controls. RNA was extracted from 50 × 20 μM sections using a RNeasy isolation kit (Qiagen, Hilden, Germany). DNA was extracted from 25 × 20 μM sections using the Genome-tip 20/G isolation kit and Proteinase K solution (Qiagen). This study was performed in accordance with the Dutch code and Leiden University Medical Center guidelines on leftover material. Patients provided informed consent in accordance with the Declaration of Helsinki.
Detection of somatic mutations
In 13 cases of PCLBCL, leg type, and 19 cases of PCFCL, mutations in BCL6, PAX5, RhoH/TTF, and MYC were investigated. Mutation analysis was restricted to regions previously shown to contain more than 90% of mutations in systemic DLBCL.2 Polymerase chain reaction (PCR) amplification of DNA was performed using previously described primer pairs covering a part of the promoter region, the first exon and intron of BCL6, PAX5, RhoH/TTF, and MYC.11,20 PCR products were purified and sequenced as described.21
Quantitative real-time PCR
In 10 cases of PCLBCL, leg type, and 8 cases of PCFCL, quantitative real-time PCR for AID was performed. cDNA synthesis, real-time PCR, and data analysis were performed as described previously.21 For the amplification of AID (AID, accession no. NM_020 661) and the housekeeping gene small nuclear ribonucleoprotein particle U1A (U1A, accession no. X06 347) the following primers were used: AID-F, 5′-AGA GGC GTG ACA GTG CTA CA-3′; AID-R, 5′-TGT AGC GGA GGAAGA GCAAT-3′; U1A-F, 5′-GCA GCT TAT GCC AGC ACA GAT-3′; and U1A-R, 5′-TTG GTG AGG AAC AAG ATG TGA TTC-3′.
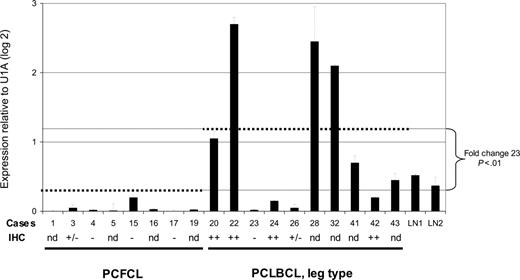
Expression of AID mRNA in PCFCL and PCLBCL, leg type. Expression of AID mRNA was measured in triplicate using quantitative real-time PCR and normalized to the expression of the housekeeping gene small nuclear ribonucleoprotein particle U1A (U1A). Error bars indicate standard deviations. Case numbers correspond to cases presented in Tables 1-2. dotted lines indicate the average AID mRNA expression within the group of PCFCL and PCLBCL, leg type; – indicates no tumor cells stained; +/– indicates less than 10% of tumor cells stained; and ++ indicates more than 50% of tumor cells stained. LN indicates lymph nodes; IHC, immunohistochemical staining; and nd, not determined.
Expression of AID mRNA in PCFCL and PCLBCL, leg type. Expression of AID mRNA was measured in triplicate using quantitative real-time PCR and normalized to the expression of the housekeeping gene small nuclear ribonucleoprotein particle U1A (U1A). Error bars indicate standard deviations. Case numbers correspond to cases presented in Tables 1-2. dotted lines indicate the average AID mRNA expression within the group of PCFCL and PCLBCL, leg type; – indicates no tumor cells stained; +/– indicates less than 10% of tumor cells stained; and ++ indicates more than 50% of tumor cells stained. LN indicates lymph nodes; IHC, immunohistochemical staining; and nd, not determined.
Immunohistochemical stainings for AID
In 12 cases of PCLBCL, leg type, and 8 cases of PCFCL, immunohistochemical stainings for AID, using antibody Eh2-5G9, were performed as previously described.18
Results and discussion
This study demonstrates the presence of SHM in BCL6 in 8 (62%) of 13 patients with PCLBCL, leg type, and 7 (37%) of 19 patients with PCFCL, and aberrant SHM in PAX5, RhoH/TTF, and/or MYC in 7 (54%) of 13 patients with PCLBCL, leg type, and 10 (53%) of 19 patients with PCFCL (Tables 1 and 2). The molecular profile of the detected mutations was similar in PCFCL and PCLBCL, leg type, and resembled the characteristics of mutations induced by the physiologic SHM process (Table S1, available on the Blood website; see the Supplemental Tables link at the top of the online article).3-5 These data provide additional evidence that PCLBCL, leg type, and PCFCL are derived from germinal center experienced B cells.
Overview of BCL6, PAX5, RhoH/TTF, and MYC mutations in PCFCL
. | Survival . | . | . | Mutation, position* . | . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | BCL6 . | . | PAX5 . | . | RhoH/TTF . | . | MYC . | . | ||||||||
| Case . | Status . | Time, mo . | AID . | No. . | Mutations . | No. . | Mutations . | No. . | Mutations . | No. . | Mutations . | ||||||||
| 1 | D0 | 28 | ND | 1 | g.751C>A | 0 | NA | 0 | NA | 1 | g.2480G>T | ||||||||
| 2 | A0 | 78 | ND | 1 | g.842T>C | 3 | g.739C>T, g.846C>T, g.848C>T | 3 | g.744C>G, g.816G>A, g.901T>C | 0 | NA | ||||||||
| 3 | A0 | 264 | Occasional | 0 | NA | 2 | g.1082C>T, g.1187C>T | 0 | NA | 0 | NA | ||||||||
| 4 | A+ | 86 | Negative | 0 | NA | 1 | g.714_715insT | 0 | NA | 0 | NA | ||||||||
| 5 | A0 | 35 | ND | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 6 | A0 | 30 | ND | 1 | g.683-695del | 1 | g.839-855del | 0 | NA | 1 | g.2739G>T | ||||||||
| 7 | A0 | 28 | Negative | 1 | g.713G>A | 0 | NA | 2 | g.469A>G, g.609G>A | 1 | g.2710C>T | ||||||||
| 8 | A0 | 23 | ND | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 9 | A0 | 42 | ND | 1 | g.798G>A | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 10 | A0 | 74 | ND | 2 | g.766C>A, g.809G>A | 7 | g.893C>T, g.1046C>T, g.1206G>A, g.1223G>A, g.1338C>G, g.1357C>G, g.1435G>C | 0 | NA | 1 | g.2541C>A | ||||||||
| 11 | A0 | 63 | Occasional | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 12 | A0 | 57 | Negative | 0 | NA | 0 | NA | 0 | NA | 1 | g.2466A>G | ||||||||
| 13 | A0 | 62 | ND | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 14 | A0 | 69 | ND | 1 | g.759G>A | 0 | NA | 0 | NA | 1 | g.2366C>T | ||||||||
| 15 | D0 | 70 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 16 | A0 | 52 | ND | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 17 | A0 | 40 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 18 | D+ | 130 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 19 | A0 | 113 | ND | 0 | NA | 0 | NA | 0 | NA | 1 | g.2616C>G | ||||||||
. | Survival . | . | . | Mutation, position* . | . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | BCL6 . | . | PAX5 . | . | RhoH/TTF . | . | MYC . | . | ||||||||
| Case . | Status . | Time, mo . | AID . | No. . | Mutations . | No. . | Mutations . | No. . | Mutations . | No. . | Mutations . | ||||||||
| 1 | D0 | 28 | ND | 1 | g.751C>A | 0 | NA | 0 | NA | 1 | g.2480G>T | ||||||||
| 2 | A0 | 78 | ND | 1 | g.842T>C | 3 | g.739C>T, g.846C>T, g.848C>T | 3 | g.744C>G, g.816G>A, g.901T>C | 0 | NA | ||||||||
| 3 | A0 | 264 | Occasional | 0 | NA | 2 | g.1082C>T, g.1187C>T | 0 | NA | 0 | NA | ||||||||
| 4 | A+ | 86 | Negative | 0 | NA | 1 | g.714_715insT | 0 | NA | 0 | NA | ||||||||
| 5 | A0 | 35 | ND | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 6 | A0 | 30 | ND | 1 | g.683-695del | 1 | g.839-855del | 0 | NA | 1 | g.2739G>T | ||||||||
| 7 | A0 | 28 | Negative | 1 | g.713G>A | 0 | NA | 2 | g.469A>G, g.609G>A | 1 | g.2710C>T | ||||||||
| 8 | A0 | 23 | ND | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 9 | A0 | 42 | ND | 1 | g.798G>A | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 10 | A0 | 74 | ND | 2 | g.766C>A, g.809G>A | 7 | g.893C>T, g.1046C>T, g.1206G>A, g.1223G>A, g.1338C>G, g.1357C>G, g.1435G>C | 0 | NA | 1 | g.2541C>A | ||||||||
| 11 | A0 | 63 | Occasional | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 12 | A0 | 57 | Negative | 0 | NA | 0 | NA | 0 | NA | 1 | g.2466A>G | ||||||||
| 13 | A0 | 62 | ND | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 14 | A0 | 69 | ND | 1 | g.759G>A | 0 | NA | 0 | NA | 1 | g.2366C>T | ||||||||
| 15 | D0 | 70 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 16 | A0 | 52 | ND | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 17 | A0 | 40 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 18 | D+ | 130 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 19 | A0 | 113 | ND | 0 | NA | 0 | NA | 0 | NA | 1 | g.2616C>G | ||||||||
A0 indicates alive without disease; A+, alive with disease; D0, death unrelated to lymphoma; D+, death related to lymphoma; negative, no tumor cells stained; occasional, less than 10% of tumor cells stained; NA, not applicable; and ND, not determined.
Overview of BCL6, PAX5, RhoH/TTF, and MYC mutations in PCLBCL, leg type
. | Survival . | . | . | Mutation, position* . | . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | BCL6 . | . | PAX5 . | . | RhoH/TTF . | . | MYC . | . | ||||||||
| Case . | Status . | Time, mo . | AID . | No. . | Mutations . | No. . | Mutations . | No. . | Mutations . | No. . | Mutations . | ||||||||
| 20 | A0 | 75 | Double-positive | 1 | g.835T>C | 2 | g.883C>T, g.1070G>A | 2 | g.574T>G, g.589C>G | 0 | NA | ||||||||
| 21 | D+ | 73 | Double-positive | 1 | g.704G>C | 1 | g.1334C>T | 1 | g.912G>A | 0 | NA | ||||||||
| 22 | D+ | 29 | Double-positive | 2 | g.545G>T, g.661G>A | 0 | NA | 0 | NA | 1 | g.2405delC | ||||||||
| 23 | A0 | 72 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 24 | D+ | 20 | Double-positive | 1 | g.799C>A | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 25 | D+ | 12 | Occasional | 2 | g.552A>C, g.845C>T | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 26 | D0 | 10 | Occasional | 1 | g.837T>G | 0 | NA | 0 | NA | 1 | g.4835G>C, p.Ser146Leu | ||||||||
| 27 | A0 | 31 | Double-positive | 0 | NA | 1 | g.1347G>A | 0 | NA | 0 | NA | ||||||||
| 28 | D+ | 26 | ND | 2 | g.782C>G, g.794G>A | 0 | NA | 2 | g.368G>A, g.682C>T | 0 | NA | ||||||||
| 29 | D+ | 3 | ND | 2 | g.778C>T, g.779G>C | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 30 | D+ | 9 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 31 | A0 | 76 | Double-positive | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 32 | D+ | 38 | ND | 0 | NA | 0 | NA | 1 | g.306T>C | 0 | NA | ||||||||
. | Survival . | . | . | Mutation, position* . | . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | BCL6 . | . | PAX5 . | . | RhoH/TTF . | . | MYC . | . | ||||||||
| Case . | Status . | Time, mo . | AID . | No. . | Mutations . | No. . | Mutations . | No. . | Mutations . | No. . | Mutations . | ||||||||
| 20 | A0 | 75 | Double-positive | 1 | g.835T>C | 2 | g.883C>T, g.1070G>A | 2 | g.574T>G, g.589C>G | 0 | NA | ||||||||
| 21 | D+ | 73 | Double-positive | 1 | g.704G>C | 1 | g.1334C>T | 1 | g.912G>A | 0 | NA | ||||||||
| 22 | D+ | 29 | Double-positive | 2 | g.545G>T, g.661G>A | 0 | NA | 0 | NA | 1 | g.2405delC | ||||||||
| 23 | A0 | 72 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 24 | D+ | 20 | Double-positive | 1 | g.799C>A | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 25 | D+ | 12 | Occasional | 2 | g.552A>C, g.845C>T | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 26 | D0 | 10 | Occasional | 1 | g.837T>G | 0 | NA | 0 | NA | 1 | g.4835G>C, p.Ser146Leu | ||||||||
| 27 | A0 | 31 | Double-positive | 0 | NA | 1 | g.1347G>A | 0 | NA | 0 | NA | ||||||||
| 28 | D+ | 26 | ND | 2 | g.782C>G, g.794G>A | 0 | NA | 2 | g.368G>A, g.682C>T | 0 | NA | ||||||||
| 29 | D+ | 3 | ND | 2 | g.778C>T, g.779G>C | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 30 | D+ | 9 | Negative | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 31 | A0 | 76 | Double-positive | 0 | NA | 0 | NA | 0 | NA | 0 | NA | ||||||||
| 32 | D+ | 38 | ND | 0 | NA | 0 | NA | 1 | g.306T>C | 0 | NA | ||||||||
A0 indicates alive without disease; A+, alive with disease; D0, death unrelated to lymphoma; D+, death related to lymphoma; negative, no tumor cells stained; occasional, less than 10% of tumor cells stained; positive, 10% to 50% of tumor cells stained; double-positive, more than 50% of tumor cells stained; and ND, not determined.
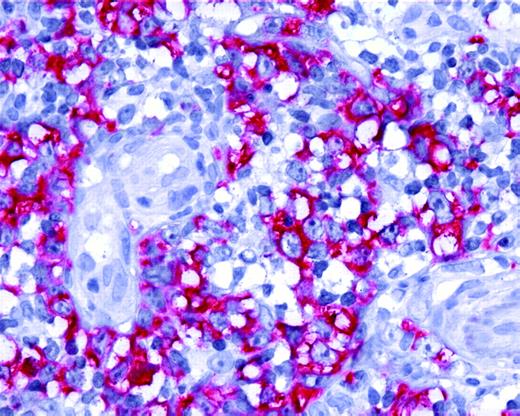
Expression of AID by tumor cells in PCLBCL, leg type. Neoplastic B cells of primary cutaneous large B-cell lymphoma, leg type, show positive staining for AID protein (original magnification ×40). Images were captured with a Leica DMLB microscope (Leica, Bensheim, Germany), using a Leica PL FLUOTAR objective lens (40×/0.70) and a Leica DC200 camera. Images were directly imported into Power-point (Microsoft, Redmond, WA), using the Leica DC200 camera software (version 2.51).
Expression of AID by tumor cells in PCLBCL, leg type. Neoplastic B cells of primary cutaneous large B-cell lymphoma, leg type, show positive staining for AID protein (original magnification ×40). Images were captured with a Leica DMLB microscope (Leica, Bensheim, Germany), using a Leica PL FLUOTAR objective lens (40×/0.70) and a Leica DC200 camera. Images were directly imported into Power-point (Microsoft, Redmond, WA), using the Leica DC200 camera software (version 2.51).
Similar to studies of systemic DLBCL, mutations were most frequently demonstrated in BCL6, and the mutation frequency for aberrant SHM in PAX5, MYC, and RhoH/TTF was generally lower than that observed for SHM of BCL6 sequences in the same cases (Tables S2-S3).2,11 A higher mutation frequency for BCL6 was observed in PCLBCL, leg type (P < .05, Student t test). Because one-third of patients with PCLBCL, leg type, harbor translocations affecting BCL6, whereas this chromosomal aberration is not observed in PCFCL, these data are in accordance with previous studies that suggested that SHM could play a role in the induction of chromosomal translocations.2,22 The functional consequences of the mutations detected in this study include potential deregulated gene transcription resulting from mutations in the 5′ regulatory regions of BCL6, PAX5, RhoH/TTF, and MYC. In addition, in one patient with PCLBCL, leg type, a mutation in exon 2 of MYC, representing a mutational hotspot in translocated MYC alleles in Burkitt lymphoma and mouse plasmacytoma, was found.23,24
Based on observations in systemic B-cell lymphomas, it was postulated that aberrant SHM is a selective feature of aggressive B-cell lymphomas.2,11 In our study, however, we demonstrate aberrant SHM in PCFCLs that are characterized by an indolent behavior and PCLBCL, leg type, with an intermediate prognosis. Also within the group of PCLBCL, leg type, the prevalence of aberrant SHM did not correlate with survival.
The mean expression level of AID mRNA, normalized to the housekeeping genes, was significantly higher in PCLBCL, leg type, than in PCFCL (P < .01, Student t test; Figure 1). Expression of AID protein was confined to the cytoplasm of neoplastic cells (Figure 2) and was demonstrated in 10 (83%) of 12 patients with PCLBCL, leg type (with more than 50% of tumor cells staining in 6 patients), whereas in PCFCL, AID protein was not detected in 6 of 8 patients and was expressed by less than 10% of tumor cells in 2 cases (Tables 1-2). A close correlation between AID transcripts and protein expression was found in the 10 patients in whom both quantitative real-time PCR and immunohistochemical stainings were performed (Figure 1). No correlation was found between expression of AID and occurrence of SHM, which is in line with previous observations.15,19
Because PCLBCL, leg type, and PCFCL display an ABC-like and GCB-like gene expression profile, respectively,25 our observations are in line with studies in nodal DLBCL, which demonstrated that AID protein expression is tendentially higher in the ABC-like DLBCL.19
In conclusion, we demonstrate that in the majority of patients with PCFCL and PCLBCL, leg type, the proto-oncogenes BCL6, PAX5, RhoH/TTF, and, to a lesser extent, MYC are affected by (aberrant) SHM. In addition, we show that within the group of PCLBCLs, expression of AID is found in the vast majority of cases of PCLBCL, leg type, but is rarely detected in PCFCL. These results demonstrate that aberrant SHM is not restricted to B-cell non-Hodgkin lymphoma with an aggressive behavior, and suggest that persistent expression of AID may contribute to the lymphomagenesis of PCLBCL, leg type.
Prepublished online as Blood First Edition Paper, February 28, 2006; DOI 10.1182/blood-2005-08-3443.
Supported by grants from The Netherlands Organisation for Health Research and Development (no. 907-00-066; M.H.V.).
R.D., C.P.T., R.W., and M.H.V. designed the study; R.D., C.P.T., and M.B. performed the research; R.D., C.P.T., G.N., M.B., R.W., and M.H.V. analyzed the data; R.D., C.P.T., R.W., and M.H.V. wrote the paper; and all authors checked the final version of the manuscript.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Wim Zoutman and Jacqueline van Harmelen for their excellent technical assistance. We would like to thank Elisabeth Kremmer for providing the antibody EK2-5G9.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal