Abstract
The role of signal transducers and activators of transcription 5 (STAT5) in chronic myelogenous leukemia (CML) is controversial. To clarify the role of STAT5 signaling in P210BCR/ABL leukemogenesis, P210 was introduced into primary murine STAT5A-deficient (STAT5A–/–) bone marrow (BM) cells, which, unlike STAT5A/5B double knockout BM cells, have no major intrinsic hematopoietic defects. Interestingly, only 21% of mice reconstituted with P210-transduced STAT5A–/– BM cells developed classic CML, compared with 80% to 100% of P210/STAT5A+/+ and P210/STAT5A+/–-reconstituted animals. The remainder of P210/STAT5A–/– animals died from an acute B-cell lymphoblastic leukemia (ALL)–like disease (32%) or a CML/ALL mix (47%), reflecting impairment in the induction and maintenance of CML, which normally predominates in this mouse model. Of mice that ultimately developed CML, P210/STAT5A–/– animals had prolonged survival and increased myeloid immaturity. Importantly, reconstitution of wild-type mice with BM cells coexpressing P210 and dominant-negative STAT5 also profoundly reduced the incidence of CML, without impairing the induction of ALL. Altogether, these findings indicate that STAT5 and STAT5A play an important role in the pathogenesis of the CML-like disease in mice. A greater understanding of the STAT5 target genes involved in CML induction may lead to new therapeutic targets that influence CML progenitor cell biology.
Introduction
The non–receptor tyrosine kinase Bcr/Abl activates a complex array of signaling molecules that alter cellular survival, proliferation, differentiation, genomic stability, and adhesion.1-5 The central role of Bcr/Abl in the pathogenesis of Philadelphia chromosome–positive (Ph+) hematologic malignancy has been recently illustrated by the success of Bcr/Abl-targeted therapy with the tyrosine kinase inhibitor imatinib mesylate.6-9 However, cure of Ph+ leukemia with imatinib mesylate has not yet been established. Moreover, the growing problem of imatinib-resistant chronic myelogenous leukemia (CML)10-14 emphasizes the need to better understand the role of downstream signaling events in Bcr/Abl-induced leukemia. The constitutive activation of signal transducers and activators of transcription 5 (STAT5) in Bcr/Abl-expressing cells has stimulated considerable interest because it implicates deregulation of a transcription factor prominent in hematopoietic cytokine signaling with a leukemia-associated oncogene.15-19 Indeed, a variety of studies have demonstrated that interference with STAT5 activation negatively impacts the survival and proliferation of Bcr/Abl-expressing cells.20-23 Moreover, STAT5 activation has been a useful surrogate marker of Bcr/Abl kinase activity in vivo,24 and has been implicated in tyrosine kinase inhibitor–mediated CML cell apoptosis.22,25
The role of STAT5 in Bcr/Abl-induced leukemia in vivo, however, remains incompletely defined. In 32D cells, expression of a Bcr/Abl mutant defective in STAT5 activation, or coexpression of dominant-negative (DN) STAT5 with wild-type P210, inhibited leukemogenesis in immunodeficient mice.20 DN STAT5 also inhibited the P210-dependent transformation of primary murine bone marrow (BM) cells,20 and the growth of the CML cell line K562 in semisolid medium.21 In Ba/F3 P210 cells, STAT5 mediated the expression of the survival factor Bcl-XL, and played an important role in Bcr/Abl cellular viability and chemosensitivity.23,26 However, STAT5A- and STAT5B-deficient (STAT5A/B–/–) mouse BM cells were still capable of generating fatal leukemia in a murine P210 BM transduction and transplantation model, suggesting a nonessential role of STAT5 in Bcr/Abl-induced leukemia.27 Nonetheless, in that study the relative proportion of the pure CML-like myeloproliferative disorder was 4-fold lower in P210/STAT5A/B–/–-reconstituted mice, suggesting a potentially important role of STAT5 in CML pathogenesis. Interpretation of these results has been complex, however, since BM cells from STAT5A/B–/– mice form approximately 50% fewer interleukin (IL)–3, granulocyte-macrophage colony-stimulating factor–, and granulocyte colony–stimulating–induced colonies than wild-type BM cells28 ; show decreased hematopoietic repopulating ability29-31 ; and may express truncated forms of STAT5.32,33 Thus, decreased CML in this knockout mouse strain could reflect an important role for STAT5 in the efficient generation of CML, or an intrinsic hematopoietic defect in the STAT5A/B–/– CML progenitor population.
To clarify the role of STAT5 signaling in P210BCR/ABL leukemogenesis, we have expressed Bcr/Abl in primary murine STAT5A–/– BM cells, which have no demonstrable defects in bone marrow colony formation.28 In leukemogenesis assays, STAT5A–/– BM cells were defective in inducing the CML-like illness in recipient mice, shifting the Bcr/Abl leukemic spectrum toward lymphoid malignancy, which was not impaired by the absence of STAT5. In a complementary approach, coexpression of P210 and dominant negative STAT5 in wild-type murine hematopoietic cells prolonged survival and profoundly decreased the incidence of the CML-like disease. Taken together, these findings demonstrate an important role for STAT5 in the efficient generation of the pure CML-like illness in mice, and suggest that STAT5 target genes may influence CML progenitor cell function.
Materials and methods
Cell lines
293T cells were grown in DME (high-glucose) medium supplemented with 10% fetal calf serum, penicillin/streptomycin, 2 mm glutamine, and nonessential amino acids at 37°C at 5% CO2. The murine cytokine–dependent cell lines 32D and Ba/F3 were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, penicillin/streptomycin, 2 mm glutamine, and 10% WEHI-3B–conditioned medium as a source of IL-3. NIH 3T3 cells were propagated in DME (high-glucose) medium supplemented with 10% bovine calf serum.
Plasmids and retroviral vectors
In the STAT5A knockout and control animals, the murine CML-like myeloproliferative disorder was induced by expressing P210 in the retroviral vectors MSCV/neo34 or MSCV/IRES/GFP.35 For leukemogenesis experiments in the strain Balb/c, P210 was coexpressed with DN STAT5 or a control gene using the retroviral vector MINV.36 The construction and characterization of the pCDNA3 STAT5A mutants STAT5A/Δ53C and STAT5A-EE (E436E to AA) have been previously described elsewhere.36 The parent vector MINV/IRES/P210 was cloned by a 3-part ligation using EcoRI/BamHI (all 5′ to 3′) and BclI/EcoRI fragments derived from MINV/IRES/P190 (kindly provided by Rick Van Etten, Tufts–New England Medical Center, Boston, MA), and a BamHI/BclI fragment derived from MSCV/P210. To construct STAT5A/IRES/P210, the STAT5A mutant cDNA inserts were liberated from pCDNA3 vectors as blunted HindIII/NotI fragments and cloned into the HpaI and NotI sites of MINV/IRES/P210. Neo/IRES/P210 was constructed by partial digestion of MINV/IRES/P210 with SalI, followed by digestion with EcoRI to obtain IRES/P210. This fragment was then ligated with an EcoRI/SalI neomycin resistance gene cassette obtained from the vector pGD/P210.37 The integrity of all constructs was confirmed by restriction enzyme digestion and DNA sequencing.
Retrovirus generation and characterization
Helper virus–free, replication-defective retroviral supernatants were obtained by transient transfection of 293T cells with the indicated retroviral plasmid and an ecotropic packaging construct38 as previously described.36,39 For the experiments in the STAT5A knockout strain, retroviral supernatants were titered by fluorescence-activated cell sorting (FACS; MSCV/IRES/GFP) or NIH 3T3 cell–based neomycin-resistant colony formation assays (MSCV/neo). Because the STAT5 mutant/P210 bicistronic retroviral vectors lacked a selectable marker, retroviral supernatants were titered by anti-Abl immunoblot to ensure equivalent potency. Briefly, NIH 3T3 cells, plated at a density of 5 × 105 per 10 cm2 plate the night before, were incubated with 3 mL bicistronic retroviral supernatant for 3 hours at 37°C in the presence of 8 μg/mL hexadimethrine bromide (polybrene; Sigma, St Louis, MO). After incubation, fresh medium was applied and the cells were allowed to grow to confluence (approximately 48 hours). Protein lysates were then prepared and analyzed by anti-Abl western immunoblot to confirm equivalent expression of P210, and antihemagglutin (HA) immunoblot to confirm expression of the HA-tagged STAT5A mutants.
STAT5A-deficient mice
The generation and initial characterization of STAT5A-deficient mice have been previously described elsewhere.40 Mice (B6;129S6-Stat5atm1Mam/J) with homozygous (–/–) or heterozygous (+/–) deficiency of STAT5A were purchased from Jackson Laboratory (Bar Harbor, Maine). All subsequent STAT5A-deficient or STAT5A wild-type (+/+) mice used in this study were generated from matings with littermates. The STAT5A genotype was determined by a polymerase chain reaction (PCR)–based assay using primer pairs directed against the neomycin-resistance gene targeting construct40 and STAT5A (Figure 1A). The primers (all from 5′ to 3′) used were STAT5A (290 bp) CTGGATTGACGTTTCTTACCTG (sense), TGGAGTCAACTAGTCTGTCTCT (antisense); Neo (480 bp) AGAGGCTATTCGGCTATGACTG (sense), TTCGTCCAGATCATCCTGATC (antisense), using the conditions of 94°C × 30 seconds, 52°C × 45 seconds, and 72°C × 45 seconds for a total of 30 cycles.40 The average white blood cell (WBC) and cell differential of STAT5A–/– and STAT5A+/+ donor mice was determined from peripheral blood samples (n = 3; 600 cells counted per smear). Peripheral blood cytospins were prepared to illustrate baseline morphology (Figure 2C). To determine the percentage of MAC1+ and B220+ cells, BM cells were obtained from STAT5A-null and STAT5A wild-type BM donors at baseline and after priming with 5-fluorouracil (5-FU; n = 3), and analyzed by FACS.
Murine Bcr/Abl leukemogenesis assay
All mouse leukemia experiments were performed according to established animal protocols approved by the Institutional Animal Use and Care Committee at the University of Texas Southwestern Medical Center. STAT5A wild-type (+/+) mice were used as BM recipients for all experiments, while BM donors were divided into STAT5A–/– or STAT5A+/+ cohorts. As a control, STAT5A+/– donors were used in some experiments. The methodologic details of the murine BM retroviral transduction and transplantation model of CML have been extensively described elsewhere.41 Briefly, murine BM cells were harvested from 5-FU–primed donor mice, transduced with a replication-defective P210BCR/ABL retrovirus by serial spinfection (centrifugation of cells in retroviral supernatant), and then injected into lethally irradiated syngeneic recipient animals by tail-vein injection (average of 5 × 105 total BM cells per mouse). STAT5A–/– and STAT5A+/+ BM donor cells were each transduced with the same P210 retroviral stock. Animals were serially monitored for signs of leukemia according to established protocols. Mice showing sufficient signs of pain and suffering were killed, and a complete necropsy was performed. Final diagnoses were established by clinical, pathologic, morphologic, and FACS analysis, as previously described.42 In general, a diagnosis of CML/ALL was made when B-lymphoblasts comprised at least 25% of the peripheral blood or BM cellularity in the setting of otherwise classic murine CML features, including marked splenomegaly and granulocytosis. Photomicrographs of hematoxylin and eosin–stained smears were prepared using a Nikon Eclipse E800 microscope (Nikon, Melville, NY) and a 40 × /0.95 numeric aperture (NA) (Figure 1C) or a 100 × /1.4 NA (Figures 2B-C, 4) Plan Apo objective lens. Images were captured using a Nikon DXM 1200 digital camera and Nikon Act-1 software version 2.51, and were processed using Adobe Photoshop (Adobe Systems, San Jose, CA). Representative examples of the leukemia diagnoses are shown in Figure 1C. For the survival curve analysis, statistical significance was evaluated using the log-rank test. For comparisons of leukemia lineage proportions, a 2-tailed Fisher exact test was performed (Graphpad Prism version 4.0 for Macintosh, San Diego, CA).
Antibodies and immunoblot
Protein lysates were prepared from the indicated cell lines or primary murine tissues with RIPA buffer supplemented with 1% vol/vol aprotinin, 1 mM sodium orthovanadate, 25 mM sodium fluoride, 5 μg/mL leupeptin, 1 μg/mL pepstatin, 2 mM Pefabloc SC (Roche Diagnostics, Mannheim, Germany), and 1 mM phenylmethylsulfonyl fluoride. Lysates were normalized by OD595 (Bio-Rad Protein assay; Bio-Rad Laboratories, Hercules, CA), and resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using 7% or 12% Laemmli polyacrylamide gels. Gels were transferred by electrophoresis overnight at 4°C to a nitrocellulose membrane, and analyzed by total anti-STAT5 (C-17; Santa Cruz Biotechnology, Santa Cruz, CA), anti-HA (Covance Research Products, Denver, PA), antiphosphotyrosine (Upstate Biotechnology, Lake Placid, NY), anti-Abl (EMD Biosciences, San Diego, CA), or anti-STAT5A (L-20; Santa Cruz Biotechnology) antibodies, and detected by enhanced chemiluminescence (Amersham, Arlington Heights, IL). For FACS, anti-GR1 (positive in the CML-like disease), anti-MAC1 (positive in CML and macrophage leukemia), and anti-B220 (positive in CML/ALL and ALL) antibodies were used (all from BD PharMingen, San Diego, CA).
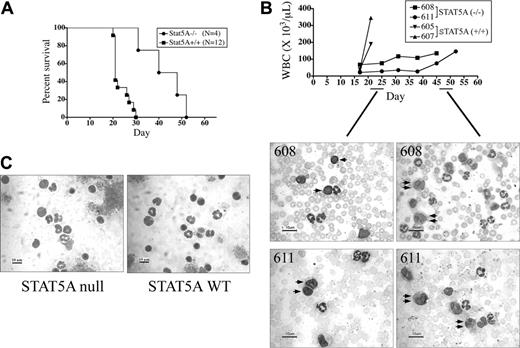
Mice reconstituted with P210-transduced STAT5A-null BM cells have a significantly lower incidence of the pure CML-like disease. (A) The STAT5A genotype of B6;129S6 mice was determined by PCR, detecting the presence or absence of genomic sequences from the mouse STAT5A gene (290-bp fragment), and the neomycin (neo) phosphotransferase gene (480-bp) used in the original targeting construct.40 STAT5A genomic and water (–) controls are shown at right. (B) Lethally irradiated STAT5A+/+ mice were reconstituted with P210-transduced BM cells from 5-FU–primed STAT5A–/– or STAT5A+/+ donors and assessed for survival by the method of Kaplan and Meier. The number of mice (N) in each cohort is shown at right. The median survival of P210/STAT5A–/–-reconstituted mice was 31 days, compared with 21 days for P210/STAT5A+/+-reconstituted animals (P = .13). (C) Photomicrographs of blood smears (original magnification, × 40; scale bar = 50 μM) of representative animals with CML, CML/ALL mix, and ALL. Note that CML/ALL is comprised of a fairly equal distribution of granulocytes (single arrow) and lymphoblasts (double arrow). The respective MAC1/B220 FACS profiles are shown beneath each disease photomicrograph panel. Isotype control peaks are shown at left in gray, and quantitation of the number of MAC1 (myeloid)– or B220 (B-lymphoid)–positive cells is shown at the top of each panel. (D) Leukemia subtype of mice reconstituted with P210-transduced STAT5A–/–, STAT5A+/+, or STAT5A+/– BM cells. NE indicates nonevaluable; other, macrophage-monocytic/basophil leukemia. The incidence of CML was significantly decreased in P210/STAT5A–/–-reconstituted mice (P = .001 compared with STAT5A+/+ mice; Fisher exact test).
Mice reconstituted with P210-transduced STAT5A-null BM cells have a significantly lower incidence of the pure CML-like disease. (A) The STAT5A genotype of B6;129S6 mice was determined by PCR, detecting the presence or absence of genomic sequences from the mouse STAT5A gene (290-bp fragment), and the neomycin (neo) phosphotransferase gene (480-bp) used in the original targeting construct.40 STAT5A genomic and water (–) controls are shown at right. (B) Lethally irradiated STAT5A+/+ mice were reconstituted with P210-transduced BM cells from 5-FU–primed STAT5A–/– or STAT5A+/+ donors and assessed for survival by the method of Kaplan and Meier. The number of mice (N) in each cohort is shown at right. The median survival of P210/STAT5A–/–-reconstituted mice was 31 days, compared with 21 days for P210/STAT5A+/+-reconstituted animals (P = .13). (C) Photomicrographs of blood smears (original magnification, × 40; scale bar = 50 μM) of representative animals with CML, CML/ALL mix, and ALL. Note that CML/ALL is comprised of a fairly equal distribution of granulocytes (single arrow) and lymphoblasts (double arrow). The respective MAC1/B220 FACS profiles are shown beneath each disease photomicrograph panel. Isotype control peaks are shown at left in gray, and quantitation of the number of MAC1 (myeloid)– or B220 (B-lymphoid)–positive cells is shown at the top of each panel. (D) Leukemia subtype of mice reconstituted with P210-transduced STAT5A–/–, STAT5A+/+, or STAT5A+/– BM cells. NE indicates nonevaluable; other, macrophage-monocytic/basophil leukemia. The incidence of CML was significantly decreased in P210/STAT5A–/–-reconstituted mice (P = .001 compared with STAT5A+/+ mice; Fisher exact test).
RNA isolation and RT-PCR
Total RNA was prepared from the peripheral blood, BM, and spleen of leukemic mice using the Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After the concentration and integrity of the RNA was confirmed by agarose gel electrophoresis, complementary (c) DNA was performed using the Superscript First Strand Synthesis System (Invitrogen, Carlsbad, CA). The primers used for semi-quantitative reverse transcription (RT)–PCR expression of STAT5 target gene expression were (all 5′ to 3′): Bcl-XL (324 bp) CCGGAGAGCGTTCAGTGATC (sense), and TCAGGAACCAGCGGTTGAAG (antisense);26 oncostatin M (450 bp) GGGCCAGAGTACCAGGACCCAGTAT (sense), GCCGGGCCATGCAGAAAACA (antisense). The ribosomal-associated protein L7 (202 bp) was used as an integrity control.43
Results
STAT5A–/– bone marrow cells are inefficient in generating the mouse CML-like illness
Lethally irradiated STAT5A+/+ mice were reconstituted with equivalent numbers of P210BCR/ABL-transduced BM cells obtained from 5-FU–primed STAT5A–/– or STAT5A+/+ donor mice, following the well-established BM retroviral transduction and transplantation model of CML.35,37,41 The percentage of mature (MAC1+/GR1+) and immature (MAC1+/GR1–) myeloid populations and B-lymphoid (B220+) cells were similar in STAT5A-null and STAT5A wild-type BM (Table 1; non–5-FU). As expected, STAT5A-null and STAT5A wild-type BM donors had higher ratios of immature to mature myeloid cells after priming with 5-FU (Table 1; 5-FU). Consistent with the results observed in the mouse strain Balb/c, all animals reconstituted with P210-transduced BM cells died from Bcr/Abl-induced leukemia.35,37,44 Mice reconstituted with P210/STAT5A+/+ BM cells had a median survival of 21 days, while P210/STAT5–/–-reconstituted mice had a median survival of 31 days (P = .13; Figure 1B).
STAT5A–/– and STAT5A+/+ donor bone marrow populations at baseline (non–5-FU) and after priming with 5-FU
. | STAT5A-/-,% . | . | . | STAT5A+/+,% . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | MAC1+* . | . | . | MAC1+ . | . | . | ||||
. | GR1- . | GR1+ . | B220 . | GR1- . | GR1+ . | B220 . | ||||
| 5-FU | 82 ± 3 | 18 ± 3 | 42 ± 1 | 86 ± 5 | 14 ± 5 | 55 ± 1 | ||||
| Non—5-FU | 32 ± 4 | 68 ± 4 | 32 ± 4 | 32 ± 2 | 68 ± 4 | 33 ± 3 | ||||
. | STAT5A-/-,% . | . | . | STAT5A+/+,% . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | MAC1+* . | . | . | MAC1+ . | . | . | ||||
. | GR1- . | GR1+ . | B220 . | GR1- . | GR1+ . | B220 . | ||||
| 5-FU | 82 ± 3 | 18 ± 3 | 42 ± 1 | 86 ± 5 | 14 ± 5 | 55 ± 1 | ||||
| Non—5-FU | 32 ± 4 | 68 ± 4 | 32 ± 4 | 32 ± 2 | 68 ± 4 | 33 ± 3 | ||||
Results expressed as percent ± SE; n = 3 for each STAT5A genotype.
Immature (MAC1+/GR1-) and mature (MAC1+/GR1+) myeloid cells are expressed as a percentage of total MAC1+ cells.
To explore the reasons why P210/STAT5A–/–-reconstituted mice tended to survive longer than P210/STAT5A+/+-reconstituted animals, leukemia cells were analyzed by morphology and FACS to establish the cause of death. As in Balb/c mice, most (80%) of P210/STAT5A+/+-reconstituted mice died from the CML-like myeloproliferative disorder, characterized by a marked expansion of MAC1+ granulocytes in the peripheral blood (Figure 1C, left).35,45 One animal (7%) died of an acute leukemia on day 19 that was likely CML (splenomegaly, pulmonary hemorrhage), but was scored as nonevaluable (NE) because of suboptimal material for diagnosis. Two P210/STAT5A+/+-reconstituted animals (13%) died on days 50 and day 52 of a monocyte-macrophage/basophil–mix leukemia (Figure 1D; other). In contrast, only 21% of P210/STAT5–/–-reconstituted died from the CML-like illness, almost 4-fold less than control animals (P < .001, Fisher exact test). Approximately one-third (32%) of P210/STAT5A–/–-reconstituted mice died from the B-ALL–like disease, with replacement of the peripheral blood and BM with immature B-220+ lymphoblasts (Figure 1C, right), and lymphadenopathy or hind-limb paralysis.37,42 Nearly half (47%) of the P210/STAT5A–/–-reconstituted animals died from a CML/ALL overlap illness,27 characterized by a mixture of MAC1+ myeloid cells and B220+ B-ALL–like cells in the peripheral blood and BM (Figure 1C, middle). None of the P210/STAT5A+/+-reconstituted mice died from CML/ALL or B-ALL. Recipients reconstituted with P210-transduced STAT5A+/– BM cells all died from CML (Figure 1D), and had a survival similar to P210/STAT5+/+ mice (data not shown).
P210/STAT5A–/– CML mice have prolonged survival due to slower CML disease kinetics. (A) The survival of mice with the pure CML-like disease resulting from the transplantation of P210/STAT5A–/– or P210/STAT5A+/+ BM cells was analyzed by Kaplan-Meier survival curve. The median survival of P210/STAT5A–/– CML animals was 44 days, compared with 21 days for P210/STAT5A+/+ CML animals (P < .001, log-rank test). (B) Serial peripheral WBC counts from mice dying of the pure CML-like illness. Note that P210/STAT5A–/–-reconstituted CML animals (no. 608 and no. 611) had WBC counts of 76 × 109/L (76 000/μL) and 28 × 109/L (28 000/μL), respectively, on day 25 after BM reconstitution, comprised mostly of maturing granulocytic lineage cells, intermixed with some B lymphoblasts (left panels, single arrows). Relatively little change was noted in serial WBC counts until days 45 and 52, when the P210/STAT5A–/– mice died from the CML-like disease with WBC counts of 135 × 109/L (135 000/μL) and 146 × 109/L (146 000/μL), respectively, composed entirely of granulocytes, including relatively immature myeloid cells (right panels, double arrows). For comparison, P210/STAT5A+/+-reconstituted CML mice (no. 605 and no. 607) had WBC counts of 40 × 109/L (40 000/μL) and 38.5 × 109/L (38 500/μL), respectively, on day 17, and died 4 days later from CML with WBC counts of 190 × 109/L (190 000/μL) and 345 × 109/L (345 000/μL). (C) Peripheral blood cytospins from nonleukemic STAT5A-null and STAT5A wild-type (WT) mice are shown for comparison, composed of lymphocytes and mature granulocytes (average 80% and 20%, and 79% and 21%, respectively). A 10-μm size bar is shown at bottom left of each photomicrograph.
P210/STAT5A–/– CML mice have prolonged survival due to slower CML disease kinetics. (A) The survival of mice with the pure CML-like disease resulting from the transplantation of P210/STAT5A–/– or P210/STAT5A+/+ BM cells was analyzed by Kaplan-Meier survival curve. The median survival of P210/STAT5A–/– CML animals was 44 days, compared with 21 days for P210/STAT5A+/+ CML animals (P < .001, log-rank test). (B) Serial peripheral WBC counts from mice dying of the pure CML-like illness. Note that P210/STAT5A–/–-reconstituted CML animals (no. 608 and no. 611) had WBC counts of 76 × 109/L (76 000/μL) and 28 × 109/L (28 000/μL), respectively, on day 25 after BM reconstitution, comprised mostly of maturing granulocytic lineage cells, intermixed with some B lymphoblasts (left panels, single arrows). Relatively little change was noted in serial WBC counts until days 45 and 52, when the P210/STAT5A–/– mice died from the CML-like disease with WBC counts of 135 × 109/L (135 000/μL) and 146 × 109/L (146 000/μL), respectively, composed entirely of granulocytes, including relatively immature myeloid cells (right panels, double arrows). For comparison, P210/STAT5A+/+-reconstituted CML mice (no. 605 and no. 607) had WBC counts of 40 × 109/L (40 000/μL) and 38.5 × 109/L (38 500/μL), respectively, on day 17, and died 4 days later from CML with WBC counts of 190 × 109/L (190 000/μL) and 345 × 109/L (345 000/μL). (C) Peripheral blood cytospins from nonleukemic STAT5A-null and STAT5A wild-type (WT) mice are shown for comparison, composed of lymphocytes and mature granulocytes (average 80% and 20%, and 79% and 21%, respectively). A 10-μm size bar is shown at bottom left of each photomicrograph.
At time of retroviral transduction, 5-FU–primed STAT5A–/– donor BM contained an average of 42% B220+ cells, compared with 55% from STAT5A+/+ BM donors (Table 1). Thus, the increase in B-ALL in P210/STAT5A–/–-reconstituted mice was not associated with increased availability of B220+ progenitors. These findings demonstrate that STAT5A-deficient BM cells are inefficient in generating the classic murine CML-like myeloproliferative disorder.
CML-related survival is prolonged in P210/STAT5A–/–-reconstituted mice
To determine whether the absence of STAT5A influenced the pathogenesis of the murine CML-like disease per se, attention was focused on the P210/STAT5A–/– and P210/STAT5A+/+ CML cohorts. P210/STAT5A+/+-reconstituted CML animals had a median survival of 21 days, consistent with previous reports.35,44 In contrast, the median survival of P210/STAT5A–/–-reconstituted CML animals was 44 days, more than 2-fold longer than P210/STAT5A+/+-reconstituted mice (Figure 2A; P < .001, log-rank test). Consistent with these findings, serial WBC analysis of leukemic animals revealed a more gradual clinical progression of CML in P210/STAT5A–/–-reconstituted mice. For example, P210/STAT5A–/– mice no. 608 and no. 611 had peripheral WBC counts of 76 × 109/L (76 000/μL) and 28 × 109/L (28 000/μL) on day 25 after BM reconstitution (Figure 2B, top). Despite hematologic evidence of leukemia, the peripheral WBC count was relatively stable for 3 to 4 weeks until the animals succumbed to the CML-like disease on days 45 and 52, with WBC counts of 135 × 109/L (135 000/μL) and 146 × 109/L (146 000/μL), respectively. In contrast, P210/STAT5A+/+ mice no. 605 and no. 607 had WBC counts of 40 × 109/L (40 000/μL) and 38.5 × 109/L (38 500/μL) on day 17, and died only 4 days later from CML with WBC counts of 190 × 109/L (190 000/μL) and 345 × 109/L (345 000/μL), respectively. In P210/STAT5A–/– CML mice, the peripheral blood smear on day 25 revealed mainly maturing myeloid cells, intermixed with some B-lymphoblasts (Figure 2B, left, single arrows), suggesting an early CML-ALL competition. However, 3 to 4 weeks later at autopsy the peripheral blood was comprised entirely of granulocytes, diagnostic of the pure CML-like disease.
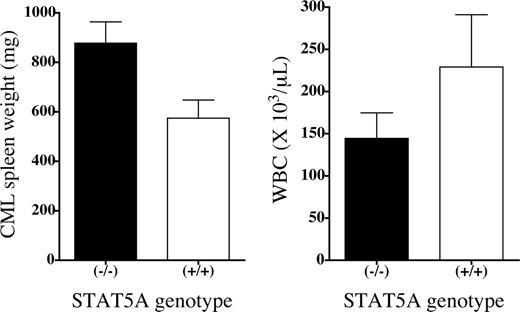
At autopsy, P210/STAT5A–/– CML mice had a mean WBC count of 145 × 109 ± 30 × 109/L (145 000 ± 30 000/μL) compared with 229 × 109 ± 63 109/L (229 000 ±63 000/μL) for P210/STAT5A+/+ CML animals, but this difference was not statistically significant (P = .38; Figure 3, right). Interestingly, P210/STAT5A–/– CML mice had greater splenomegaly than P210/STAT5A+/+ CML animals, with mean spleen weights of 880 (± 86) mg and 570 (± 74) mg, respectively (P = .046; Figure 3, left), perhaps as a consequence of longer survival with the CML disease. For comparison, the average spleen weight for this mouse strain was 70 mg and the average WBC count for nonleukemic STAT5A–/– and STAT5A+/+ mice was 4.6 ×109/L (4600/μL) and 5.5 × 109/L (5500/μL), respectively.
Greater splenomegaly in P210/STAT5A–/– CML mice. The average spleen weight (left panel) and peripheral WBC count (right panel) at autopsy is depicted by bar graph according to the STAT5A genotype of the leukemia cells. The difference in splenomegaly between P210/STAT5A-null (n = 4) and P210/STAT5A wild-type (n = 12) CML mice was statistically significant (P = .046), but the difference in WBC counts (n = 4 and n = 8, respectively) was not (P = .38). The error bars represent the standard error. For comparison, the average spleen weight of normal mice was 70 mg, and the average WBC counts of nonleukemic STAT5A–/– and STAT5A+/+ mice was 4.6 × 109/L (4600/μL) and 5.5 × 109/L (5500/μL), respectively.
Greater splenomegaly in P210/STAT5A–/– CML mice. The average spleen weight (left panel) and peripheral WBC count (right panel) at autopsy is depicted by bar graph according to the STAT5A genotype of the leukemia cells. The difference in splenomegaly between P210/STAT5A-null (n = 4) and P210/STAT5A wild-type (n = 12) CML mice was statistically significant (P = .046), but the difference in WBC counts (n = 4 and n = 8, respectively) was not (P = .38). The error bars represent the standard error. For comparison, the average spleen weight of normal mice was 70 mg, and the average WBC counts of nonleukemic STAT5A–/– and STAT5A+/+ mice was 4.6 × 109/L (4600/μL) and 5.5 × 109/L (5500/μL), respectively.
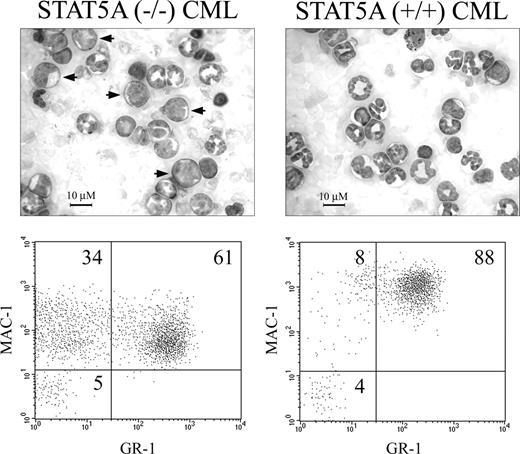
Besides a more protracted clinical course, the CML-like disease in STAT5A-null BM recipients demonstrated an increased proportion of immature myeloid cells (Figure 2B, right, double arrows). To determine if the absence of STAT5A altered the degree of myeloid leukemia differentiation, peripheral blood leukemia samples from additional cohorts of P210/STAT5A–/–- and P210/STAT5A+/+-reconstituted CML mice were analyzed by FACS (n = 4; representative animals shown in Figure 4). An average of 72% (± 6%) of P210/STAT5A+/+ CML cells were MAC1 and GR1 double-positive, reflecting the predominance of morphologically mature granulocytic cells (Figure 4, top right) observed in the CML-like myeloproliferative disorder.35,44 An average of 16% (± 4%) P210/STAT5A+/+ CML cells were MAC1+/GR1–, a marker of less mature granulocytic and monocytic cells.46 In P210/STAT5A–/– CML, the average percentage of immature MAC1+/GR1– cells was 32% (± 2%), 2-fold higher than in P210/STAT5A+/+ CML (P = .01).
Increased myeloid immaturity in STAT5A–/– CML. Photomicrographs and FACS plots of representative mice with the CML-like illness induced by P210/STAT5A–/– or P210/STAT5A+/+ BM cells. Note the increased proportion of morphologically immature myeloid cells (arrows) in P210/STAT5A–/– CML, associated with an increase in MAC1+ cells that lack expression of the mature granulocytic marker GR1. Cell percentage is depicted in the top right portions of each grid. Similar results were seen in 3 additional mice from each cohort (average values in “CML-related survival is prolonged in P210/STAT5A–/–-reconstituted mice.”
Increased myeloid immaturity in STAT5A–/– CML. Photomicrographs and FACS plots of representative mice with the CML-like illness induced by P210/STAT5A–/– or P210/STAT5A+/+ BM cells. Note the increased proportion of morphologically immature myeloid cells (arrows) in P210/STAT5A–/– CML, associated with an increase in MAC1+ cells that lack expression of the mature granulocytic marker GR1. Cell percentage is depicted in the top right portions of each grid. Similar results were seen in 3 additional mice from each cohort (average values in “CML-related survival is prolonged in P210/STAT5A–/–-reconstituted mice.”
This finding was associated with a constitutive increase in morphologically immature myeloid cells in P210/STAT5A–/– CML, including some myeloblasts (Figure 4, left, arrows). The average percentage of MAC1/GR1 double-positive cells was 49% (± 5%) in P210/STAT5A–/– CML, about a third lower than in P210/STAT5A+/+ CML. There was no evidence of baseline myeloid immaturity in STAT5A-null or STAT5A wild-type donor mice, with average peripheral blood neutrophil/band and lymphocyte differentials of 20% and 80%, and 21% and 79%, respectively (representative peripheral blood cytospins are shown in Figure 2C). Thus, despite slower clinical progression, STAT5A–/– CML had a less mature myeloid phenotype, suggesting that the absence of STAT5A constitutively altered the balance of myeloid proliferation and differentiation.
No difference in CML clonality in P210/STAT5A–/– and P210/STAT5A+/+ mice
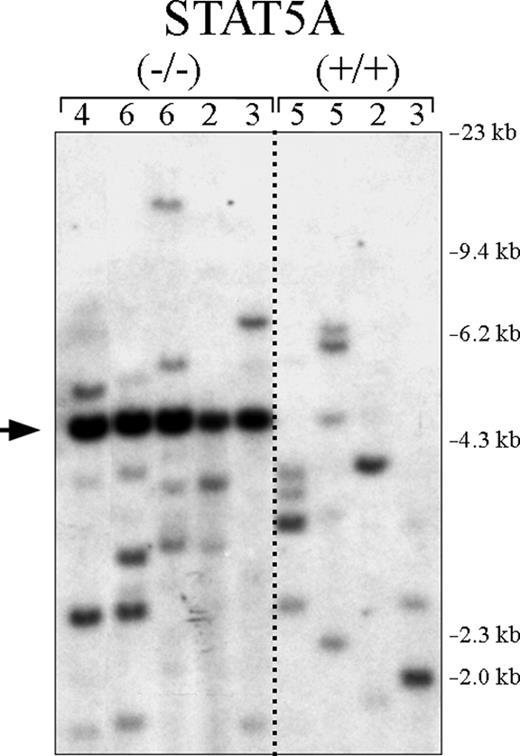
In Balb/c mice, primary CML is a polyclonal disease, generally with a range of 4 to 12 unique clones by Southern blot.42,44 To determine if the low incidence of CML in P210/STAT5A–/– BM cell recipients was due to fewer available CML progenitor cell targets, leukemia cells from P210/STAT5A–/– and P210/STAT5A+/+-reconstituted CML mice were examined by Southern blot to assess P210 proviral clonality.44 Control P210/STAT5A+/+ CML mice had an average of 3.8 leukemic clones (Figure 5, right), slightly lower than that typically observed in the strain Balb/c, the mouse strain most susceptible to Abl-induced malignancy.47 P210/STAT5A–/–-induced CML had an average of 4.2 unique clones, the same as control CML animals. Because a fragment of the neomycin resistance gene was used as the probe for these experiments, an additional band (Figure 5, arrow) was detected in all P210/STAT5A–/– leukemia samples (but not P210/STAT5A+/+ samples), confirming the presence of the neomycin resistance gene-targeting construct used to generate the STAT5A knockout strain.40 These findings suggest that the lower incidence of CML in P210/STAT5A–/–-reconstituted mice was not due to insufficient numbers of BCR/ABL cellular targets.
The absence of STAT5A in hematopoietic progenitors does not reduce CML clonality. Genomic DNA was prepared from P210/STAT5A–/– and P210/STAT5A+/+ leukemia cells isolated from mice with the CML-like disease, digested with the restriction enzyme Bgl II, and analyzed by Southern blot to assess the clonality of proviral integration. The number of unique leukemic clones is shown at the top for each CML animal. Size markers are shown at far right. Note that the neomycin resistance gene probe also detects the neomycin resistance cassette present in the STAT5A targeting vector (arrow).
The absence of STAT5A in hematopoietic progenitors does not reduce CML clonality. Genomic DNA was prepared from P210/STAT5A–/– and P210/STAT5A+/+ leukemia cells isolated from mice with the CML-like disease, digested with the restriction enzyme Bgl II, and analyzed by Southern blot to assess the clonality of proviral integration. The number of unique leukemic clones is shown at the top for each CML animal. Size markers are shown at far right. Note that the neomycin resistance gene probe also detects the neomycin resistance cassette present in the STAT5A targeting vector (arrow).
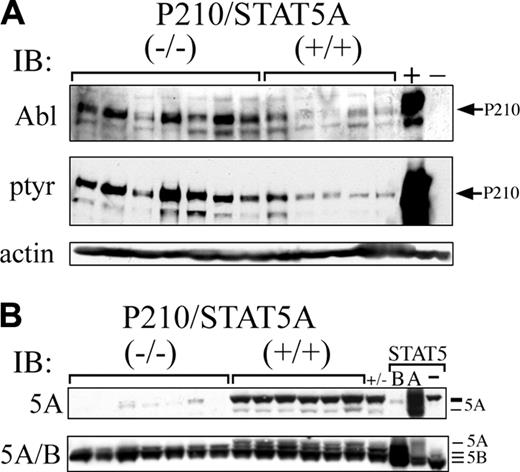
Bcr/Abl protein expression is constitutively increased in P210/STAT5A–/– CML. (A) Leukemia cells from P210/STAT5A–/– or P210/STAT5A+/+ mice with CML or CML/ALL were analyzed by anti-Abl or antiphosphotyrosine immunoblot. Ba/F3 cells expressing P210 (+) or not (–) are shown at far right as controls. Actin serves as a loading control (bottom panel). (B) Leukemia cells were analyzed by western immunoblot using an antibody specific for STAT5A (5A), or an antibody recognizing both STAT5A/B. Controls for STAT5A/5B and their migration positions are shown at far right. Note that the STAT5A-specific antibody (top panel) cross-reacts slightly with the slower migrating form of STAT5B.
Bcr/Abl protein expression is constitutively increased in P210/STAT5A–/– CML. (A) Leukemia cells from P210/STAT5A–/– or P210/STAT5A+/+ mice with CML or CML/ALL were analyzed by anti-Abl or antiphosphotyrosine immunoblot. Ba/F3 cells expressing P210 (+) or not (–) are shown at far right as controls. Actin serves as a loading control (bottom panel). (B) Leukemia cells were analyzed by western immunoblot using an antibody specific for STAT5A (5A), or an antibody recognizing both STAT5A/B. Controls for STAT5A/5B and their migration positions are shown at far right. Note that the STAT5A-specific antibody (top panel) cross-reacts slightly with the slower migrating form of STAT5B.
Increased P210 protein expression in P210/STAT5A–/– leukemia cells
P210/STAT5+/+ CML mice constitutively expressed a range of P210 protein (Figure 6A, top), consistent with previous findings.44 Interestingly, P210/STAT5–/– CML or CML/ALL mice generally expressed higher constitutive levels of P210 protein than P210/STAT5+/+ animals. Six of 7 leukemia samples from CML or CML/ALL P210/STAT5–/– mice had P210 protein expression greater or equal to the highest expresser in the P210/STAT5+/+ CML cohort (Figure 6, top left). P210 tyrosine phosphorylation was also generally increased in P210/STAT5–/– mice, paralleling the levels of P210 protein expression (Figure 6A, middle). These results suggest that increased P210 expression might have been able to compensate, in part, for the absence of STAT5A signaling in CML hematopoietic progenitors.
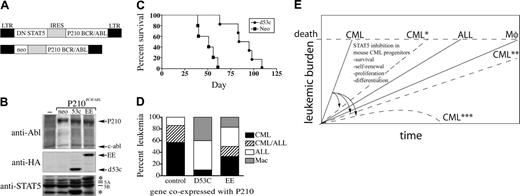
Prolonged survival and decreased CML in mice reconstituted with BM cells coexpressing P210 and DN STAT5. (A) Schematic depicting a bicistronic retroviral vector used to co-express DN STAT5 and Bcr/Abl in primary murine BM cells. (B) Retroviral supernatants were titered on NIH 3T3 fibroblast cells by anti-Abl and antihemagglutinin (HA) Western immunoblot (recognizing epitope-tagged STAT5 mutants). The positions of the STAT5 mutants were confirmed by anti-STAT5A/5B immunoblot (bottom panel). The carboxy-terminal deletion mutant DN STAT5A/Δ53C (D53c) migrates below endogenous STAT5A/5B (bottom asterisk). P210 bicistronic retroviruses coexpressing neomycin (neo) or a STAT5A point mutant with limited DN STAT5 activity (STAT5A-EE; top asterisk), or sham-transduced cells (–), are shown as expression controls. (C) Survival curves of mice reconstituted with BM cells transduced by DNSTAT5A/Δ53C/P210 (n = 6), or neo/P210 (n = 5). The survival difference between DN STAT5A/Δ53C/P210- and neo/P210-reconstituted animals was statistically significant (P < .001, log-rank test). The results depicted are representative of at least duplicate experiments using equivalent titer bicistronic retroviruses, as described in “Materials and methods.” (D) A bar graph depicting the leukemia phenotype of STAT5A/Δ53C/P210 (n = 10), STAT5A-EE/P210 (n = 6), or control (neo)/P210 (n = 7) mice. (E) A schematic depicting the kinetics of this mouse Bcr/Abl leukemia model, in which the CML-like disease reaches a lethal leukemic burden before the later onset Bcr/Abl ALL and macrophage/monocyte leukemias can develop. By a not-yet fully understood mechanism, STAT5 inhibition has a greater impact on CML progenitors than other BCR/ABL hematopoietic targets, shifting the kinetics and/or spectrum of leukemia by delaying (*), impairing (**), or preventing CML (***).
Prolonged survival and decreased CML in mice reconstituted with BM cells coexpressing P210 and DN STAT5. (A) Schematic depicting a bicistronic retroviral vector used to co-express DN STAT5 and Bcr/Abl in primary murine BM cells. (B) Retroviral supernatants were titered on NIH 3T3 fibroblast cells by anti-Abl and antihemagglutinin (HA) Western immunoblot (recognizing epitope-tagged STAT5 mutants). The positions of the STAT5 mutants were confirmed by anti-STAT5A/5B immunoblot (bottom panel). The carboxy-terminal deletion mutant DN STAT5A/Δ53C (D53c) migrates below endogenous STAT5A/5B (bottom asterisk). P210 bicistronic retroviruses coexpressing neomycin (neo) or a STAT5A point mutant with limited DN STAT5 activity (STAT5A-EE; top asterisk), or sham-transduced cells (–), are shown as expression controls. (C) Survival curves of mice reconstituted with BM cells transduced by DNSTAT5A/Δ53C/P210 (n = 6), or neo/P210 (n = 5). The survival difference between DN STAT5A/Δ53C/P210- and neo/P210-reconstituted animals was statistically significant (P < .001, log-rank test). The results depicted are representative of at least duplicate experiments using equivalent titer bicistronic retroviruses, as described in “Materials and methods.” (D) A bar graph depicting the leukemia phenotype of STAT5A/Δ53C/P210 (n = 10), STAT5A-EE/P210 (n = 6), or control (neo)/P210 (n = 7) mice. (E) A schematic depicting the kinetics of this mouse Bcr/Abl leukemia model, in which the CML-like disease reaches a lethal leukemic burden before the later onset Bcr/Abl ALL and macrophage/monocyte leukemias can develop. By a not-yet fully understood mechanism, STAT5 inhibition has a greater impact on CML progenitors than other BCR/ABL hematopoietic targets, shifting the kinetics and/or spectrum of leukemia by delaying (*), impairing (**), or preventing CML (***).
STAT5A and STAT5B are encoded by separate, highly homologous genes that are responsible for both unique and overlapping cellular functions,27,28,40,48-51 but their relative roles in CML are unknown. To determine if the development of the CML-like disease in some P210/STAT5A–/–-reconstituted mice reflected compensatory changes in STAT5B protein expression, peripheral blood or BM leukemia cells from mice with CML or CML/ALL were examined by Western immunoblot. As expected, leukemia cells from mice reconstituted with P210/STAT5–/– BM cells lacked STAT5A protein expression by STAT5A-specific or total anti-STAT5 (5A/B) immunoblot (Figure 6B). However, there was no difference in the constitutive expression of STAT5B protein in P210/STAT5–/– and P210/STAT5+/+ leukemia cells (Figure 6B, bottom).
Coexpression of P210 and DN STAT5 in primary murine leukemia cells interferes with the generation of the mouse CML-like illness
Although BM cells from mice with homozygous deficiency of STAT5A have generally normal hematopoietic colony formation,28 it is possible that the impaired generation of the CML-like disease reflected occult STAT5A–/–-related defects in CML progenitor cell population(s). To address this possibility, a complementary approach was devised in which wild-type Balb/c BM cells were transduced with a bicistronic retrovirus co-expressing P210 and the DN STAT5 mutant STAT5A/Δ53C.36 In this vector, an HA-tagged STAT5A/Δ53C mutant was expressed from the first position of the bicistronic retrovirus MINV, and P210 was expressed downstream from the internal ribosome entry site (IRES) (Figure 7A). To titer retroviral supernatants, and to confirm simultaneous expression of P210 and STAT5 mutant proteins in recipient cells, NIH 3T3 cells were transduced by STAT5A/Δ53C/P210 or control (neo) retroviral supernatants and analyzed 48 hours later by Western immunoblot. The STAT5A/Δ53C mutant protein was expressed at levels similar to endogenous STAT5A and migrated below endogenous STAT5B (Figure 7B, lower asterisk), as confirmed by anti-HA immunoblot (Figure 7B, middle panel). The levels of Bcr/Abl protein were the same in DNSTAT5/P210 and control neo/P210-transduced cells, reflecting equivalent retroviral titer (Figure 7B, top panel).
Lethally irradiated wild-type Balb/c mice were reconstituted with equivalent numbers of STAT5A/Δ53C/P210- or neo/P210-transduced BM cells and clinically monitored for signs of leukemia. Mice reconstituted with neo/P210-transduced cells had a median survival of 51 days, while STAT5A/Δ53C-reconstituted mice lived almost twice as long, with a median survival of 97 days (Figure 7C; P < .001, log-rank test). Most (86%) of the neo/P210-reconstituted mice died from the CML-like myeloproliferative disorder or a mixture of CML and B-cell ALL (Figure 7D). STAT5A/Δ53C/P210-reconstituted mice (d53c), in contrast, rarely developed the CML-like myeloproliferative disorder, with only one mouse (10%) developing the CML-like disease (P = .004, compared with CML plus CML/ALL neo/P210 animals). This CML animal died on day 60 after BM reconstitution with a WBC count of 115 × 109/L (115 000/μL) and a spleen weight of 1180 mg (normal, 70 mg44 ). Half (50%) of STAT5A/Δ53C/P210 mice died from B-cell ALL between days 72 and 95 after BM reconstitution, and 40% died from P210-induced macrophage/monocytic leukemia37,52,53 between days 83 and 109. As an additional control for specificity, mice reconstituted with BM cells transduced with a bicistronic retroviral vector coexpressing P210 and STAT5A/EE (Figure 7B, right), a STAT5 point mutant with only modest DN STAT5 activity,36 had a leukemia spectrum intermediate between STAT5A/Δ53C/P210 and neo/P210 animals. Half (50%) of STAT5A-EE/P210 animals died from CML or CML/ALL (P = .27, compared with neo/P210), while the remainder died from either B-ALL or macrophage/monocytic leukemia. Thus, the DN STAT5 mutant STAT5A/Δ53C interfered with the ability of P210 to induce the CML-like disease, but did not interfere with the induction of the ALL or macrophage/monocytic leukemia.
Discussion
The tyrosine kinase inhibitor imatinib mesylate has become the standard of care in newly diagnosed CML, but the persistence of BCR/ABL progenitors and emergence of imatinib-resistant CML suggest combination therapeutic approaches may be required for pharmacologic cure. Thus, the identification of additional signaling pathways important in the pathogenesis of CML remains an important goal. In the case of STAT5 and Bcr/Abl, reaching firm conclusions has been difficult. A wealth of studies have shown that STAT5 mediates signals important for Bcr/Abl survival20,22,23,25,26 and cellular transformation.20,54 In STAT5A/B-deficient mice, the CML-like illness still occurred, but it was more commonly manifested as a CML/ALL overlap disease,27 suggesting the absence of STAT5 impaired Bcr/Abl-induced myeloid leukemogenesis. However, given the hematopoietic repopulation and BM colony formation defects of STAT5A/5B knockout mice,28-31 and the possibility of residual STAT5 activity in these mice,32,33 the role of STAT5 in CML has remained unresolved.
The results of our study demonstrate that STAT5A (STAT5) plays an important role in supporting the mouse CML-like illness. Recipients of P210/STAT5A–/– BM cells experienced a 4-fold decrease in the pure CML disease because of a shift in the spectrum of leukemia toward B-cell ALL and a CML/ALL mix. The increase in Bcr/Abl-induced ALL is particularly significant considering the customary dominance of myeloid leukemia in this Bcr/Abl leukemia model. In fact, the addition of murine CML cells can suppress even established B-ALL in this mouse Bcr/Abl leukemia model.45 Thus, the increased incidence of ALL and CML/ALL in P210/STAT5A–/– recipients reflected a STAT5A-related defect in myeloid leukemogenesis, which translated into a more protracted CML illness and prolonged CML-related survival. Even the P210/STAT5A–/– animals that ultimately died from pure CML did so only after an antecedent, protracted CML/ALL competition not observed in mice reconstituted with P210/STAT5A+/+ cells (Figure 2B).
Besides prolonged survival, the attenuated STAT5A–/– CML disease was associated with greater splenomegaly, likely because the slower progression of pulmonary hemorrhage (the usual proximate cause of death) allowed more time for progression of leukemic organomegaly. There was also a significant increase in myeloid immaturity in P210/STAT5A–/–-reconstituted mice, consistent with previous findings implicating STAT5 in myeloid differentiation.36 This suggests that STAT5A loss may have impaired the differentiation of early P210-expressing myeloid cells, manifested by a constitutive increase in MAC1+/GR1– leukemia cells. In chicken myeloblasts expressing DN STAT and cytokine-stimulated murine STAT5A/5B-null BM cells, there was increased apoptosis during myeloid differentiation that was rescued by over-expression of Bcl-2 or Bcl-XL.55 In our study, Bcl-XL and oncostatin M mRNA expression was constitutively decreased in some, but not all P210/STAT5A–/– CML animals (data not shown), raising the possibility that slower STAT5A–/– CML progression and decreased myeloid maturity might have reflected increased myeloid cell turnover during differentiation. However, by FACS, there was no evidence of increased cell death in P210/STAT5A–/– leukemia samples (data not shown). Because samples were prepared from animals with advanced CML, it is possible that compensatory mechanisms or clonal selection had obscured early STAT5A-null–related detrimental effects. Interestingly, P210 expression was generally increased in P210/STAT5A–/– CML, consistent with recent findings implicating Bcr/Abl overexpression in CML progression and imatinib mesylate resistance.11,12,56-58 Despite the increased number of myeloblasts in mouse P210/STAT5A–/– CML, the clinical and hematologic picture was more consistent with the loss of myeloid differentiation seen in human CML in accelerated phase, rather than the more fulminant CML blast crisis. Perhaps further study of STAT5 signaling in early- and late-stage human CML will provide novel insights in the mechanisms responsible for the loss of hematopoietic differentiation associated with CML progression.
Although STAT5A knockout BM cells function normally in colony formation assays,28 there is a possibility that CML impairment reflected a cell extrinsic phenomenon, namely that STAT5A–/– donor animals lacked sufficient CML progenitors. Several of our findings argue against this possibility, however. First, clonality was not decreased in STAT5A–/– CML, suggesting preserved availability of CML-generating hematopoietic cells. Second, evidence of CML was detectable at some point, albeit partially, in most P210/STAT5A–/–-reconstituted mice, suggesting that the absence of STAT5A impaired the ability of P210-expressing progenitors to sustain CML. Third, we obtained similar results when P210 was coexpressed with DN STAT5 in a wild-type inbred (Balb/c) mouse strain. In those experiments, the pure CML illness was almost 6-fold lower in P210/STAT5A/Δ53C mice than control animals. Further, coexpression of a STAT5A mutant with only modest DN STAT5 activity (STAT5A/EE) had an intermediate effect on suppressing CML, further supporting a specific role for STAT5 in the murine CML-like illness.
Overall, our findings support the hypothesis that decreased CML in P210/STAT5A–/– and DN STAT5/P210 BM recipients is the consequence of inhibiting STAT5 in distinct Bcr/Abl hematopoietic cellular targets. In this model, when the effect on CML progenitors is modest (due to preserved STAT5B, increased P210 expression, or other compensatory mechanisms), CML can still occur but survival is prolonged (Figure 7E, CML*). In other cases, loss of STAT5-related signaling leads to imbalances in CML progenitor proliferation, differentiation, self-renewal, and/or viability, eliminating the biologic superiority of CML over the other leukemia subtypes (Figure 7E, CML**). More severe adverse effects on CML progenitor function may block CML induction at a very early stage (Figure 7E, CML***). In the case of DN STAT5, CML and CML/ALL were both rare, likely because DN STAT interfered with both STAT5A and STAT5B function.36 In contrast to the STAT5A-null experiments, a fairly large proportion (40%) of DN STAT5/P210 animals died of the macrophage/monocytic leukemia, suggesting the possibility that interference with STAT5A and STAT5B might have slightly favored the macrophage/monocyte leukemia progenitor, which has not been well studied.
Interestingly, in 32D cells, DN STAT5A/Δ53C profoundly blocked myeloid differentiation, with many cells displaying a monocytic/macrophage-like morphology.36
In summary, murine STAT5A-null BM cells were inefficient in generating and maintaining the pure CML-like disease, suggesting an important role of STAT5 in the pathogenesis of CML and implying that STAT5B cannot fully replace STAT5A-mediated leukemogenic signals. Perhaps future experiments with the recently described genomic loxP-STAT5–engineered mice32 will shed additional light into whether the role of STAT5 is relative or absolute for CML induction. Nonetheless, targeting STAT5 or pivotal downstream genes may provide an opportunity to disrupt leukemogenic signaling in the earliest CML progenitors, which may otherwise escape Abl kinase inhibitor–based therapy.
Prepublished online as Blood First Edition Paper, March 7, 2006; DOI 10.1182/blood-2005-10-4110.
Supported by National Institutes of Health grant HL61764 (R.L.I.).
D.Y., N.W., L.L., and S.Z. performed the research and analyzed the data; and R.L.I. developed the overall research plan, reviewed the data, and wrote the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Bonnie Darnell and Angela Mobley for assistance with the FACS mouse leukemia phenotyping, Rance Berg and Rick Van Etten for helpful technical advice, James A. Richardson and the University of Texas Southwestern Molecular Pathology Core Facility for assistance with the analysis of mouse leukemia samples, and Richard Gaynor for critical review of the manuscript prior to publication.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal