Abstract
The proteome is determined by rates of transcription, translation, and protein turnover. Definition of stem cell populations therefore requires a stem cell proteome signature. However, the limit to the number of primary cells available has restricted extensive proteomic analysis. We present a mass spectrometric method using an isobaric covalent modification of peptides for relative quantification (iTRAQ), which was employed to compare the proteomes of approximately 1 million long-term reconstituting hematopoietic stem cells (Lin–Sca+Kit+; LSK+) and non–long-term reconstituting progenitor cells (Lin–Sca+Kit–; LSK–), respectively. Extensive 2-dimensional liquid chromatography (LC) peptide separation prior to mass spectrometry (MS) enabled enhanced proteome coverage with relative quantification of 948 proteins. Of the 145 changes in the proteome, 54% were not seen in the transcriptome. Hypoxia-related changes in proteins controlling metabolism and oxidative protection were observed, indicating that LSK+ cells are adapted for anaerobic environments. This approach can define proteomic changes in primary samples, thereby characterizing the molecular signature of stem cells and their progeny.
Introduction
Definition of cell form and function is derived from the transcription of specific sets of genes, followed by their translation and post-translational regulation. Cellular development alters pheno-type via such changes in transcription and translation, and in this respect post-translational control of protein levels is key in many cell regulatory pathways, such as cell cycling.1
Stem cell commitment to differentiation is a critical step in development, where proteomic changes will be observed as a cell undergoes successive commitment and developmental steps. The precise nature of the elements of gene expression that contribute to stem cell characteristics have been assessed by transcriptional profiling and the elements of a stem cell signature refined via comparative expression analyses.2-4 Alternatively, the critical stem cell characteristics can be defined by analysis of the function of specific genes (in the hematopoietic system, SCL, HOX-B4, and BMI-15-7 ). Many key regulators are transcriptional activators or suppressors that will inevitably affect the transcriptomic profile of a cell. However, this process is known to be subject to further regulation via post-translational mechanisms.8,9
Lin–Sca+Kit+ (LSK+) cells and Lin–Sca+Kit– (LSK–) cells display differential abilities to reconstitute hematopoiesis, and the latter is a more mature cell type than the former, only able to support hematopoiesis in the short term.10,11 Comparison of these 2 cell populations can help us derive knowledge of the intracellular systems that regulate the ability to maintain pluripotency and long-term self-renewal. Transcriptomic analysis, previously used to define a stem cell “signature,” will not detail the true differences between stem cells and their progeny at the proteome level because of the importance of post-translational regulation of protein levels.9
We therefore sought to establish a method for defining the proteome of stem cell populations where only limited sample is available. Furthermore, the method should not rely on transcriptome data to infer changes at the protein level. Given the limited material (approximately 1 million LSK+ and LSK– cells, respectively, per experiment), we developed a procedure employing isobaric tags for relative quantification (iTRAQ). This allows 4 samples to be analyzed simultaneously, giving relative quantification12,13 on hundreds of proteins at any one time. Samples are separately proteolytically digested, and 4 isotope-coded, isobaric reagents (using differential stable isotope distribution between reporter group and balance group moieties) label all available peptides via free amine groups. The labeled samples are mixed, permitting concurrent delivery of all 4 differentially tagged isobaric peptides to a mass spectrometer. Peptide ions, efficiently separated in time using extensive liquid chromatography procedures, are fragmented, allowing peptide sequence to be inferred. Since the tags are of equal mass, identical peptides from 4 samples provide a combined single signal, thus increasing sensitivity, and the “reporter group” fragment from the isobaric tag formed in tandem mass spectrometry (MS) gives relative quantification on each peptide from the 4 samples.12,13
Here we demonstrate that extensive 2-dimensional liquid chromatography separation of tryptic peptides, plus a relative quantification tandem mass spectrometry methodology, gives relative quantification on hundreds of proteins from fewer than 2 million flow cytometrically enriched cells. By comparing these data to transcriptome analyses, the shift in proteome from LSK+ cells to LSK– cells was shown to be associated with post-transcriptional control of protein levels. Many of these were associated with hypoxia, demonstrating the potential influence of environment on LSK+ cells and a major shift away from anaerobic metabolism as more mature progeny are formed.
Materials and methods
Enrichment of hematopoietic cells
Primitive hematopoietic cells were isolated from murine bone marrow from C57Bl/6 mice. Femoral bone marrow was harvested by flushing and mononuclear cells prepared by density gradient centrifugation (Optiprep; Axis-Shield; PoC AS, Nycomed, Norway). Mature hematopoietic lineage-depleted (Lin–) cells were obtained using a cocktail of lineage-marker antibodies and sheep antirat magnetic beads (Dynal; Wirral, United Kingdom). Lineage (CD2, CD4, CD5, CD8, CD45R, GR-1, Mac-1, Ter-119)–depleted cells were then labeled with goat anti–rat Ig phycoerythrin, biotin anti–mouse Ly6A/E (Sca1), followed by avidin fluorescein isothiocyanate (FITC) and allophycocyanin anti–mouse CD117 (c-Kit). Cells were labeled with propidium iodide (Molecular Probes, Leiden, The Netherlands) to gate out dead cells. All antibodies were obtained from Becton Dickinson, Oxford, United Kingdom. The lineage-depleted cells were then sorted on a FACS Aria flow cytometer to prepare either lineage-marker–negative Sca-1–expressing (Sca+) cells expressing c-Kit (LSK+ cells) or not (LSK–). In a standard preparation 40 mice were employed to yield approximately 5 × 105 LSK+ cells, equating to approximately 10 μg of total protein. We are aware that further enrichment of stem cell populations can be achieved with other markers (eg, rhodamine 123 or CD34). Presently, our MS technology cannot perform incisive studies with further enrichment based on these parameters; the cell yield would be too low.
Mass spectrometry
LSK+ or LSK– cells (1 × 106) were lysed in 200 μL of 0.5 M triethylammonium bicarbonate (Sigma, Poole, Dorset, United Kingdom) plus 0.05% (wt/vol) sodium dodecyl sulfate (SDS) on ice for 20 minutes. The lysate was centrifuged at 10 000g for 20 minutes at 4°C, supernatant removed, and protein quantified (EZQ assay, Molecular Probes). Protein (20 μg) in 50 μL 0.5 M triethylammonium bicarbonate/0.05% SDS was reduced by addition of 5 μL 50 mM TCEP (tris(hydroxymethyl)aminomethane)-(2-carboxyethyl) phosphine and incubation at 60°C for 1 hour. Reduced cysteine residues were then blocked by addition of 2.5 μL of 200 mM methylmethanethiosulphate in isopropanol and incubation at room temperature for a further 10 minutes. Protein was then digested by addition of 4 μL of trypsin at 0.5 μg/μL and incubated at 37°C overnight. Peptides were labeled with iTRAQ reagent (Applied Biosystems, Framingham, MA), as previously described.13 Reactions were checked for complete labeling using mass spectrometry and then pooled prior to analysis.
Peptide fractionation and mass spectrometry
For separation prior to RP-LC-MS/MS, peptides were fractionated off-line using an SCX cation exchange column (10 cm × 2.1 cm PolyLC Polysulfoethyl A column, 5 μm beads, 200 Å micron pore size, Hichrom Ltd, Reading, Berks, United Kingdom) on a Dionex LC Ultimate nanoflow LC system. The gradient was formed using, initially, 10 mM KH2PO4, 20% acetonitrile (ACN), pH 2.7 with KCl concentration increasing from 0 to 250 mM KCl over a 45-minute period, and then 250 mM to 500 mM over 10 minutes, followed by 1 M for 5 minutes. One-minute fractions were collected. Each salt fraction was then concentrated and dried down.
Dried peptide fractions were resuspended in 180 μL of 2% (vol/vol) acetonitrile/0.1% (vol/vol) formic acid. For analysis, about 30% of the peptide sample was loaded onto a 75 μm internal diameter (i.d.) × 15 cm column, packed with C18 PepMap100, 3 μm, 100 Å using an LC Packings (Amsterdam, The Netherlands) UltiMate pump and separated over a 120-minute solvent gradient from 5% (vol/vol) acetonitrile/0.1% (vol/vol) formic acid to 40% (vol/vol) acetonitrile/0.1% (vol/vol) formic acid on-line to a QSTAR XL mass spectrometer (Applied Biosystems), as previously described.13 Data were acquired using an independent data acquisition (IDA) protocol where, for each cycle, the 2 most abundant multiply charged peptides (2+ to 4+) in the MS scan with m/z between 480 and 1500 were selected for MS/MS. Each peptide was selected twice, and then dynamically excluded (± 50 μm) for 60 seconds.
Transcriptome analysis
RNA from LSK+ and LSK– cells was prepared in triplicate using Qiagen RNeasy kit (Qiagen, Crawley, United Kingdom), as per manufacturer's instructions. Transcriptome analysis was undertaken by the Cancer Research UK (CRUK) microarray facility, Paterson Institute (Manchester, United Kingdom). Details of protocols can be found at http://bioinformatics.picr.man.ac.uk. Total RNA was subject to amplification using the “Gene chip eukaryotic small sample target labeling assay version II” developed by Affymetrix and gene expression profile assessed using Mouse430-2 chips in triplicate. Transcriptome data are available in a Minimum Information About a Microarray Experiment (MIAME)–compliant format.
Data analysis
Proteome data were searched against a mouse KBMS3.0 protein database from the Celera Discovery System (Applied Biosystems) using ProQUANT v1.1 (Applied Biosystems) as previously described.13 The following modifications were allowed for iTRAQ reagent labeling at N-terminal residues and internal K and Y residues: methylmethanethiosulphate-modified cysteine as fixed modifications, plus one missed cleavage. Search parameters within ProQUANT were set with an MS tolerance of 0.15 Da, an MS/MS tolerance of 0.1 Da, and a minimum confidence score of 70. ProGROUP (Applied Biosystems) software was used to pool data from all experiments and assign a single accession number for each protein, allowing comparison of replicate datasets. For a protein to be considered significantly different between LSK+ and LSK– cells, at least 3 peptides had to be identified and quantified, the fold change had to be greater than 1.2, the protein had to have a P < .05 from a Student t test for 5 of 6 pairwise analyses (LSK+1 vs LSK–1 and LSK+2 vs LSK–1 for each replicate), and the LSK+1 versus LSK+2 and LSK–1 versus LSK–2 ratios had to be between 0.92 and 1.10 in all replicates where data were obtained for that protein.
Transcriptome data were analyzed by submitting each replicate pair individually to GCOS v1.2 software (Affymetrix) with default settings, with the exception of the target (TGT) normalization value, which was set to 100. A transcript was determined to be “significantly differentially expressed” if it was called “present” and “increased” or “decreased” in all 3 replicate arrays.
To compare transcriptome and proteome data, all proteins with a SWISS-PROT, TrEMBL, or RefSeq accession number were selected, and these accessions were used to identify the relevant probeset identifiers from the microarray dataset using the NetAffx tool (www.affymetrix.com) (C.L.W., R.D.U., Y. Connolly, S. Pepper, R. Clarke, C.J.M., and A.D.W., manuscript in preparation).
Glucose transporter protein expression, lactate, and transwell migration assays
Cytospin slides of LSK+ and LSK– cells were prepared as previously described and immunostained for GLUT1.14 For imaging, 20 optical sections were captured at 0.2-μm intervals with a Delta Vision system (Applied Precision, Issaquah, WA) comprising an Olympus IX70 microscope linked to a CCD camera and processed using the manufacturer's deconvolution software. Image J (http://rsb.info.nih.gov/ij/) was used to quantify cell surface fluorescence from images captured from 3 separate experiments. Lactate assays were performed using a kit provided by Trinity Biotech (Wicklow, Ireland). Transwell migration assays were performed as described.15
Results
Relative quantification of multiple proteins from primary enriched LSK+ and LSK– hematopoietic cells
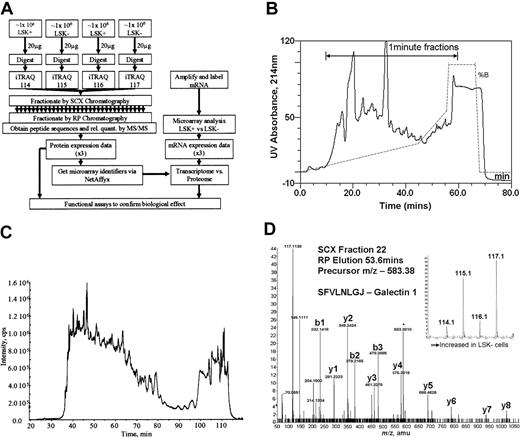
Four equal samples (2 from LSK+ cell lysates and 2 from LSK– cell lysates, approximately 1 million cells per sample) were trypsinized and each labeled with a specific isobaric iTRAQ reagent (Figure 1A). The samples were then mixed. Using strong cation exchange (SCX) liquid chromatography, the derivatized peptides were extensively separated into approximately 50 fractions, employing a gradient designed to give equal peptide distribution per fraction (Figure 1B). These samples were then further fractionated using reverse-phase liquid chromatography (Figure 1C) online to the tandem mass spectrometer to derive spectra for peptide sequence determination (Figure 1D). Our yield of protein from LSK+ cells was 10 to14 μg from an enrichment procedure, taking 5 to 6 hours of flow cytometric sorting time. Protein content per cell was the same in both LSK+ and LSK– cell populations.
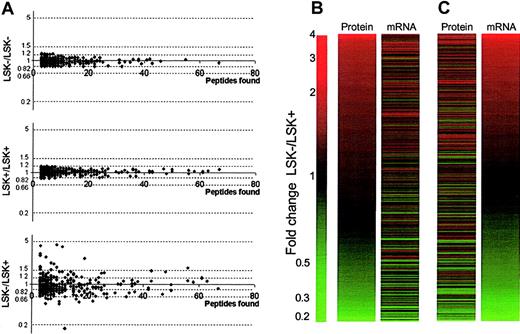
The experimental design allows for replicate samples (Figure 1). We have shown that iTRAQ internal replicates give a high confidence, so differences greater than 1.2-fold can be considered significant.13 Fold differences for all proteins where more than 3 peptides were characterized was less than 1.2 in replicate samples (Figure 2A top and middle panels). This was not the case for LSK+ versus LSK– ratios from the same experiment (Figure 2A bottom panel). Thus, the replication in our approach is reliable, and we can proceed to examine with confidence stem cell proteomes using small quantities of cells and protein.
Overall experimental design. (A) Flow diagram showing the steps involved in determining relative quantities of proteins between LSK+ cells and LSK– cells and the comparison of these data to transcriptome data obtained on the same samples. (B) UV absorbance spectrum (214 nm) showing fractionation of iTRAQ-labeled peptides by strong cation exchange chromatography. The trace shows the elution profile of the cell lysates. The gradient is illustrated by the dotted line, and the range during which 1-minute fractions were collected is marked with an arrow. (C) Separation of a single SCX fraction (fraction 22) on a reverse phase gradient on-line to a mass spectrometer. The trace shows the total ion chromatograph (TIC) indicated as intensity on the y-axis, showing the elution profile of peptides from the reverse phase high-performance liquid chromatography (HPLC) column into the mass spectrometer. (D) Tandem mass spectrum showing the sequence of a peptide of mass: charge 583.37 eluting from the reverse phase column at 53.6 minutes. The labeled y-ion series allows calculation of the peptide sequence (SFVLNLGJ, where J denotes a lysine with an iTRAQ-labeled side-chain); the iTRAQ reporter ions (inset) give relative quantification data for this peptide.
Overall experimental design. (A) Flow diagram showing the steps involved in determining relative quantities of proteins between LSK+ cells and LSK– cells and the comparison of these data to transcriptome data obtained on the same samples. (B) UV absorbance spectrum (214 nm) showing fractionation of iTRAQ-labeled peptides by strong cation exchange chromatography. The trace shows the elution profile of the cell lysates. The gradient is illustrated by the dotted line, and the range during which 1-minute fractions were collected is marked with an arrow. (C) Separation of a single SCX fraction (fraction 22) on a reverse phase gradient on-line to a mass spectrometer. The trace shows the total ion chromatograph (TIC) indicated as intensity on the y-axis, showing the elution profile of peptides from the reverse phase high-performance liquid chromatography (HPLC) column into the mass spectrometer. (D) Tandem mass spectrum showing the sequence of a peptide of mass: charge 583.37 eluting from the reverse phase column at 53.6 minutes. The labeled y-ion series allows calculation of the peptide sequence (SFVLNLGJ, where J denotes a lysine with an iTRAQ-labeled side-chain); the iTRAQ reporter ions (inset) give relative quantification data for this peptide.
After running the experiment in triplicate, a total of 948 proteins were identified, with at least one peptide with high confidence of correct sequence assignment. Following exclusion of proteins where fewer than 3 peptides were relatively quantified, this approach generated relative quantification on 668 proteins from LSK+ and LSK– cells (Table S1, available at the Blood website; see the Supplemental Tables link at the top of the online article). Of these, 145 (21.7%) were shown to be significantly differentially expressed between LSK+ and LSK– cells.
A stem cell signature can and has been defined at the mRNA level. However, we have previously shown that the ratio of specific mRNAs in LSK+ and LSK– cells does not directly correlate to the ratio of some protein levels present.16 We therefore analyzed changes seen at the proteome and the mRNA level (generated using microarray technology, see “Materials and methods” for details). This was performed for all of the proteins where there were differences between LSK+ and LSK– cells and for 374 unchanging proteins, which were chosen based on their having been ascribed a Swiss-PROT, TrEMBL, or RefSeq database accession number by the protein analysis software.13 The results of this comparison (significant transcript differences were determined using GCOS software, see “Materials and methods”) are shown in Table 1.
Correlation of transcriptomic and proteomic comparisons of LSK+ and LSK– cells
. | mRNA (Affymetrix array) . | . | . | ||
|---|---|---|---|---|---|
| Protein (iTRAQ) . | Up . | No change . | Down . | ||
| Up | 35 | 27 | 1 | ||
| No change | 27 | 325 | 22 | ||
| Down | 0 | 47 | 27 | ||
. | mRNA (Affymetrix array) . | . | . | ||
|---|---|---|---|---|---|
| Protein (iTRAQ) . | Up . | No change . | Down . | ||
| Up | 35 | 27 | 1 | ||
| No change | 27 | 325 | 22 | ||
| Down | 0 | 47 | 27 | ||
Shown are relative changes in mRNA levels for all of the proteins shown to be differentially expressed by iTRAQ labeling and for proteins shown to have no change between the 2 cell types where more than 3 peptides were identified and where the analysis software reported a hit against the Swiss-Prot, TrEMBL, or RefSeq databases. Seven protein changes and 6 unchanged proteins were scored “absent” on the microarray analysis. One protein change could not be matched to the array data.
In respect to the stem cell markers indicated from previous microarray studies, Zhong et al4 performed a meta-analysis including data from their own and 2 other studies2,3 identifying 75 genes that were present at higher levels in stem cell populations. Our microarray data showed 73% overlap with this list, indicating that our transcriptome data are comparable with previous studies and, therefore, that our proteome versus transcriptome comparison has value for the definition of stem cell populations from other groups.
Overview of proteomic and transcriptomic data. (A) Scatter plots examining the reproducibility of the iTRAQ quantitation procedure on stem cells. Top and middle panels show plotted ratios from proteins using the internal duplicates LSK–/LSK– and LSK+/LSK+, respectively, from one experiment. No ratios are seen greater than 1.2-fold, giving confidence in the cut-off value chosen. The bottom panel shows plotted ratios from proteins for one of the LSK–/LSK+ datasets, clearly demonstrating differences in the protein expression profiles between the 2 cell types. (B,C) Expression profiles of LSK– cells versus LSK+ cells. Panel B shows fold changes in 704 proteins in rank order between LSK– and LSK+ cells alongside corresponding expression ratios for transcripts of these proteins obtained by microarray analysis. Panel C shows the same expression data, sorted in rank order of fold change in the mRNA level, alongside corresponding protein expression ratios.
Overview of proteomic and transcriptomic data. (A) Scatter plots examining the reproducibility of the iTRAQ quantitation procedure on stem cells. Top and middle panels show plotted ratios from proteins using the internal duplicates LSK–/LSK– and LSK+/LSK+, respectively, from one experiment. No ratios are seen greater than 1.2-fold, giving confidence in the cut-off value chosen. The bottom panel shows plotted ratios from proteins for one of the LSK–/LSK+ datasets, clearly demonstrating differences in the protein expression profiles between the 2 cell types. (B,C) Expression profiles of LSK– cells versus LSK+ cells. Panel B shows fold changes in 704 proteins in rank order between LSK– and LSK+ cells alongside corresponding expression ratios for transcripts of these proteins obtained by microarray analysis. Panel C shows the same expression data, sorted in rank order of fold change in the mRNA level, alongside corresponding protein expression ratios.
Of the 145 proteins shown to be differentially expressed, one could not be matched to the microarray data, and 7 were reported as “absent” according to the microarray data, despite the detection of protein. Of the remaining 137 proteins, 62 (45.3%) followed the same trend at the transcriptome level. The remaining inferred protein level changes showed no significant difference (P > .05) between LSK+ and LSK– cells at the transcriptome level. The fold change diagrams where proteins (Figure 2B) or mRNA (Figure 2C) are ranked by fold difference between LSK– and LSK+ cells show a trend of correlation between protein and mRNA changes, but there are many exceptions (Table 1 and Table S1).
LSK+ cells express relatively high levels of glycolytic and oxidative repair proteins
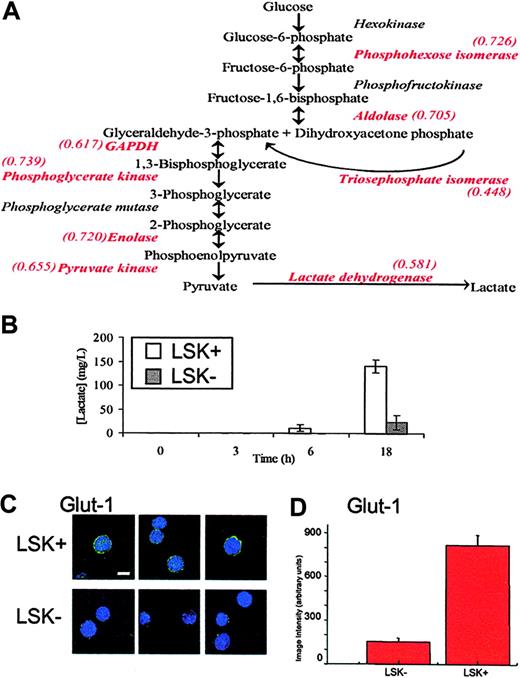
Although many of the changes in protein levels appear to be fairly small, it is important to keep in mind the observation that relatively small changes in several components in metabolic pathways can synergistically affect flux, or outcome.17 Therefore, to identify key biologic differences between LSK+ cells and LSK– populations, the list of protein differences was scrutinized to identify changes occurring along the same pathway. A significant feature of this dataset is that, in the more mature LSK– cells, there is a decrease in the expression of every glycolytic enzyme identified (Figure 3A). The details of these proteins are found in Table 2; notably, only 2 of 8 change significantly at the transcript level. Since a list of protein changes remains largely suggestive with respect to biologic relevance, it is important to extend these observations to a biologic setting. In this case, we performed an assay for lactate production, the end product of anaerobic glycolysis, by LSK+ and LSK– cells. This assay supported the inference from proteomic analysis that LSK+ cells would display a higher rate of anaerobic glycolysis and as such have a greater rate of lactate production (Figure 3B). LSK+ cells also show increased expression of lactoylglutathione lyase, which detoxifies methylglyoxal, a side product of triosephosphate isomerase catalysis, which (as part of the glycolytic pathway) also is increased in LSK+ cells.
Differences between LSK+ and LSK– cells in proteins that are involved in glycolysis and associated pathways and in protection against DNA damage caused by the production of reactive oxygen species
Type, accession number . | Name . | Peptides found . | LSK-/LSK+ protein ratio . | LSK-/LSK+ mRNA ratio . | Gene symbol . |
|---|---|---|---|---|---|
| Glycolytic pathway | |||||
| P45376 | Aldose reductase | 16 | 0.717 | 1.196 | Akr1b1 |
| P06745 | Glucose phosphate isomerase 1 | 29 | 0.726 | 1.017 | Gpi |
| P05064 | Fructose-bisphosphate aldolase A | 30 | 0.705 | 1.069 | Aldoa |
| P17751 | Triose-phosphate isomerase | 32 | 0.448 | 0.687* | Tpi1 |
| P16858 | Glyceraldehyde 3-phosphate dehydrogenase | 73 | 0.739 | 1.250 | Gapd |
| P09411 | Phosphoglycerate kinase 1 | 29 | 0.617 | 0.560* | Pgk1 |
| P17182 | Alpha enolase | 53 | 0.720 | 0.979 | Enoa |
| P52480 | Pyruvate kinase, M2 isozyme | 64 | 0.655 | 0.834 | Kpy2 |
| P16125 | Lactate dehydrogenase 2, B chain | 9 | 0.581 | 0.824 | Ldhb |
| TCA cycle, respiratory chain | |||||
| Q80Y13 | Malate dehydrogenase 1 (TCA cycle) | 42 | 0.707 | 1.006 | Mdh1 |
| Q9EQK1 | NADP+-specific isocitrate dehydrogenase | 24 | 0.692 | 0.676 | Idh2 |
| Q9CQB4 | Ubiquinol-cytochrome c reductase binding protein | 3 | 1.695 | 0.935 | Uqcrb |
| Pentose-phosphate shunt and purine biosynthesis | |||||
| Q9CQ60 | 6-Phosphogluconolactonase | 11 | 1.638 | 2.062* | Pgls |
| P40142 | Transketolase | 48 | 0.722 | 0.895 | Tkt |
| Q80UH0 | IMP cyclohydrolase | 13 | 0.603 | 0.684* | Atic |
| P50247 | Adenosylhomocysteinase | 16 | 0.679 | 0.565* | Ahcy |
| Q64737 | Trifunctional purine biosynthetic protein adenosine-3 | 4 | 0.784 | 0.743* | Gart |
| Protection against oxidative damage | |||||
| P08228 | Cu/Zn superoxide dismutase | 36 | 0.695 | 0.975 | Sod1 |
| P34884 | Macrophage migration inhibitory factor | 3 | 0.377 | 0.974 | Mif |
| P09671 | Mn superoxide dismutase | 3 | 0.655 | 0.900 | Sod2 |
| P10649 | Glutathione S-transferase Mu 1 | 17 | 0.232 | 0.670* | Gstm1 |
| O09131 | Glutathione transferase omega 1 | 7 | 0.608 | 0.604* | Gsto1 |
| O08709 | Peroxiredoxin 6 | 22 | 0.428 | 1.041 | Prdx6 |
| Q9CPU0 | Lactoylglutathione lyase | 3 | 0.607 | 0.771* | Glo1 |
| P28352 | DNA-(apurinic or apyrimidinic site) lyase | 11 | 0.660 | 0.763* | Apex1 |
| Protection against oxidative damage and alkylating agents | |||||
| P47738 | Aldehyde dehydrogenase | 23 | 0.460 | 0.770* | Aldh2 |
Type, accession number . | Name . | Peptides found . | LSK-/LSK+ protein ratio . | LSK-/LSK+ mRNA ratio . | Gene symbol . |
|---|---|---|---|---|---|
| Glycolytic pathway | |||||
| P45376 | Aldose reductase | 16 | 0.717 | 1.196 | Akr1b1 |
| P06745 | Glucose phosphate isomerase 1 | 29 | 0.726 | 1.017 | Gpi |
| P05064 | Fructose-bisphosphate aldolase A | 30 | 0.705 | 1.069 | Aldoa |
| P17751 | Triose-phosphate isomerase | 32 | 0.448 | 0.687* | Tpi1 |
| P16858 | Glyceraldehyde 3-phosphate dehydrogenase | 73 | 0.739 | 1.250 | Gapd |
| P09411 | Phosphoglycerate kinase 1 | 29 | 0.617 | 0.560* | Pgk1 |
| P17182 | Alpha enolase | 53 | 0.720 | 0.979 | Enoa |
| P52480 | Pyruvate kinase, M2 isozyme | 64 | 0.655 | 0.834 | Kpy2 |
| P16125 | Lactate dehydrogenase 2, B chain | 9 | 0.581 | 0.824 | Ldhb |
| TCA cycle, respiratory chain | |||||
| Q80Y13 | Malate dehydrogenase 1 (TCA cycle) | 42 | 0.707 | 1.006 | Mdh1 |
| Q9EQK1 | NADP+-specific isocitrate dehydrogenase | 24 | 0.692 | 0.676 | Idh2 |
| Q9CQB4 | Ubiquinol-cytochrome c reductase binding protein | 3 | 1.695 | 0.935 | Uqcrb |
| Pentose-phosphate shunt and purine biosynthesis | |||||
| Q9CQ60 | 6-Phosphogluconolactonase | 11 | 1.638 | 2.062* | Pgls |
| P40142 | Transketolase | 48 | 0.722 | 0.895 | Tkt |
| Q80UH0 | IMP cyclohydrolase | 13 | 0.603 | 0.684* | Atic |
| P50247 | Adenosylhomocysteinase | 16 | 0.679 | 0.565* | Ahcy |
| Q64737 | Trifunctional purine biosynthetic protein adenosine-3 | 4 | 0.784 | 0.743* | Gart |
| Protection against oxidative damage | |||||
| P08228 | Cu/Zn superoxide dismutase | 36 | 0.695 | 0.975 | Sod1 |
| P34884 | Macrophage migration inhibitory factor | 3 | 0.377 | 0.974 | Mif |
| P09671 | Mn superoxide dismutase | 3 | 0.655 | 0.900 | Sod2 |
| P10649 | Glutathione S-transferase Mu 1 | 17 | 0.232 | 0.670* | Gstm1 |
| O09131 | Glutathione transferase omega 1 | 7 | 0.608 | 0.604* | Gsto1 |
| O08709 | Peroxiredoxin 6 | 22 | 0.428 | 1.041 | Prdx6 |
| Q9CPU0 | Lactoylglutathione lyase | 3 | 0.607 | 0.771* | Glo1 |
| P28352 | DNA-(apurinic or apyrimidinic site) lyase | 11 | 0.660 | 0.763* | Apex1 |
| Protection against oxidative damage and alkylating agents | |||||
| P47738 | Aldehyde dehydrogenase | 23 | 0.460 | 0.770* | Aldh2 |
Protein accession numbers refer to SWISS-PROT or TrEMBL entries. Confidence score (a percentage measure of the confidence of the protein identification13 ) for all proteins in the table was 99%. Peptides found denote the sum of all different peptides found and relatively quantified from the proteins shown in 3 replicate experiments. All proteins are significantly different (P < .05) between LSK+ cells and LSK- cells.
Significantly different mRNA ratios (P < .05).
Macrophage migration inhibitory factor (MIF) is also down-regulated at the protein, but not the transcriptome, level. This cytokine is relatively poorly understood but promotes the proinflammatory responses of immune cells.18 It is unique in also possessing thiol-protein oxidoreductase activity and has pleiotropic effects, including regulation of cellular redox homeostasis.19 Thus, regulation of oxidative events is potentially of key importance in LSK+ cells. A marker for stem cells, aldehyde dehydrogenase (Table 2) plays a critical role in their protection from alkylating agents20 and is also highly expressed in our proteomic study, validating the value of this proteomic approach.
It is noteworthy that many proteins expressed more highly in LSK+ cells are regulated by hypoxia, such as several glycolytic enzymes,21 Hsp60,22 and nucleophosmin.23 Hypoxia also decreases levels of some proteins, and we see several of these reduced in the LSK+ cells, for example, gamma actin,24 RhoA,25,26 and vimentin.27 To further analyze the potential role of the micro-environment in defining the LSK+ proteome, we assayed levels of the hypoxic state marker, GLUT1 (Figures 3C,D21 ). The immunostaining approach (for a protein not found in our MS analyses) clearly shows that the LSK+ cells express GLUT1 at much higher levels that LSK– cells, although there is no change at the transcriptome level (data not shown). GLUT1 was presumably not identified in our proteome screen as its expression level is too low. This supports the hypothesis, generated by the systematic proteomics approach, that gene/protein expression in stem cells is regulated by the proposed (hypoxic) microenvironment of the endosteal region where LSK+ cells, but not LSK– cells, reside.28,29 That in vitro hypoxic conditions favor the survival and expansion of severe combined immunodeficiency disease (SCID)–repopulating human cells30 and that bone marrow in humans is hypoxic relative to the peripheral circulation argue for the importance of the pO2 in LSK+ cell biology.31
Glycolytic flux in LSK+ and LSK– cells. (A) A schematic diagram of the glycolytic pathway. Enzymes identified in the iTRAQ experiment are shown in red type, along with their relative expression ratio as LSK–/LSK+, showing that all enzymes identified in this experiment are found at higher levels in LSK+ cells. (B) LSK+ and LSK– cells were cultured for 3, 6, and 18 hours. Lactate production was measured in the medium and after 6 and 18 hours and was significantly different between LSK+ and LSK– cells (P < .05). Error bars show standard error of the mean (n = 3). (C) Cytospin slides of LSK+ or LSK– hematopoietic cells were immunostained for GLUT1. Single deconvolved images of hematopoietic cells stained for GLUT1 (green) and the nucleus (DAPI) are shown. Bar shown is 10 μM. (D) Image intensity analysis of cells showed LSK+ cells were significantly different from those of LSK– (P < .05) for GLUT1 expression.
Glycolytic flux in LSK+ and LSK– cells. (A) A schematic diagram of the glycolytic pathway. Enzymes identified in the iTRAQ experiment are shown in red type, along with their relative expression ratio as LSK–/LSK+, showing that all enzymes identified in this experiment are found at higher levels in LSK+ cells. (B) LSK+ and LSK– cells were cultured for 3, 6, and 18 hours. Lactate production was measured in the medium and after 6 and 18 hours and was significantly different between LSK+ and LSK– cells (P < .05). Error bars show standard error of the mean (n = 3). (C) Cytospin slides of LSK+ or LSK– hematopoietic cells were immunostained for GLUT1. Single deconvolved images of hematopoietic cells stained for GLUT1 (green) and the nucleus (DAPI) are shown. Bar shown is 10 μM. (D) Image intensity analysis of cells showed LSK+ cells were significantly different from those of LSK– (P < .05) for GLUT1 expression.
An increase in proteins involved in protection against oxidative damage and DNA repair also was observed (Table 2), including 2 forms of glutathione transferase, 2 forms of superoxide dismutase (which converts reactive oxygen species to H2O2), and peroxiredoxin (which converts H2O2 to H2O). Aldehyde dehydrogenase (Table 2) also has a role in cellular defense mechanisms against oxidative damage.32 LSK+ cells are a population that requires long-term maintenance to provide blood cells throughout the lifetime of the organism. We show for the first time that they preferentially express antioxidant enzymes compared to the LSK– cells; committed progenitor cells will give rise to cells that will undergo clonal extinction by forming mature postmitotic hematopoietic cells. We conclude that protection for stem cells is mediated by systematic elevation of protection from oxygen radicals.
LSK– cells show an increase in the expression of cytoskeleton associated proteins, leading to an increase in motility
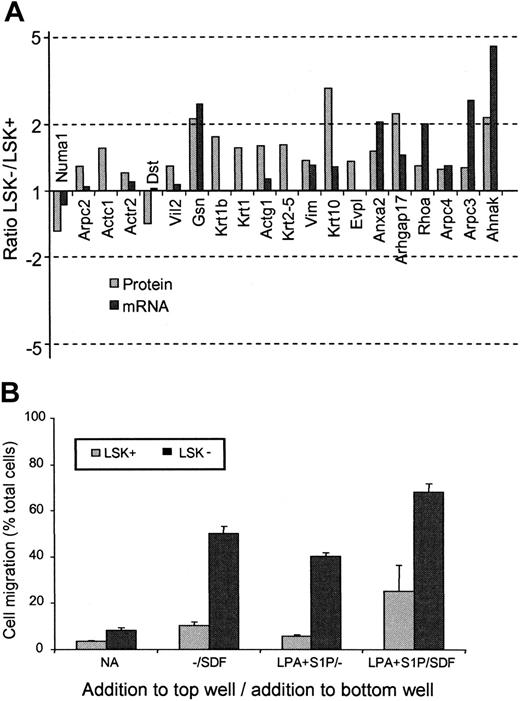
Another feature of the proteomes of LSK+ and LSK– cells was the difference in proteins involved in cytoskeleton and motility pathways. In every case except 2, the protein levels were observed to be higher in the LSK– than the LSK+ cells (Table 3). Noticeably, of the 20 motility-associated proteins shown to change, only 5 were altered significantly according to the mRNA analysis (Figure 4A). Altered proteins include key motility factors such as Rho, Nadrin 2 (a Rho-GAP), actin polymerizing proteins such as ARP2/3 complex members, and gelsolin, plus cytoskeletal components such as actin, keratin, and vimentin. Two cell motility–related proteins are lower in LSK– cells. One of these, dystonin, is known to stabilize the actin cytoskeleton33 and thereby can be envisaged to decrease actin filament turnover, which is essential in motility. The niche theory of stem cell development34 would argue for increased motile function of a committed progenitor cell (such as an LSK– cell), thus allowing cells to move to marrow microenvironments favoring development.35,36
Differences between LSK+ and LSK-cells in proteins that are involved in the regulation of cell structure and motility
Accession no. . | Name . | Confidence . | Peptides found . | LSK-/LSK+ protein ratio . | LSK-/LSK+ mRNA ratio . | Gene symbol . |
|---|---|---|---|---|---|---|
| Q9CXK3 | Actin, alpha, cardiac | 99 | 168 | 1.563 | A | Actc1 |
| P63260 | Actin, gamma, cytoplasmic | 99 | 91 | 1.601 | 1.131 | Actg1 |
| P61161 | Actin-related protein (ARP) 2 | 99 | 19 | 1.204 | 1.099 | Actr2 |
| P59999 | ARP2/3 complex 20-kDa subunit | 99 | 15 | 1.242 | 1.295 | Arpc4 |
| Q9JM76 | ARP2/3 complex p21-Arc (subunit 3) | 99 | 9 | 1.261 | 2.572* | Arpc3 |
| Q9CVB6 | ARP2/3 complex, subunit 2 | 99 | 20 | 1.293 | 1.045 | Arpc2 |
| Q6UL10 | AHNAK | 99 | 29 | 2.151 | 4.530* | Ahnak |
| P07356 | Annexin A2 | 99 | 16 | 1.500 | 2.048* | Anxa2 |
| Q91ZU6 | Dystonin isoform a | 87 | 9 | 0.703 | 1.029 | Dst |
| Q9D952 | Envoplakin | 95 | 22 | 1.354 | A | Evpl |
| P26040 | Ezrin (cytovillin) (villin 2) | 99 | 25 | 1.294 | 1.064 | Vil2 |
| P13020 | Gelsolin (actin depolymerizing factor) | 99 | 9 | 2.122 | 2.490* | Gsn |
| Q920F2 | Keratin 5, type 2 | 99 | 7 | 1.608 | A | Krt2-5 |
| P04104 | Keratin, type 2 cytoskeletal 1 | 99 | 14 | 1.559 | A | Krt1 |
| Q6IFZ6 | Keratin complex 2, basic, gene 17 | 99 | 4 | 1.751 | A | Krt1b |
| P02535 | Keratin, type 1 cytoskeletal 10 | 99 | 4 | 2.911 | 1.276 | Krt10 |
| Q99N39 | Nadrin2 | 99 | 3 | 2.238 | 1.453 | Arhgap17 |
| Q80Y35 | Nuclear mitotic apparatus protein 1 | 99 | 17 | 0.654 | 0.859 | Numa1 |
| Q9QUI0 | RhoA | 99 | 16 | 1.288 | 2.001* | Rhoa |
| P20152 | Vimentin | 99 | 123 | 1.373 | 1.299* | Vim |
Accession no. . | Name . | Confidence . | Peptides found . | LSK-/LSK+ protein ratio . | LSK-/LSK+ mRNA ratio . | Gene symbol . |
|---|---|---|---|---|---|---|
| Q9CXK3 | Actin, alpha, cardiac | 99 | 168 | 1.563 | A | Actc1 |
| P63260 | Actin, gamma, cytoplasmic | 99 | 91 | 1.601 | 1.131 | Actg1 |
| P61161 | Actin-related protein (ARP) 2 | 99 | 19 | 1.204 | 1.099 | Actr2 |
| P59999 | ARP2/3 complex 20-kDa subunit | 99 | 15 | 1.242 | 1.295 | Arpc4 |
| Q9JM76 | ARP2/3 complex p21-Arc (subunit 3) | 99 | 9 | 1.261 | 2.572* | Arpc3 |
| Q9CVB6 | ARP2/3 complex, subunit 2 | 99 | 20 | 1.293 | 1.045 | Arpc2 |
| Q6UL10 | AHNAK | 99 | 29 | 2.151 | 4.530* | Ahnak |
| P07356 | Annexin A2 | 99 | 16 | 1.500 | 2.048* | Anxa2 |
| Q91ZU6 | Dystonin isoform a | 87 | 9 | 0.703 | 1.029 | Dst |
| Q9D952 | Envoplakin | 95 | 22 | 1.354 | A | Evpl |
| P26040 | Ezrin (cytovillin) (villin 2) | 99 | 25 | 1.294 | 1.064 | Vil2 |
| P13020 | Gelsolin (actin depolymerizing factor) | 99 | 9 | 2.122 | 2.490* | Gsn |
| Q920F2 | Keratin 5, type 2 | 99 | 7 | 1.608 | A | Krt2-5 |
| P04104 | Keratin, type 2 cytoskeletal 1 | 99 | 14 | 1.559 | A | Krt1 |
| Q6IFZ6 | Keratin complex 2, basic, gene 17 | 99 | 4 | 1.751 | A | Krt1b |
| P02535 | Keratin, type 1 cytoskeletal 10 | 99 | 4 | 2.911 | 1.276 | Krt10 |
| Q99N39 | Nadrin2 | 99 | 3 | 2.238 | 1.453 | Arhgap17 |
| Q80Y35 | Nuclear mitotic apparatus protein 1 | 99 | 17 | 0.654 | 0.859 | Numa1 |
| Q9QUI0 | RhoA | 99 | 16 | 1.288 | 2.001* | Rhoa |
| P20152 | Vimentin | 99 | 123 | 1.373 | 1.299* | Vim |
Protein accession numbers refer to SWISS-PROT or TrEMBL entries. For explanation of confidence and peptides found, see Table 2. All proteins are significantly different (P > .05) between LSK+ cells and LSK- cells.
A indicates this mRNA was called “absent” in the microarray experiments.
Significantly different mRNA ratios (P < .05).
To confirm that these protein changes were indeed reflecting an increase in motile potential in the cell populations prepared for the proteomics experiments, a transwell migration assay was performed to assess both intrinsic motility and the response of the cells to chemokinetic factors for these cells (lysophosphatidic acid [LPA] and sphingosine-1-phosphate [S1P]) in the presence or absence of the chemotactic agent stromal-derived factor (SDF-1) that we have previously shown to act on both of these cell populations15 (Figure 4B). The LSK– cells showed greater chemokinesis in response to LPA plus S1P. SDF-1 stimulated chemotaxis in LSK+ and LSK– cells,15 but this was markedly enhanced in the presence of LPA + S1P and to a greater degree in LSK– cells. We can correlate enhanced chemokinesis and chemotaxis with altered protein expression in LSK+ and LSK– cells, respectively.
Aside from the proteins described, other proteins were found to alter in their expression between LSK+ and LSK– cells. These can be further separated into functional activities, including changes in protein folding/transport, protein degradation, nucleolar architecture, transcription, and several proteases and signaling proteins. These proteins are listed in Table S2. The full list of proteins detected is shown as Table S1.
Discussion
Clearly many proteomic changes would not therefore be reported as “changes” in a stand-alone microarray experiment. While transcriptome analysis can be said to generate a cell-specific signature, the addition of a proteome analysis and combination of the 2 datasets adds penetration and biologic information. We conclude stem cell definition, as required for benchmarking and systematic analysis projects in stem cell biology,3 will benefit from proteomic analyses. This is especially the case as increased proteomic penetration can be achieved with larger quantities of starting material. For example, nuclear preparations of an embryonic stem cell line have yielded 1925 protein identifications (with relative quantification) from 8 million cells (D.L.S. and A. Williamson, unpublished observations, July 2005).
iTRAQ-based proteomics is a valuable tool for the definition of relatively scarce cell populations
We have used recent developments in mass spectrometry and advanced liquid chromatography, plus a method for the relative quantification of protein expression, to analyze the proteome of primitive cell populations from in vivo sources. Such populations have never before yielded such large quantities of proteomic information. As far as we are aware, no other readily accessible approach could have gained statistically significant data on the relative levels of more than 650 proteins (imposing a cut-off of 3 peptides identified and quantified) and determined the presence of a further 300, given the quantity of material available from primary cells.37 The technique we describe therefore offers greater insight into the biochemistry of any cell population. The use of iTRAQ reagents also gives an advantage over other stable isotope-labeling techniques, as all peptides are labeled, giving more data and greater confidence where several peptides report the same ratios from the same protein from 4 samples. In addition, since the tags are isobaric, the signal from the same peptide from 4 samples is effectively pooled, theoretically allowing good-quality MS data to be obtained from lower copy number proteins. The Stable Isotope Labelling in Tissue Culture (SILAC) method, with its many advantages,38 cannot be employed for primary samples such as those we have employed here, as the isotope coding requires in vitro culture for a minimum of 5 cell cycles.
Based on these data, there is a requirement to define stem cell populations by proteomics for a systematic understanding of changes occurring during development. Several important points underpin this statement. First is the fact that, albeit in a small number of cases, a protein is clearly present while its mRNA is recorded as absent. Protein levels are controlled not only by the amount of mRNA and its rate of translation, but also the rate of protein degradation. These proteins may have a long half life or, alternatively, the transcript data may not reflect true mRNA levels. Second, less than half of the changes seen in the proteome between LSK+ cells and LSK– cells are reflected in the mRNA data. It is clear that had this study been performed using microarray technology alone, several key molecular changes would not have been observed, including changes in several of the glycolytic enzymes (contributing to major differences in anaerobic glycolysis rates, as shown in Figure 3A) and motility-associated proteins. In addition, there are some changes seen at the mRNA level that are not reflected at the protein level. This analysis clearly demonstrates the value of performing both transcriptome and proteome analysis, a case in point being that many of the proteins that protect against damage are post-translationally regulated (Table 2).
LSK+ cells and LSK– cells show differential expression of motility-associated proteins. (A) Histogram showing the relative protein (▦) and mRNA (▪) expression ratios of cell-motility associated proteins between LSK+ and LSK– cells. For gene name definitions, see Table 3. All proteins were significantly different (P < .05) between the 2 cell types. (B) Transwell migration assay to compare the motile responses of LSK+ and LSK– cells. Basal motility and chemokinetic responses to LPA and S1P were assessed in the presence or absence of the chemotactic factor SDF-1.15 Results shown are the mean values for the numbers of cells that migrated to the bottom chamber, expressed as a percentage of total cells present (n = 3). Significantly different responses were seen between LSK+ cells and LSK– cells for all conditions (P < .05, n = 3).
LSK+ cells and LSK– cells show differential expression of motility-associated proteins. (A) Histogram showing the relative protein (▦) and mRNA (▪) expression ratios of cell-motility associated proteins between LSK+ and LSK– cells. For gene name definitions, see Table 3. All proteins were significantly different (P < .05) between the 2 cell types. (B) Transwell migration assay to compare the motile responses of LSK+ and LSK– cells. Basal motility and chemokinetic responses to LPA and S1P were assessed in the presence or absence of the chemotactic factor SDF-1.15 Results shown are the mean values for the numbers of cells that migrated to the bottom chamber, expressed as a percentage of total cells present (n = 3). Significantly different responses were seen between LSK+ cells and LSK– cells for all conditions (P < .05, n = 3).
However, the value of any new technology is based upon provision of new insights into the biology of the system under investigation. The LSK+ cell proteome complement is geared to protect from oxidative damage by enabling the cell to concentrate on anaerobic glycolysis via relatively high expression of glycolytic enzymes, glucose transporter, and antioxidant proteins. Can this contribute directly to the self-renewal of stem cells? The answer is yes, as shown by the work of Ito et al.39 LSK+ cells have an Atm protein-dependent pathway for the regulation of oxidative damage. Aging Atm-null mice showed progressive bone marrow failure resulting from a defect in LSK+ function associated with elevated reactive oxygen intermediates. If Atm is not present, the LSK+ lose the ability to self-renew, initiated via activation of the p16INK4a/retinoblastoma protein pathway. Thus, LSK+ cells reside in a hypoxic environment and employ an Atm-mediated pathway to limit oxidative damage, permitting self-renewal while expressing relatively high levels of antioxidant enzymes and employing anaerobic glycolysis. Using the approaches we describe, this and other aspects of stem cell physiology can be further elucidated when combined with transgenic approaches.
Prepublished online as Blood First Edition Paper, February 28, 2006; DOI 10.1182/blood-2005-12-4995.
Supported by the Leukaemia Research Fund (United Kingdom); the Biological Sciences and Biotechnology Research Council, United Kingdom; and the Wellcome Trust, United Kingdom.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank Yuning Lu for help with cell preparations.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal