Abstract
Fc-receptor homolog 5 (FcRH5) is a recently identified B-cell membrane protein of unknown function. In Burkitt lymphoma cell lines with chromosome 1q21 abnormalities, FcRH5 expression is deregulated, implicating FcRH5 in lymphomagenesis. Epstein-Barr virus infects and immortalizes B cells, and is implicated in the etiology of several tumors of B-cell origin. Overexpression of genes located on 1q21-25 has been proposed as a surrogate for Epstein-Barr virus in Burkitt lymphoma. We now report that Epstein-Barr virus nuclear antigen 2 (EBNA2) markedly induces the expression of the FcRH5 gene, encoded on chromosome 1q21. Induction occurred in the absence of other viral proteins and did not require de novo protein synthesis. EBNA2 lacks a DNA-binding domain and can target responsive genes through the host DNA binding protein CBF1. We show that induction of FcRH5 by EBNA2 is strictly CBF1 dependent, as it was abolished in CBF1-deficient cells. Accordingly, EBNA2 targeted CBF1 binding sites present in the FcRH5 promoter in vivo, as detected by chromatin immunoprecipitation. These results identify FcRH5 as a novel, direct target of EBNA2 that may contribute to the development of Epstein-Barr virus–associated tumors.
Introduction
Epstein-Barr virus (EBV) is a γ-herpes virus with a widespread distribution in human populations.1 EBV infects and immortalizes B cells and persists in the vast majority of individuals as a lifelong latent infection of the resting memory B-cell pool.2 EBV is implicated in the etiology of a diverse range of malignancies, including Burkitt lymphoma, Hodgkin disease, and lymphomas that arise in immunosuppressed individuals. In addition, EBV is implicated in the development of a number of autoimmune diseases, including systemic lupus erythematosus.3
EBNA2, together with EBNA–leader protein (LP), is the first viral gene expressed following infection and is absolutely required for B-cell transformation by initiating and maintaining proliferation. EBNA2 is a transcription factor with a potent transactivator domain, but without intrinsic DNA binding activity. EBNA2 is tethered to responsive viral and cellular promoters through host DNA-binding proteins, including CBF1 (also called RBP-Jκ),4-7 PU.1,8 and hnRNP D.9 EBNA2 initiates the transcription of a cascade of primary and secondary target genes that ultimately govern B-cell transformation. The relatively small group of proposed direct cellular target genes of EBNA2 includes CD23,10 CD21,11-13 C-MYC,14 CCR7,15 AML-2,16 BATF,17 and HES-1.18 Of these, only CD23, AML-2, and C-MYC were conclusively demonstrated to be direct targets of EBNA2, as these genes did not require new protein synthesis for induction. Using a CBF1-deficient cell line, CD21 and CCR7 have recently been shown to strictly depend on CBF1 for induction by EBNA2.19 AML-2, c-myc, BATF, and Hes-1 are all transcription factors, indicating there are likely to be numerous indirect targets of EBNA2. Indeed, among other genes, EBNA2 induces the tumor necrosis factor-α (TNF-a), cyclin D2, and interleukin-18 (IL-18) receptor genes indirectly.20,21 In contrast, EBNA2 suppresses the transcription of the immunoglobulin (Ig) μ gene via CBF1-independent mechanisms.19,22 EBNA2 is considered a viral analog of the cellular protein Notch. Accordingly, EBNA2 can functionally replace activated Notch.23 All 4 Notch proteins interact with CBF1, and similarly to EBNA2 function by masking the transcriptional repressor domain of CBF1.23-25 Not surprisingly, the target genes of EBNA2, at least partially, overlap with those of activated Notch.17,26,27
Fc-receptor homolog (FcRH; also called immunoglobulin superfamily receptor translocation associated [IRTA]) comprises a family of 5 recently identified genes contiguously encoded on human chromosome 1q21.28-30 Three mouse FcRHs have subsequently been identified, but there is considerable divergence from the human genes.31 FcRH5 has no mouse homolog. The human FcRH genes are differentially expressed by specific B-cell subpopulations.32,33 These genes encode transmembrane proteins with 3 to 9 Ig-like extracellular domains. All FcRH contain immunoreceptor tyrosine–based inhibitory motif (ITIM) and/or immunoreceptor tyrosine–based activation motif (ITAM) in their cytoplasmic portions in different combinations suggesting a functional role as signaling receptors. Accordingly, FcRH4 has been shown to deliver a potent inhibitory signal to B cells,34 whereas FcRH1, expressed by naive B cells, has been demonstrated to coactivate B cells.35 No ligand for any FcRH has thus far been identified. Due to their restricted expression on specific subpopulations of B cells, FcRHs are potential new tumor markers and candidate targets of tumor immunotherapy.
FcRH5 (IRTA2) is comprised of 9 Ig-like extracellular domains and a cytoplasmic domain with several predicted ITIM and 1 potential ITAM domain. Hatzivassiliou et al originally identified FcRH5 by sequencing the translocation breakpoint in a multiple myeloma cell line with (1;14) (q21:q23) chromosomal translocation.28 In the same paper, FcRH5 expression was shown to be up-regulated on average 10-fold in Burkitt lymphoma cell lines with 1q21 abnormalities. FcRH5 has also been identified as a gene induced following B-cell activation by anti-IgM.30 FcRH5 mRNA is expressed by minor subsets of B cells such as follicular mantle naive B cells, B cells in the light zone of germinal centers, and in intrafollicular and intraepithelial regions of lymph nodes.28,36 FcRH5 protein was found to be highly expressed by malignant B cells of hairy-cell leukemia patients, but undetectable in peripheral B cells of healthy donors.37 The physiologic functions of FcRH5 are unknown.
In the present study we demonstrate direct up-regulation of FcRH5 expression by EBNA2.
Materials and methods
Cells and cell culture
Three EBV-negative human B-cell lines, the lymphoblastoid cell line BJAB-K3 and the Burkitt lymphoma lines BL41-K3 and DG75 (clone SM295), were engineered to express transgenic EBNA2 fused to the hormone-binding domain of the estrogen receptor.19,22,38 In a novel derivative of the DG75 cell line (clone SM296) the CBF1 gene was inactivated by homologous recombination.19 As controls, we used the parent cell lines BJAB,39,40 BL41, and DG75, which do not contain EBNA2-estrogen receptor transgene. Regarding the status of 1q21 aberrations, BJAB is not known to have any,28 while BL41 does have limited 1q abnormalities which do not appear to include 1q21-25;41 the status of 1q21 in DG75 cells is unknown. All cells were cultured in RPMI-1640 (HyClone, Logan, UT) containing 10% fetal bovine serum (HyClone), 2 μM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and nonessential amino acids. To activate EBNA2, estrogen (β-estradiol) was added to a final concentration of 1 μM. In some experiments, cells were pretreated with 5 μg/mL of cyclohexamide to block protein synthesis.
RNA and cDNA preparation and real-time PCR
Total RNA was extracted using Qiagen shredder columns and Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA). RNA was DNase treated using Turbo DNA-free kit (Ambion, Austin, TX). RNA was quantitated using a spectrophotometer. The same amount of RNA, ranging in individual experiments from 0.5 to 2 μg, was used for all samples to make cDNA using random decamers as primers, according to the protocol of Ambion's RETROscript kit. The cDNA samples were then analyzed by real-time polymerase chain reaction (PCR), using specific primers (Table 1), Sybr Green mastermix (Applied Biosystems, Foster City, CA), and an ABI Prizm 7900HT detection system (Applied Biosystems). The PCR conditions to amplify the FcRH5 and EBNA2 mRNA, and 18S rRNA, were the following: 95°C for 10 minutes followed by 45 cycles of 95°C for 30 seconds, 64°C for 30 seconds, and 72°C for 30 seconds. PCR conditions to amplify the CD21 mRNA were the same, except the annealing step was performed at 60°C. Following amplification, the dissociation plot of the product was recorded to ensure that a single product with the expected melting temperature had been amplified. Results were normalized to 18S rRNA, and fold increases in mRNA content were calculated, considering every 1 cycle difference between the threshold cycles a 2-fold difference in mRNA content.
Sequences of real-time PCR primers
Primer . | Sequence, 5′-3′ . |
|---|---|
| FcRH5, forward | GGTGCATATCGCTGTACTGGA |
| FcRH5, reverse | AATGGCTCTTGGACTTGGATTTTGAC |
| EBNA2, forward | GGTTGCTGGAGAGGGCAAGG |
| EBNA2, reverse | GCCCAGAGGCTCCCATTCTC |
| 18S rRNA, forward | CACGGCCGGTACAGTGAAAC |
| 18S rRNA, reverse | CCCGTCGGCATGTATTAGCT |
| CD21, forward | TGTCTTCGGGAAAATGGAGT |
| CD21, reverse | ACCTTCCCATTGGGAAATCGT |
| FcRH5, ChIP no. 1, forward | GGGACAGGAATTGACAAGGT |
| FcRH5, ChIP no. 1, reverse | TCCTCATGCCAATACCACAC |
| FcRH5, ChIP no. 2, forward | TCTAGCAGAACGGCCAGATT |
| FcRH5, ChIP no. 2, reverse | GAGTGGAAGTCCTGGCTCAT |
| FcRH5, ChIP no. 3, forward | CGCATATGCCCCAATAAATG |
| FcRH5, ChIP no. 3, reverse | GCCCATACCTTGGTCTTCCT |
| FcRH5, ChIP no. 4, forward | ACTCCTCGTGGCAGCTAATG |
| FcRH5, ChIP no. 4, reverse | TTCCTGGAAAGTACACACAAAAA |
| FcRH5, ChIP no. 5, forward | TCTTCCCTTCTTGGCATGTT |
| FcRH5, ChIP no. 5, reverse | TCACAGTTTCCTACCCGATG |
| FcRH5, ChIP no. 6, forward | ACGGGTACTCCCACTTCCTT |
| FcRH5, ChIP no. 6, reverse | CACAGCAGCATGAAGACCTG |
| CD23, ChIP, forward | AACTCCAGGCCGTCCTTCTA |
| CD23, ChIP, reverse | ACCTAAATATGGGCTTGCCA |
Primer . | Sequence, 5′-3′ . |
|---|---|
| FcRH5, forward | GGTGCATATCGCTGTACTGGA |
| FcRH5, reverse | AATGGCTCTTGGACTTGGATTTTGAC |
| EBNA2, forward | GGTTGCTGGAGAGGGCAAGG |
| EBNA2, reverse | GCCCAGAGGCTCCCATTCTC |
| 18S rRNA, forward | CACGGCCGGTACAGTGAAAC |
| 18S rRNA, reverse | CCCGTCGGCATGTATTAGCT |
| CD21, forward | TGTCTTCGGGAAAATGGAGT |
| CD21, reverse | ACCTTCCCATTGGGAAATCGT |
| FcRH5, ChIP no. 1, forward | GGGACAGGAATTGACAAGGT |
| FcRH5, ChIP no. 1, reverse | TCCTCATGCCAATACCACAC |
| FcRH5, ChIP no. 2, forward | TCTAGCAGAACGGCCAGATT |
| FcRH5, ChIP no. 2, reverse | GAGTGGAAGTCCTGGCTCAT |
| FcRH5, ChIP no. 3, forward | CGCATATGCCCCAATAAATG |
| FcRH5, ChIP no. 3, reverse | GCCCATACCTTGGTCTTCCT |
| FcRH5, ChIP no. 4, forward | ACTCCTCGTGGCAGCTAATG |
| FcRH5, ChIP no. 4, reverse | TTCCTGGAAAGTACACACAAAAA |
| FcRH5, ChIP no. 5, forward | TCTTCCCTTCTTGGCATGTT |
| FcRH5, ChIP no. 5, reverse | TCACAGTTTCCTACCCGATG |
| FcRH5, ChIP no. 6, forward | ACGGGTACTCCCACTTCCTT |
| FcRH5, ChIP no. 6, reverse | CACAGCAGCATGAAGACCTG |
| CD23, ChIP, forward | AACTCCAGGCCGTCCTTCTA |
| CD23, ChIP, reverse | ACCTAAATATGGGCTTGCCA |
ChIP indicates chromatin immunoprecipitation.
Northern blotting
Total RNA was isolated using the peqGOLD TriFast kit (peqLAB, Erlangen, Germany) following the manufacturer's protocol. RNA (5 μg) was separated on 1.2% formaldehyde agarose gels and transferred to Hybond-N+ membrane (Amersham, Piscataway, NJ). Probes were digoxigenin labeled by PCR. The filters were hybridized with 10 ng/mL probe and detected as described.42 The probe to detect the FcRH5 mRNA was produced by PCR with forward primer TGGTGGTCCAGGTCTTCAT and reverse primer AAGTGCACAGGGCTACTGAGA, which amplify a 310-bp product corresponding to nucleotides –17/+293 of the coding region of the FcRH5 mRNA.
Western blotting
Cells were lysed in RIPA buffer supplemented with protease inhibitors. The same amount of total protein, ranging in individual experiments from 20 to 40 μg, was separated on 10% SDS polyacrylamide gels (Invitrogen, Carlsbad, CA) and transferred to PVDF membranes (Amersham, Piscataway, NJ). FcRH5 was detected by 0.6 μg/mL FcRH5-specific mouse monoclonal antibody (mAb) F2537 (provided by Dr Ira Pastan, Bethesda, MD). Immunolabeling was carried out using horseradish peroxidase–labeled Ab (Amersham), and enzymatic chemiluminescence (ECL) Plus Reagents (Amersham). Blots were stripped and reblotted with antiactin Ab (Santa Cruz Biotechnology, Santa Cruz, CA). A FluorChem 8900 Imaging System from Alpha Innotech Corporation (San Leandro, CA) was used to scan the X-ray film images, without any gamma adjustment.
Flow cytometry
Flow cytometry was carried out using standard protocols on a FACSCalibur (Becton Dickinson, San Jose, CA) and data analyzed using Cell Quest Pro (Becton Dickinson). FcRH5 was detected by incubation with 0.5 μg per sample of mAb F56 (from Dr Ira Pastan).37
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described,43 based on established protocols.44 Briefly, estrogen stimulated or unstimulated BJAB-K3 cells were fixed with 1% formaldehyde at 37°C for 10 minutes, then washed, lysed, and sonicated for a total of 2.5 minutes. Extracts obtained from 30 × 106 cells per immunoprecipitation reaction were precleared for 0.5 hours with Protein A/G PLUS-agarose (Santa Cruz Biotechnology). The precleared samples were immunoprecipitated with 6.5 μg murine EBNA2–specific mAb (DAKO, Glostrup, Denmark), 6.5 μg murine CBF1–specific Ab (clone C-20, Santa Cruz Biotechnology), or 5 μg murine hnRNP D–specific mAb 5B9 (gift of Dr Gideon Dreyfuss, Philadelphia, PA) at 4°C overnight, or processed without Ab. The immune complexes were recovered by adding Protein A/G PLUS-agarose, which had been treated with sonicated salmon sperm DNA for 1 hour. Precipitates were washed a total of 8 times. The samples were subsequently digested with proteinase K and the crosslink was reversed by heating the samples at 65°C overnight. Recovered DNA (3%) was used per real-time PCR reaction as template. The PCR conditions were the following: 95°C for 10 minutes followed by 45 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. All primers are listed in Table 1.
Results
EBNA2 induces FcRH5 expression in the absence of other EBV proteins
To determine whether EBNA2 regulates FcRH5 expression we used 3 EBV-negative human B-cell lines, BJAB-K3, BL41-K3, and DG75, that express functional EBNA2 protein in an inducible manner. All 3 cell lines were engineered to express transgenic EBNA2 fused to the hormone-binding domain of the estrogen receptor.19,38 The EBNA2-estrogen receptor fusion protein is unable to enter the nucleus in the absence of estrogen. EBNA2 activity is induced by estrogen, resulting in the translocation of the fusion protein into the nucleus, where EBNA2 can exert its effects on gene expression. These cell lines have been used extensively to study regulation of host gene expression by EBNA2.15,19,22
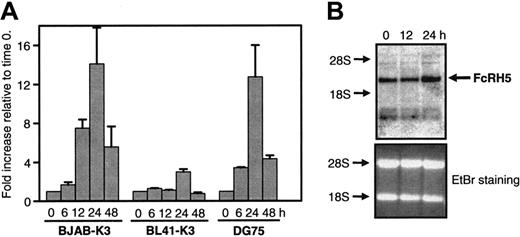
First, we measured the levels of FcRH5 mRNA upon activating EBNA2 using quantitative real-time PCR. FcRH5 mRNA levels were elevated in all 3 cell lines after inducing EBNA2 (Figure 1A). In BJAB-K3 cells FcRH5 mRNA levels were elevated 12 hours after activating EBNA2. After 24 hours we measured on average 14-fold induced FcRH5 mRNA (P = .04, as calculated using 2-tailed paired t test). In BL41-K3 cells we detected 3-fold elevated FcRH5 mRNA 24 hours after activating EBNA2 (P = .02). In DG75 cells we detected elevated FcRH5 mRNA levels as early as 6 hours after stimulation, and after 24 hours we measured on average 13-fold induced expression. The minimum times required to induce FcRH5 in the 3 cell lines indicate cell type–specific differences in their responses to EBNA2. Nevertheless, FcRH5 mRNA levels consistently peaked 24 hours after activating EBNA2 in all cell lines, and dropped thereafter. In contrast, CD21 mRNA levels continued to increase after 24 hours (data not shown). To exclude the possibility that estrogen itself induced FcRH5 expression via EBNA2-independent mechanisms, such as estrogen responsive promoter elements, we stimulated the BJAB and BL41 parent cell lines that do not express EBNA2-estrogen receptor fusion protein, with estrogen. We did not detect any elevation of FcRH5 mRNA or protein expression 6 to 48 hours after estrogen stimulation, indicating that activation of EBNA2 is required for enhanced FcRH5 expression (data not shown). Northern blots were performed in BJAB-K3 cells with probes specific to FcRH5 to substantiate the results obtained using real-time PCR. We detected a single FcRH5 mRNA corresponding to the reported 5.4-kb transmembrane form of FcRH5 mRNA.36 The intensity of the specific FcRH5 band was clearly elevated at 24 hours, but not 12 hours, after induction of EBNA2 activity (Figure 1B). The amount of CD21 mRNA, measured as a positive control, was greatly elevated as early as 12 hours after activating EBNA2 (data not shown). These results demonstrate that EBNA2 induces FcRH5 mRNA expression.
EBNA2 induces the expression of the FcRH5 gene. EBNA2 activity was induced in BJAB-K3, BL41-K3, and DG75 (clone SM295) cells by estrogen. RNA samples were collected before induction (time 0), and 6 to 48 hours after inducing EBNA2, as indicated. (A) cDNA were synthesized and used as template in quantitative real-time PCR with FcRH5 specific primers. Fold increases in mRNA content relative to the time 0 sample are shown. Mean and SEM of 2 to 5 independent experiments are shown. (B) Northern blots were performed using RNA samples obtained from BJAB-K3 cells, and a FcRH5-specific probe. On the right, the arrow indicates the position of the FcRH5 mRNA. On the left, the positions of the 18S and 28S rRNAs are indicated based on ethidium bromide staining. The bottom panel shows ethidium bromide staining of the gel. A representative experiment is shown.
EBNA2 induces the expression of the FcRH5 gene. EBNA2 activity was induced in BJAB-K3, BL41-K3, and DG75 (clone SM295) cells by estrogen. RNA samples were collected before induction (time 0), and 6 to 48 hours after inducing EBNA2, as indicated. (A) cDNA were synthesized and used as template in quantitative real-time PCR with FcRH5 specific primers. Fold increases in mRNA content relative to the time 0 sample are shown. Mean and SEM of 2 to 5 independent experiments are shown. (B) Northern blots were performed using RNA samples obtained from BJAB-K3 cells, and a FcRH5-specific probe. On the right, the arrow indicates the position of the FcRH5 mRNA. On the left, the positions of the 18S and 28S rRNAs are indicated based on ethidium bromide staining. The bottom panel shows ethidium bromide staining of the gel. A representative experiment is shown.
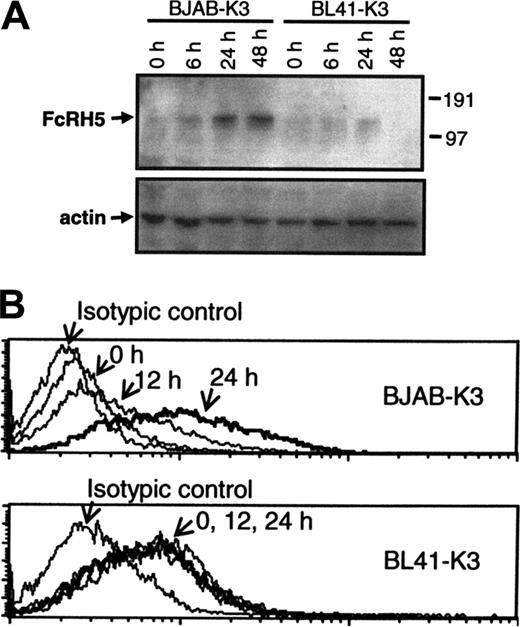
Next, we examined if elevated mRNA expression resulted in elevated protein expression. Protein samples obtained from induced and uninduced BJAB-K3 and BL41-K3 cells were subjected to Western blot analysis using FcRH5-specific mAb. In uninduced BJAB-K3 cells hardly any protein was detected, but marked FcRH5 protein expression was seen 24 and 48 hours after stimulation of EBNA2 (Figure 2A). In BL41-K3 cells we did not detect any increase in FcRH5 protein levels in repeated experiments. Assessment of cell-surface expression of FcRH5 by flow cytometry, using a different FcRH5 mAb, produced results similar to those observed by Western blotting. In BJAB-K3 cells we detected robust induction of surface FcRH5 protein expression that was evident as early as 12 hours after induction of EBNA2 (Figure 2B, top panel). The mean fluorescence intensity increased 1.8-fold by 12 hours, and 4.5-fold by 24 hours after stimulation. In contrast, we did not detect any enhancement of FcRH5 expression in BL41-K3 cells in repeated experiments (Figure 2B, bottom panel).
EBNA2 directly induces FcRH5 expression by targeting the promoter via CBF1
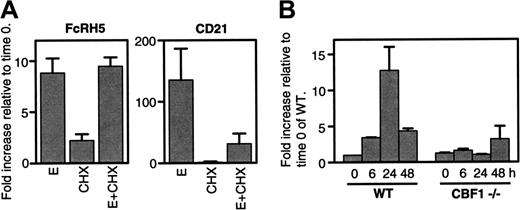
To study the mechanism of induced FcRH5 expression we first investigated whether EBNA2 activates the FcRH5 gene directly or through another transcription factor induced by EBNA2. Blocking new protein synthesis with cyclohexamide did not affect EBNA2-induced enhancement of FcRH5 mRNA levels in BJAB-K3 cells (Figure 3A), suggesting that newly synthesized protein is not required to induce FcRH5 expression, and EBNA2 acts directly. In contrast, EBNA2-induced expression of the CD21 gene was strongly inhibited by cyclohexamide, suggesting that new protein synthesis is required for the induction of CD21 by EBNA2, although cyclohexamide might impact mRNA levels through other mechanisms such as affecting mRNA stability.
EBNA2 is recruited to some responsive genes by interacting with the host DNA-binding protein CBF1. We examined the role of CBF1 in the induced expression of FcRH5 using a novel derivative of the DG75 B-cell line, in which the CBF1 gene is inactivated by homologous recombination.19 As control we used the parent DG75 cells that contain a functional CBF1 gene. Both the CBF1-positive and -negative DG75 cells carry the same EBNA2-estrogen receptor transgene. We detected markedly induced FcRH5 expression by EBNA2 in CBF1-positive cells, but no enhancement of FcRH5 expression in cells lacking CBF1 (Figure 3B). These findings imply that CBF1 is absolutely required for the EBNA2-induced expression of FcRH5.
EBNA2 enhances FcRH5 protein levels. EBNA2 activity was induced in BJAB-K3 and BL41-K3 cells with estrogen for 6 to 48 hours, as indicated. (A) Proteins were purified and analyzed by Western blotting using FcRH5 and actin-specific Ab. (B) Membrane FcRH5 protein expression was analyzed by flow cytometry using FcRH5 specific Ab and isotypic control Ab. In each panel, the 24-hour samples are shown as a thick line. Representatives of 3 experiments are shown.
EBNA2 enhances FcRH5 protein levels. EBNA2 activity was induced in BJAB-K3 and BL41-K3 cells with estrogen for 6 to 48 hours, as indicated. (A) Proteins were purified and analyzed by Western blotting using FcRH5 and actin-specific Ab. (B) Membrane FcRH5 protein expression was analyzed by flow cytometry using FcRH5 specific Ab and isotypic control Ab. In each panel, the 24-hour samples are shown as a thick line. Representatives of 3 experiments are shown.
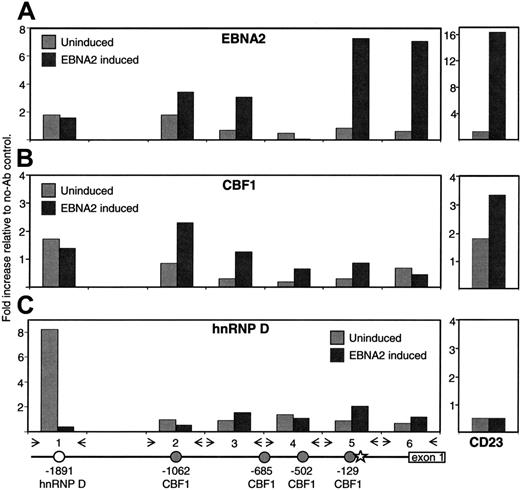
Finally, we studied the recruitment of EBNA2 to the endogenous FcRH5 gene in BJAB-K3 cells using chromatin immunoprecipitation. We identified 4 potential consensus CBF1-binding sites at –1062, –685, –502, and –129 in the FcRH5 promoter, relative to the transcription start site (Table 2), using a computer algorithm.45 Furthermore, a potential hnRNP D binding site was identified at –1891 relative to the transcription start site (Table 2). As EBNA2 has been reported to interact with both CBF1 and hnRNP D, the binding of both proteins to the FcRH5 promoter was monitored. Importantly, no binding sites for PU.1, another transcription factor that can interact with EBNA2, or c-myc, an EBNA2-induced transcription factor, were predicted using the same algorithm. BJAB-K3 cells that had been treated with estrogen for 5 hours to induce EBNA2, or left untreated, were crosslinked with formaldehyde to covalently link proteins to DNA, and proteins to proteins, within close proximity. The soluble, fragmented protein-DNA complexes were immunoprecipitated with Ab specific to EBNA2, CBF1 or hnRNP D, or processed without Ab. The precipitated DNA fragments were recovered and used in quantitative real-time PCR as templates. We used 6 PCR primer pairs to test for the enrichment of corresponding FcRH5 promoter-derived DNA fragments in the Ab-precipitated samples, relative to the sample processed without Ab. As a positive control we measured the enrichment of the best characterized EBNA2-responsive CBF1-binding site in the CD23 promoter.6 We detected no binding of any of the 3 proteins to the CD23 promoter in uninduced cells, but detected strong binding of EBNA2 (16-fold enrichment) to the CD23 promoter in cells with activated EBNA2 (Figure 4, right panels). Also, we detected modest binding of CBF1 (3.3-fold enrichment) to the CD23 promoter in induced cells. The relative efficiencies of EBNA2 and CBF1 binding, as detected by chromatin immunoprecipitation, critically depend on the qualities of Ab, and thus cannot be directly compared. Thus, our experimental system is suitable to assess in vivo recruitment of EBNA2 and CBF1. We did not detect binding of EBNA2 or CBF1 to any segment of the FcRH5 promoter in uninduced cells (Figure 4A-B, left panels). However, in cells in which EBNA2 had been induced for 5 hours we detected strong EBNA2 binding to the FcRH5 promoter (Figure 4A-B). The strongest signals were generated with primers 5 and 6 (7-fold enrichment), suggesting that EBNA2 was binding around the TATA box. In addition, primers 2 and 3 produced smaller but clear signals, suggesting that EBNA2 targeted a second, upstream promoter element. We detected modest binding of CBF1 using primer 2 (2.2-fold enrichment), which was comparable in magnitude to what we measured for the CD23 promoter site. Interestingly, we detected strong binding of hnRNP D around the predicted hnRNP D binding site in uninduced cells, but not in cells in which EBNA2 had been activated (Figure 4C).
Predicted CBF1 and hnRNP D binding sites in the FcRH5 promoter, compared with known sites
Site . | Promoter . | Position* . | Strand . | Sequence . |
|---|---|---|---|---|
| CBF1, no. 1 | FcRH5 | -1062 | + | GTGGGAA |
| CBF1, no. 2 | FcRH5 | -685 | + | GTGAGAA |
| CBF1, no. 3 | FcRH5 | -502 | - | GTGGGAA |
| CBF1, no. 4 | FcRH5 | -129 | - | TTGGGAA |
| CBF1 | CD23 | -207 | + | GTGGGAA |
| hnRNP D | FcRH5 | -1891 | - | TTCTTCAAAAT |
| hnRNP D | CD21 | -483 | + | TACTTCAAAAT |
Site . | Promoter . | Position* . | Strand . | Sequence . |
|---|---|---|---|---|
| CBF1, no. 1 | FcRH5 | -1062 | + | GTGGGAA |
| CBF1, no. 2 | FcRH5 | -685 | + | GTGAGAA |
| CBF1, no. 3 | FcRH5 | -502 | - | GTGGGAA |
| CBF1, no. 4 | FcRH5 | -129 | - | TTGGGAA |
| CBF1 | CD23 | -207 | + | GTGGGAA |
| hnRNP D | FcRH5 | -1891 | - | TTCTTCAAAAT |
| hnRNP D | CD21 | -483 | + | TACTTCAAAAT |
+ indicates coding strand; -, noncoding strand.
Positions of sites relative to the transcription start site are indicated.
EBNA2 induces FcRH5 through CBF1, independent of new protein synthesis. (A) BJAB-K3 cells were treated in one of the following ways; with estrogen (E), with cyclohexamide, a protein synthesis inhibitor (CHX), or with both estrogen and cyclohexamide (E + CHX). RNA samples were collected before treatment (time 0), and 12 hours later. cDNA were synthesized and used as template in quantitative real-time PCR with FcRH5- and CD21-specific primers. Fold increases in mRNA content at 12 hours relative to the time 0 samples are shown. Mean and SEM of 3 independent experiments are shown. (B) EBNA2 activity was induced in wild-type (WT) and CBF1–/– DG75 cells with estrogen for 6 to 48 hours, as indicated. RNA samples were collected and cDNA were synthesized and used as template in quantitative real-time PCR with FcRH5 specific primers. Fold increases in mRNA content relative to the time 0 sample of the wild-type DG75 cells are shown. Mean and SEM of 2 independent experiments are shown.
EBNA2 induces FcRH5 through CBF1, independent of new protein synthesis. (A) BJAB-K3 cells were treated in one of the following ways; with estrogen (E), with cyclohexamide, a protein synthesis inhibitor (CHX), or with both estrogen and cyclohexamide (E + CHX). RNA samples were collected before treatment (time 0), and 12 hours later. cDNA were synthesized and used as template in quantitative real-time PCR with FcRH5- and CD21-specific primers. Fold increases in mRNA content at 12 hours relative to the time 0 samples are shown. Mean and SEM of 3 independent experiments are shown. (B) EBNA2 activity was induced in wild-type (WT) and CBF1–/– DG75 cells with estrogen for 6 to 48 hours, as indicated. RNA samples were collected and cDNA were synthesized and used as template in quantitative real-time PCR with FcRH5 specific primers. Fold increases in mRNA content relative to the time 0 sample of the wild-type DG75 cells are shown. Mean and SEM of 2 independent experiments are shown.
Discussion
Our study establishes FcRH5 as a new, direct target of EBNA2. We show, as part of the mechanism, that EBNA2 targets the FcRH5 gene in vivo, and that the cellular protein CBF1 is absolutely required for induced FcRH5 expression.
In 3 independent human B-cell lines, each expressing an inducible EBNA2-estrogen receptor transgene, FcRH5 mRNA levels were markedly induced by EBNA2, in the absence of other EBV proteins. FcRH5 mRNA levels peaked after 24 hours of activating EBNA2, and significantly dropped thereafter, suggesting a mechanism that might ensure temporary expression of FcRH5. In BJAB-K3 cells, in which FcRH5 mRNA levels were induced on average 14-fold, we measured 4.5-fold elevated FcRH5 protein levels on the cell surface. In BL41-K3 cells, in which we detected up to 3-fold elevated FcRH5 mRNA levels, FcRH5 protein levels were unchanged. Mechanisms affecting mRNA and/or protein processing and stability may have different efficiencies in the cell lines used.46 The substantial impact of the cellular background on target gene activation has been demonstrated for a number of EBNA2-regulated genes.12,19,47 Alternatively, the lack of response in BL41-K3 cells may be secondary to aberrations of 1q21.41 Of the 3 cell lines used, BJAB is not known to have any 1q21 abnormalities;28 thus, it is a particularly suitable model to investigate the B-cell response to EBV. EBNA2 is known to down-regulate the immunoglobulin μ gene,22 and LMP2A was proposed to take over B-cell–receptor signaling,48 in which a role for induced FcRH5 remains to be established.
EBNA2 targets the endogenous FcRH5 promoter. BJAB-K3 cells that had been treated with estrogen for 5 hours to induce EBNA2, or left untreated, were crosslinked with formaldehyde. Soluble, fragmented chromatin was immunoprecipitated with (A) EBNA2, (B) CBF1, or (C) hnRNP D Ab, or without Ab. Precipitated DNA were used in quantitative real-time PCR as template. Fold increases in DNA content relative to the no-Ab controls are shown. ▦ denotes uninduced samples; ▪, samples with induced EBNA2. At the bottom is a schematic representation of the FcRH5 gene from approximately –2000 to +200, relative to the transcription start site. Potential CBF1 ( ) and hnRNP D (○) binding sites and the likely TATA box (★) are marked. Inverted arrows indicate the positions of PCR primer pairs (see Table 1 for primer sequences). At the right of each panel, binding of proteins to the known EBNA2-responsive CBF1 binding site at –207 (see Table 2) of the CD23 promoter is shown.
) and hnRNP D (○) binding sites and the likely TATA box (★) are marked. Inverted arrows indicate the positions of PCR primer pairs (see Table 1 for primer sequences). At the right of each panel, binding of proteins to the known EBNA2-responsive CBF1 binding site at –207 (see Table 2) of the CD23 promoter is shown.
EBNA2 targets the endogenous FcRH5 promoter. BJAB-K3 cells that had been treated with estrogen for 5 hours to induce EBNA2, or left untreated, were crosslinked with formaldehyde. Soluble, fragmented chromatin was immunoprecipitated with (A) EBNA2, (B) CBF1, or (C) hnRNP D Ab, or without Ab. Precipitated DNA were used in quantitative real-time PCR as template. Fold increases in DNA content relative to the no-Ab controls are shown. ▦ denotes uninduced samples; ▪, samples with induced EBNA2. At the bottom is a schematic representation of the FcRH5 gene from approximately –2000 to +200, relative to the transcription start site. Potential CBF1 ( ) and hnRNP D (○) binding sites and the likely TATA box (★) are marked. Inverted arrows indicate the positions of PCR primer pairs (see Table 1 for primer sequences). At the right of each panel, binding of proteins to the known EBNA2-responsive CBF1 binding site at –207 (see Table 2) of the CD23 promoter is shown.
) and hnRNP D (○) binding sites and the likely TATA box (★) are marked. Inverted arrows indicate the positions of PCR primer pairs (see Table 1 for primer sequences). At the right of each panel, binding of proteins to the known EBNA2-responsive CBF1 binding site at –207 (see Table 2) of the CD23 promoter is shown.
In an effort to establish the mechanism of EBNA2-induced FcRH5 expression, we first investigated whether FcRH5 is a direct target of EBNA2. This is important, because EBNA2 enhances the expression of several transcription factors that could indirectly act upon the FcRH5 gene.14,16-18 We show that EBNA2 did induce FcRH5 mRNA in the absence of new protein synthesis, strongly suggesting that EBNA2 targets the FcRH5 gene directly. Using similar experimental approaches, direct activation of the CD23, AML-2, and C-MYC genes by EBNA2 has so far been demonstrated.10,14,16 Thus, to the short list of known, genuine target genes of EBNA2 we have now added FcRH5.
EBNA2 is recruited to responsive viral genes by interacting with the host DNA-binding protein CBF1.4,5,7 The obligatory role of CBF1 in mediating the activation of cellular genes by EBNA2 is unclear. We used a novel, CBF1-deficient B-cell line to address the contribution of CBF1 to EBNA2-induced FcRH5 expression. Notably, in the absence of active EBNA2, basal FcRH5 expression levels of CBF1-deficient and wild-type cells were comparable, indicating that CBF1 did not repress FcRH5 expression in wild-type cells. Similar findings were reported for the CD21 and CCR7 genes.19 However, following activation of EBNA2, we detected complete abrogation of FcRH5 gene induction in CBF1-deficient cells, indicating that CBF1 is indispensable for EBNA2-induced FcRH5 expression.
Prompted by our findings about the involvement of CBF1, we inspected the DNA sequence of the FcRH5 promoter and first intron, and identified 4 potential consensus CBF1 binding sites in the proximal promoter. Using chromatin immunoprecipitation, we detected strong EBNA2 binding to the FcRH5 promoter region surrounding the TATA box, and modest EBNA2 binding to an upstream promoter region. Each interacting promoter region contains 1 predicted CBF1 binding site, implicating those in EBNA2 binding. We detected moderate binding of CBF1 to the upstream promoter region, but no CBF1 binding around the TATA box. It is possible that the upstream CBF1 site is the primary mediator of EBNA2 binding, and EBNA2 subsequently targets the TATA box via promoter looping, facilitated by protein-protein interactions, to communicate with the basal transcription machinery. Alternatively, proteins binding to CBF11,49 might mask the epitope recognized by our anti–CBF1 Ab. This is the first report of monitoring of EBNA2 binding to cellular genes by chromatin immunoprecipitation, as the only published study examined viral genes.50 In addition, we detected the binding of hnRNP D to a far-upstream FcRH5 promoter region, but only in cells in which EBNA2 had not been induced. HnRNP D has been proposed to repress the CD21 gene43,51 and interact with EBNA2.9 Our data are compatible with hnRNP D playing a role in repressing the FcRH5 gene, although the relative contribution of this repression is unknown, and suggest that binding of EBNA2 to hnRNP D might release repression.
Our findings implicating CBF1 in the induction of FcRH5 expression raise the possibility of other CBF1-dependent transcription factors regulating the FcRH5 gene. Activated Notch, a cellular protein, uses CBF1 to target responsive promoters through mechanisms similar to those used by EBNA2.23-25 An important role for the Notch-CBF1 signaling pathway in regulating marginal zone B-cell development has been demonstrated in mice selectively lacking CBF1 in B cells.52 FcRH5 may regulate B-cell activation and differentiation by virtue of its proposed signaling potential, potentially providing a missing link as to how Notch and CBF1 control B-cell development. NOTCH2 is overexpressed in B-cell chronic lymphocytic leukemia cells, leading to elevated levels of CD23 through CBF1-dependent mechanisms.26 Moreover, activating mutations of the NOTCH1 gene have been described in more than 50% of human T-cell acute lymphoblastic leukemia cases,53 suggesting that alterations of the Notch-CBF1 pathway significantly impact malignant transformation. Besides EBNA2 and Notch, the Kaposi sarcoma–associated herpesvirus protein RTA and the adenoviral protein 13SE1A were shown to bind CBF1.54,55 It remains to be seen whether FcRH5 is induced during these viral infections. Nevertheless, the RTA protein induces CD21 and CD23 expression similarly to EBNA2,56 indicating that the target genes of these 2 viral proteins overlap.
As for the role of FcRH5 in EBV associated B-cell malignancies, a better general understanding of how EBV contributes to tumor development is necessary. FcRH5 is overexpressed by malignant B cells of hairy-cell leukemia patients.37 Although a pathogenic role for EBV in hairy-cell leukemia has been suggested,57,58 those studies may represent a technical artifact.59 FcRH5 expression is elevated in Burkitt lymphoma cell lines with chromosome 1q21 abnormalities, implicating FcRH5 in lymphomagenesis.28 Furthermore, a genome-wide search for chromosomal abnormalities concluded that a gene important for Burkitt lymphoma pathogenesis is located in region 1q21-25, and overexpression of this gene mimics the effects of EBV.41 As FcRH5 is encoded on 1q21 and is induced by EBV, FcRH5 is a candidate to be this unidentified gene. One can speculate that perturbation of FcRH5 expression and thus function during EBV-driven B-cell proliferation might disregulate B cells, thus contributing to B-cell malignancies. Although studies to establish the functional role of FcRH5 in B cells are needed, our results place a new piece of the puzzle on the table, potentially linking EBV infection to B-cell tumors through FcRH5.
Prepublished online as Blood First Edition Paper, January 26, 2006; DOI 10.1182/blood-2005-09-3815.
Supported in part by an appointment of J.M. to the Research Fellowship Program for the Center for Drug Evaluation and Research administered by the Oak Ridge Associated Universities through a contract with the US Food and Drug Administration. This study was also supported by the Deutsche Forschungsgemeinschaft (SFB455), Deutsche Krebshilfe (grant 10-1963-Ke-I) and Wilhelm Sander-Stiftung (grant 2003.143.1).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Ira Pastan and Satoshi Nagata for providing the FcRH5 Ab and for suggestions, Dr Gideon Dreyfuss for the hnRNP D Ab, and Dr Marjorie Shapiro, Gerald Feldman, and Sean Fitzsimmons for suggestions. We thank the valuable comments of an anonymous reviewer. The opinions expressed in this article are those of the authors and not necessarily those of the Food and Drug Administration or the US government. The publication of this article should not be construed as an endorsement or approval of either the product or the company.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal