Abstract
Using an in silico database search, we identified a novel gene encoding a cell surface molecule with a thrombospondin domain, and designated the gene as transmembrane molecule with thrombospondin module (Tmtsp). Expression profiling of Tmtsp using specific monoclonal antibodies and Venus, a variant of yellow fluorescent protein knock-in mice in the Tmtsp locus, demonstrated its specific expression in hematopoietic and endothelial cells. In lymphohematopoietic cells, Tmtsp was predominantly expressed in hematopoietic stem and progenitor cells, and the level of expression gradually declined as the cells differentiated. Venus expression faithfully traced the expression of Tmtsp, and the level of Venus expression correlated well to the in vitro hematopoietic activity as well as the in vivo bone marrow repopulating capacity. Notably, Venus expression marked the development of definitive hematopoiesis in both the extraembryonic yolk sac and the intraembryonic aorta-gonadmesonephros (AGM) region and, in combination with CD41, strikingly promoted the enrichment of developing progenitors in the CD41+Venushigh fraction at embryonic day 10.5 (E10.5). In this context, Tmtsp is a novel marker gene for primitive hematopoietic cells and endothelial cells, and TmtspVenus/+ mice would serve as a valuable mouse model for the analysis of both embryonic and adult hematopoiesis, as well as for vascular biology.
Introduction
Hematopoietic stem cells (HSCs) are defined as cells that have the ability to self-renew and differentiate into all blood cell lineages. Although the frequency of HSCs is quite low, advances in cell sorting technology and an increasing number of marker antibodies enable us to access highly purified HSCs. We have previously demonstrated that mouse bone marrow (BM) HSCs are highly enriched in CD34–/low, c-Kit+, Sca-1+, and the lineage marker–negative (CD34–KSL) cell population.1 Subsequently, CD34–KSL cells with the strongest dye efflux activity (“Tip”-SP CD34–KSL) appeared to have a high marrow-homing capacity.2-4 The progress in HSC identification and purification greatly serves the prospective investigation of HSCs, as well as the molecular characterization of HSCs.
HSC functions, including self-renewal and multilineage differentiation, are precisely regulated through tight interactions with stromal cells, cytokines, and the extracellular matrix (ECM) in BM niche. A number of cell surface molecules and secreted proteins have been implicated in this process. Of note is that recent studies uncovered an essential role of osteoblasts, one of the components of the BM niche in the maintenance of HSCs. The interaction between HSCs and osteoblasts has been shown to involve the BMP, Notch, and Tie2 signals to regulate the number and quiescent state of HSCs.5-7 Moreover, several adhesion molecules have been characterized to be key molecules of “homing to niche,” namely the migration of HSCs, such as N-cadherin, ICAMs, and LFAs.8
HSCs and endothelial cells are tightly linked in their ontogeny. Many of the genes commonly expressed in HSCs and endothelial cells play important roles in the development and maintenance of both lineages. Among the transmembrane molecules, cytokine receptors including Fms-like receptor tyrosine kinase-1 (Flk-1) and endoglin have been well characterized. Mutant mice with disrupted Flk-1 (also known as vascular endothelial growth factor receptor 2 [VEGFR-2]) die at E8.5 to E9.5 from defective development of both hematopoietic and endothelial lineages, indicative of the essential roles of Flk-1 in the ontogeny of hematopoiesis and vasculogenesis.9 In the adult mouse, VEGF regulates HSC survival by an internal autocrine loop mechanism.10 Endoglin (CD105), a specific receptor for transforming growth factor-β (TGF-β), is expressed on endothelial cells from the developmental stage, and knock-out mice die by E11.5 from a defect in the remodeling of the primary endothelial network into a mature circulatory system.11 Recent reports defined endoglin as a marker for long-term repopulating (LTR) HSCs.12,13
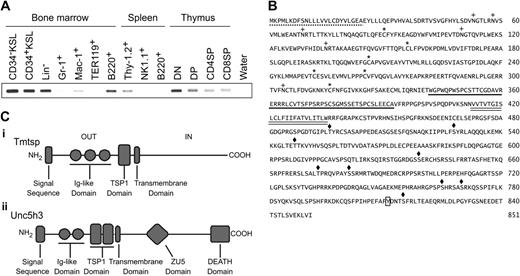
Identification of a novel cell surface molecule specific to primitive hematopoietic cells. (A) Expression of Tmtsp in hematopoietic cells. Semiquantitative RT-PCR was carried out using normalized cDNA by GAPDH. Cells analyzed are bone marrow CD34–c-Kit+Sca-1+Lineage marker– HSCs (CD34–KSL), CD34+KSL progenitors, lineage marker– cells (Lin–), Gr-1+ neutrophils, Mac-1+ monocytes/macrophages, TER119+ erythroblasts, B220+ B cells; spleen Thy-1.2+ T cells, NK1.1+ natural killer (NK) cells, and B220+ B cells; and thymic CD4–CD8– T cells (DN), CD4+CD8+ T cells (DP), CD4+CD8– T cells (CD4SP), and CD4–CD8+ T cells (CD8SP). (B) Deduced amino acid sequence and conserved domains of mouse Tmtsp. Tmtsp protein possesses several domains indicated as follows: signal sequence, dotted line; cysteine residues of Ig-like domain, asterisks; thrombospondin type I repeat (TSP1 domain), underline; transmembrane domain, double line; PKC-phosphorylation site, diamonds; N-glycosylation site, crosses; and tyrosine-phosphorylation site, box. The cDNA sequences are available from the GenBank database under the accession numbers AB039946 and AB044385 (mouse and human, respectively). (C) Schematic illustration of Tmtsp (i) and Unc5h3 proteins (ii).
Identification of a novel cell surface molecule specific to primitive hematopoietic cells. (A) Expression of Tmtsp in hematopoietic cells. Semiquantitative RT-PCR was carried out using normalized cDNA by GAPDH. Cells analyzed are bone marrow CD34–c-Kit+Sca-1+Lineage marker– HSCs (CD34–KSL), CD34+KSL progenitors, lineage marker– cells (Lin–), Gr-1+ neutrophils, Mac-1+ monocytes/macrophages, TER119+ erythroblasts, B220+ B cells; spleen Thy-1.2+ T cells, NK1.1+ natural killer (NK) cells, and B220+ B cells; and thymic CD4–CD8– T cells (DN), CD4+CD8+ T cells (DP), CD4+CD8– T cells (CD4SP), and CD4–CD8+ T cells (CD8SP). (B) Deduced amino acid sequence and conserved domains of mouse Tmtsp. Tmtsp protein possesses several domains indicated as follows: signal sequence, dotted line; cysteine residues of Ig-like domain, asterisks; thrombospondin type I repeat (TSP1 domain), underline; transmembrane domain, double line; PKC-phosphorylation site, diamonds; N-glycosylation site, crosses; and tyrosine-phosphorylation site, box. The cDNA sequences are available from the GenBank database under the accession numbers AB039946 and AB044385 (mouse and human, respectively). (C) Schematic illustration of Tmtsp (i) and Unc5h3 proteins (ii).
In this study, we attempted to identify novel transmembrane molecules expressed on HSCs by an in silico database search. By using the amino acid sequence corresponding to the transmembrane region of the interleukin-11 receptor as a bait, we identified a novel gene, transmembrane molecule with thrombospondin module (Tmtsp), that is preferentially expressed in primitive hematopoietic cells and endothelial cells. In addition, we successfully marked the primitive hematopoietic cells and endothelial cells by Venus protein, a mutant of yellow fluorescent protein in mice, by making Venus knock-in mice at the Tmtsp locus. Here, we describe the molecular characterization of Tmtsp and genetic marking of hematopoietic stem/progenitor cells and endothelial cells.
Materials and methods
Molecular cloning of Tmtsp
Human and mouse Tmtsp cDNAs were amplified by polymerase chain reaction (PCR) from human spleen and mouse total embryo (E15) phage libraries, respectively.
Culture of cell lines
COS7 and bEND.3 cells were maintained in Dulbecco modified Eagle medium (Sigma-Aldrich, St Louis, MO) supplemented with 10% heat-inactivated FCS, 2 mM l-glutamine, penicillin, and streptomycin in a 5% CO2 incubator. BW5147 cells were cultured in RPMI-1640 medium (Sigma-Aldrich) with 10% heat-inactivated FCS, 2 mM l-glutamine, penicillin, and streptomycin. To establish BW5147 cells stably expressing Tmtsp, full-length Tmtsp cDNA tagged with a FLAG epitope at the N-terminus was subcloned into pMX retrovirus vector, and infected cells were sorted with an anti-FLAG mAb (clone: M2; Sigma-Aldrich) on a FACSVantageSE (Becton-Dickinson, San Jose, CA). 293T cells were transfected by CaPO4 coprecipitation method.
Culture of ES cells and coculture with OP-9 cells
Maintenance of embryonic stem (ES) cells (E14.1), embryoid body (EB) formation, and culture of OP-9 cells were performed as previously described.14,15 For differentiation into hematopoietic or endothelial lineage cells, cells derived from EB or embryo were plated onto monolayer of mitomycin C (Sigma-Aldrich)–treated OP-9 cells in α-minimum essential medium (MEM; Invitrogen, Carlsbad, CA) supplemented with 10% FCS, 50 μM 2-mercaptoethanol, penicillin, streptomycin, and cytokines as follows: for hematopoietic differentiation, 10 ng/mL mouse stem cell factor (SCF), 50 ng/mL human thrombopoietin (TPO), 10 ng/mL human erythropoietin (EPO), and 10 ng/mL mouse interleukin-3 (IL-3; Peprotech, Rocky Hill, NJ); for endothelial differentiation, 10 ng/mL mouse vascular endothelial growth factor (VEGF; Peprotech); and for lymphatic endothelial differentiation, 20 ng/mL mouse VEGF-D (R&D System, Minneapolis, MN). Images of the EB were acquired using a Leica FW4000 microscope and a 40 × 0.60 numeric aperture (NA) objective (Leica, Heidelberg, Germany).
Cell surface biotinylation and immunoprecipitation
For cell surface biotinylation, 5 × 106 cells were biotinylated by ECL Biotinylation Module (Amersham Biosciences, Buckinghamshire, United Kingdom) as previously described.16 Immunoprecipitation (IP) was performed using protein G sepharose (Amersham Biosciences). Precipitates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto PVDF membranes (Millipore, Bedford, MA). The blot was probed with HRP-conjugated streptavidin (Amersham Biosciences). Signals were detected with SuperSignal West Pico Chemiluminescent Substrate (PIERCE, Rockford, IL).
In vitro binding assay
Binding assay between Tmtsp and netrin-1 was examined as previously described.17 Briefly, COS7 cells transiently transfected with DCC or FLAG-Tmtsp were incubated with recombinant chick netrin-1 (VI · V)–Fc protein in culture medium with 2 μg/mL heparin. After fixation, specific binding of netrin-1 (VI · V)–Fc was detected by means of double staining with the following Abs: for detection of transfected gene products, anti-FLAG mAb (M2), anti-DCC mAb (Calbiochem-Oncogene Science, Boston, MA), and Alexa-488–conjugated anti–mouse IgG (H+L) (Molecular Probe, Eugene, OR); and for detection of netrin-1-Fc protein, Cy3-conjugated anti–human IgG (H+L) (Jackson ImmunoResearch Lab, West Grove, PA). Fluorescent mounting medium was purchased from Dako (Glostrup, Denmark). Images were acquired using an Olympus BX-51 microscope system and a 40 × 0.65 NA objective (Olympus, Tokyo, Japan), and processed using Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA).
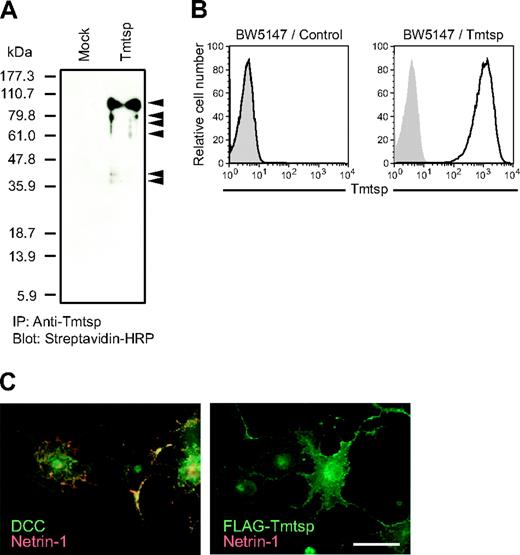
Characterization of Tmtsp protein and specific antibody. (A-B) Specificity of anti–mouse Tmtsp mAb. To verify the specificity of the anti-Tmtsp mAb, cell surface proteins of BW5147 cells infected with a Tmtsp retrovirus (BW5147/Tmtsp) and mock control cells (BW5147/control) were biotinylated, and the total cell lysates were used for immunoprecipitation with anti-Tmtsp mAb, followed by detection with streptavidin-HRP. Tmtsp protein and the putative proteolytic products are indicated by arrowheads (A). The specificity of the mAb was also confirmed by flow cytometric analysis (B). Open curve indicates anti-Tmtsp; gray shaded curve, isotype control. (C) Binding assay with recombinant netrin-1 protein. COS7 cells transiently transfected with DCC and FLAG-Tmtsp were incubated with chick netrin-1 (VI · V)–Fc. Receptor expression (green) and netrin-1-Fc (red) were merged. Scale bar: 50 μm.
Characterization of Tmtsp protein and specific antibody. (A-B) Specificity of anti–mouse Tmtsp mAb. To verify the specificity of the anti-Tmtsp mAb, cell surface proteins of BW5147 cells infected with a Tmtsp retrovirus (BW5147/Tmtsp) and mock control cells (BW5147/control) were biotinylated, and the total cell lysates were used for immunoprecipitation with anti-Tmtsp mAb, followed by detection with streptavidin-HRP. Tmtsp protein and the putative proteolytic products are indicated by arrowheads (A). The specificity of the mAb was also confirmed by flow cytometric analysis (B). Open curve indicates anti-Tmtsp; gray shaded curve, isotype control. (C) Binding assay with recombinant netrin-1 protein. COS7 cells transiently transfected with DCC and FLAG-Tmtsp were incubated with chick netrin-1 (VI · V)–Fc. Receptor expression (green) and netrin-1-Fc (red) were merged. Scale bar: 50 μm.
Production of anti-Tmtsp monoclonal antibody
The specific mAb against extracellular region of mouse Tmtsp was generated using Tmtsp-Fc fusion protein as immunogen as previously described18 and biotinylated by using EZ-Link Sulfo-NHS-LC-LC-Biotin (PIERCE).
Reverse-transcription (RT)–PCR and Northern blotting
Hematopoietic lineage cDNA panels were prepared as previously reported.19 Briefly, mononuclear cells prepared from bone marrow, spleen, and thymus were sorted into Trizol-LS (Invitrogen). Total RNA samples were reverse transcribed with ThermoScript RT-PCR System and oligo-dT primer (Invitrogen). Resultant cDNA samples were normalized along with expression of Gapdh determined by using TaqMan Rodent GAPDH Control Reagents and ABI-7900 systems (Applied Biosystem, Foster City, CA). Normalized cDNA samples were applied for conventional semiquantitative RT-PCR (cycling parameters: 38 cycles of 3-step PCR composed of 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 20 seconds) and gene-specific quantitative PCR (Q-PCR) using SYBR Green PCR master mix (Applied Biosystem) according to the manufacturer's instruction (cycling parameters: 45 cycles of 4-step PCR composed of 95°C for 15 seconds, 60°C for 15 seconds, 72°C for 20 seconds, and 83°C for 7 seconds). Relative copy number of Tmtsp transcripts is equal to the measured copy number of Tmtsp × 10 000/the copy number of GAPDH. PCR primers used are described in the supplemental methods (see the Supplemental Materials link at the top of the online article, at the Blood website). Northern blotting was performed as previously described.20 [α-32P] dCTP–labeled cDNA probe corresponding to TSP1 and transmembrane domains of mouse Tmtsp was prepared by PCR, and signals were detected by using BAS3000 system (Fuji Photo Film, Tokyo, Japan).
Immunostaining
Organs from 10-week-old male C57BL/6 mice and from embryos (E15.5) were frozen in OCT compound (Sakura Fine Technical, Tokyo, Japan) and sliced (4 μm) by Cryostat (Leica, Heidelberg, Germany). Specimens were fixed with 4% paraformaldehyde (PFA)/PBS (10 minutes, room temperature) and incubated with 10% normal serum derived from the same species as the secondary antibodies. Endogenous peroxidase activity was blocked by hydrogen peroxide/methanol treatment. To detect Tmtsp protein, specimens were incubated with anti-Tmtsp mAb (4°C for 12-16 hours) and subsequently stained with Max-PO system (HRP method; Nichirei, Tokyo, Japan) or with Cy3- or FITC-conjugated anti–rat IgG antibody (Jackson ImmunoResearch Lab) or with Alexa Fluor 647 goat anti–rat IgG antibody (Molecular Probe). Sections detected by chemiluminescence were restained with hematoxylin and eosin to visualize the structure of tissues. Antibodies against marker molecules were as follows: for primary antibodies, VE-cadherin (rat IgG; Pharmingen), VE-cadherin (rabbit IgG; Cayman Chemical, Ann Arbor, MI), CD31/PECAM-1 (BD Biosciences, San Diego, CA), and LYVE-121 ; and for secondary antibodies, Cy3-conjugated anti–rat IgG and anti–rabbit IgG (Jackson ImmunoResearch Lab) and Alexa Fluor 488 goat anti–rabbit IgG antibody (Molecular Probe). Nuclei were visualized with a standard procedure using 4′-6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Images of tissue sections detected by chemiluminescence and endothelial cells derived from TmtspVenus/+ ES cells were acquired as described in “In vitro binding assay.” Images of tissue sections detected by fluorescence and endothelial cells derived from wild-type ES cells were acquired using a Leica TCS SP2 AOBS confocal microscope and a 63 × 1.32 NA oil objective, and were processed using Adobe Photoshop 6.0. Images of whole-mount staining were acquired using a Leica FW4000 microscope and a 20 × 0.40 NA objective.
Preferential expression of Tmtsp protein on hematopoietic stem and progenitor cells. (A) Expression of Tmtsp in c-Kit+Sca-1+Lin– (KSL) fraction in the adult mouse BM. Tmtsp protein was highly expressed on both CD34–KSL HSCs and CD34+KSL multipotential progenitor cell fractions. The number in the diagram indicates the percentage of cells in the fraction. Open and shaded curves represent cells stained with the anti-Tmtsp antibody and an isotype control, respectively. (B) Expression of Tmtsp protein on lineage marker–positive hematopoietic cells. Tmtsp was not detected on lineage marker–positive cells except for CD4–CD8– (DN) cells in the thymus.
Preferential expression of Tmtsp protein on hematopoietic stem and progenitor cells. (A) Expression of Tmtsp in c-Kit+Sca-1+Lin– (KSL) fraction in the adult mouse BM. Tmtsp protein was highly expressed on both CD34–KSL HSCs and CD34+KSL multipotential progenitor cell fractions. The number in the diagram indicates the percentage of cells in the fraction. Open and shaded curves represent cells stained with the anti-Tmtsp antibody and an isotype control, respectively. (B) Expression of Tmtsp protein on lineage marker–positive hematopoietic cells. Tmtsp was not detected on lineage marker–positive cells except for CD4–CD8– (DN) cells in the thymus.
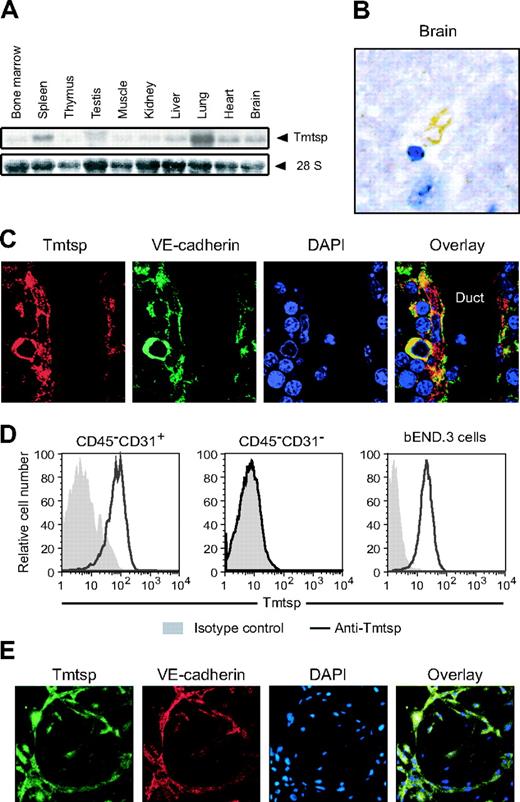
Localization of Tmtsp protein on endothelial cells. (A) Tissue distribution of Tmtsp mRNA by Northern blotting analysis. (B) Immunohistochemistry of adult mouse brain with anti-Tmtsp mAb. Tmtsp-specific signals were detected on the blood vessel–like structure in the forebrain. (C) Immunohistochemistry of the fetal liver (E15.5) with anti-Tmtsp mAb. Tmtsp protein (red) was detected in VE-cadherin–positive cells (green) with spindle nuclei located along luminal duct filled with enucleated erythrocytes and DAPI-positive blood cells. Nuclei were visualized by DAPI staining (blue). (D) Flow cytometric analysis of endothelial cells. Tmtsp protein was detected on CD45–CD31+ endothelial cells, but not on CD45–CD31– nonendothelial cells prepared from adult mouse liver and bEND.3 cells, a mouse brain–derived endothelioma cell line. (E) Double staining of cultured endothelial cells differentiated from ES cells on OP-9 stromal cells. Tmtsp protein (green) was localized on the surface of VE-cadherin–positive endothelial cells (red). Nuclei were visualized with DAPI staining (blue).
Localization of Tmtsp protein on endothelial cells. (A) Tissue distribution of Tmtsp mRNA by Northern blotting analysis. (B) Immunohistochemistry of adult mouse brain with anti-Tmtsp mAb. Tmtsp-specific signals were detected on the blood vessel–like structure in the forebrain. (C) Immunohistochemistry of the fetal liver (E15.5) with anti-Tmtsp mAb. Tmtsp protein (red) was detected in VE-cadherin–positive cells (green) with spindle nuclei located along luminal duct filled with enucleated erythrocytes and DAPI-positive blood cells. Nuclei were visualized by DAPI staining (blue). (D) Flow cytometric analysis of endothelial cells. Tmtsp protein was detected on CD45–CD31+ endothelial cells, but not on CD45–CD31– nonendothelial cells prepared from adult mouse liver and bEND.3 cells, a mouse brain–derived endothelioma cell line. (E) Double staining of cultured endothelial cells differentiated from ES cells on OP-9 stromal cells. Tmtsp protein (green) was localized on the surface of VE-cadherin–positive endothelial cells (red). Nuclei were visualized with DAPI staining (blue).
Fluorescence-activated cell sorting (FACS)
For analysis of hematopoietic cells, adult mouse tissues were homogenized and passed through the filter (pore size: 70 μm), and mononuclear cells were isolated by Lymphoprep (1.086 g/mL; Nycomed, Oslo, Norway). Cell lines, EBs, and embryos were trypsinized and passed through the filter. Prepared cells were stained with biotin-conjugated anti-Tmtsp mAb along with marker antibodies in the staining medium (5% FCS/PBS) followed by the secondary reaction with phycoerythrin (PE)–conjugated streptavidin (BD Biosciences) and enhancement of PE signal by MACS FASER-PE Kit (MACS; Miltenyi Biotech, Bergisch Gladbach, Germany). Biotinylated rat IgG2a (BD Biosciences) was used as an isotype control. Cells were analyzed on a FACSVantageSE and FlowJo software (Tree Star, Ashland, OR). Cell sorting was performed on a MoFlo (DakoCytomation, Glostrup, Denmark). Combinations of antibodies are listed in the supplemental methods.
Mice
For gene targeting, about 11 kb genomic DNA containing the translation start codon of the Tmtsp gene was isolated from 129Sv genomic library. Twelve base pairs of sequence containing the first and second ATG were replaced by the Venus and neomycin phosphotransferase cassette. The targeting construct was introduced into E14.1 embryonic stem (ES) cells by electroporation and homologous recombinants were screened by Southern blotting. Images were acquired as described in “Reverse transcription (RT)–PCR and Northern blotting.” Mice were bred and maintained at the Animal Research Center of the Institute of Medical Science, University of Tokyo (IMSUT). Animals were used under the guidelines of IMSUT.
Colony assay and competitive repopulation assay
In order to characterize Lin– cells fractionated by Venus, methylcellulose colony assay and competitive repopulation assay were performed as previously described.22 After 10 days of culture, large (diameter ≥ 1.0 mm) and small (diameter < 1.0 mm) colonies were counted to determine the numbers of high-proliferative-potential colony-forming cells (HPP-CFCs) and low-proliferative-potential colony-forming cells (LPP-CFCs), respectively.
Results
Molecular cloning and characterization of Tmtsp
In an attempt to identify novel type I cytokine receptors expressed on HSCs, we have previously performed a homology search of the EST database using the amino acid sequences spanning from the extracellular WSXWS motif to the transmembrane region of several known type I cytokine receptors as baits.23 RT-PCR analyses of identified genes were performed as the second screening. One of the identified genes showed restricted expression in undifferentiated hematopoietic cells (CD34–c-Kit+Sca-1+Lin– to Lin–) (Figure 1A). Therefore, we focused on this gene and cloned mouse and human full-length cDNAs. We designated this gene as transmembrane molecule with thrombospondin module (Tmtsp) from its putative protein structure. The obtained full-length cDNAs of the mouse and human Tmtsp were 2815-bp and 2953-bp long, and encoded polypeptides of 851 and 852 amino acids, respectively (Figure 1B). Mouse and human Tmtsp are 78.6% identical at the amino acid level. Analysis of the mouse Tmtsp peptide sequence with the SMART program (http://smart.embl-heidelberg.de) demonstrated a signal sequence, a transmembrane region, and thrombospondin type 1 repeat (TSP1) domain. PROSITE sequence analysis program (http://au.expasy.org/prosite) also displayed 6 potential N-linked glycosylation sites in the extracellular region, 9 PKC-phosphorylation sites, and a tyrosine-phosphorylation site near by its C-terminus. Although Tmtsp contained a WXXW motif in the extracellular domain, it appeared to be distinct from the WSXWS motif of type I cytokine receptors but represented a part of TSP1 domain. Multiple alignments of mouse Tmtsp protein with Unc5h3, ICAM-1, and thrombospondin-1 demonstrated that Tmtsp contains 3 Ig-like domains and consensus sequences of the TSP1 domain (ie, WXXW and CSVTCG24,25 ) in the extracellular region (Figure S1A). The domain composition of the Tmtsp extracellular region is highly homologous to that of the Unc5h proteins, whereas a domain search failed to identify any functional motifs or domains in the Tmtsp intracellular region (Figure 1C).
Gene targeting and visualization of transcription at the Tmtsp locus. (A) Schematic illustration of gene targeting construct. DNA sequence corresponding to 2 translational start codons was replaced by Venus-Neo cassette. Components of the cassette were as follows: splicing donor, D; splicing acceptor, A; enhanced mutant of yellow fluorescent protein, Venus; simian virus 40 (SV40)–derived polyadenylation signal, pA; and phosphoglycerate kinase promoter-neomycin phosphotransferase, PGK-Neo. Numbered black boxes indicate exons. Recognition site of EcoNI (E) and HindIII (H) are shown by triangles. (B) Southern blot analysis of mutant mouse. Genomic DNA digested with EcoNI and HindIII were hybridized with 5′ and 3′ probes, respectively (shown in A). (C) Image of EBs (day 4) from knock-in ES cells. (D) Correlation of Venus signal and Tmtsp protein. EBs (day 7) from knock-in ES cells were stained with anti-Tmtsp antibody and analyzed by flow cytometry. Tmtsp protein was expressed only on Venus+ (Venuslow to Venushigh) cells. (E) Immunostaining of endothelial cells on OP-9 stromal cells. After 4 days of EB formation, cells were replated on OP-9 cells and cultured for 3 days in the presence of VEGF (stained with anti-Tmtsp, VE-cadherin, and CD31/PECAM-1) or in the presence of VEGF-D (for lymphatic endothelium formation, stained with anti–LYVE-1 antibody). (F) Whole-mount immunostaining of E10.5 knock-in embryo. (i) Overview of E10.5 embryo. (ii) Venus signal (green) was highly detected in vascular network of the embryo body. (iii) Overlaid image of Venus signal and immunostaining with CD31/PECAM-1 showed expression of Venus in the capillary network beneath the body surface. Scale bar: 50 μm.
Gene targeting and visualization of transcription at the Tmtsp locus. (A) Schematic illustration of gene targeting construct. DNA sequence corresponding to 2 translational start codons was replaced by Venus-Neo cassette. Components of the cassette were as follows: splicing donor, D; splicing acceptor, A; enhanced mutant of yellow fluorescent protein, Venus; simian virus 40 (SV40)–derived polyadenylation signal, pA; and phosphoglycerate kinase promoter-neomycin phosphotransferase, PGK-Neo. Numbered black boxes indicate exons. Recognition site of EcoNI (E) and HindIII (H) are shown by triangles. (B) Southern blot analysis of mutant mouse. Genomic DNA digested with EcoNI and HindIII were hybridized with 5′ and 3′ probes, respectively (shown in A). (C) Image of EBs (day 4) from knock-in ES cells. (D) Correlation of Venus signal and Tmtsp protein. EBs (day 7) from knock-in ES cells were stained with anti-Tmtsp antibody and analyzed by flow cytometry. Tmtsp protein was expressed only on Venus+ (Venuslow to Venushigh) cells. (E) Immunostaining of endothelial cells on OP-9 stromal cells. After 4 days of EB formation, cells were replated on OP-9 cells and cultured for 3 days in the presence of VEGF (stained with anti-Tmtsp, VE-cadherin, and CD31/PECAM-1) or in the presence of VEGF-D (for lymphatic endothelium formation, stained with anti–LYVE-1 antibody). (F) Whole-mount immunostaining of E10.5 knock-in embryo. (i) Overview of E10.5 embryo. (ii) Venus signal (green) was highly detected in vascular network of the embryo body. (iii) Overlaid image of Venus signal and immunostaining with CD31/PECAM-1 showed expression of Venus in the capillary network beneath the body surface. Scale bar: 50 μm.
A BLAST search against the mouse genome database displayed that the mouse Tmtsp is composed of 5 exons and spans over 37 kbp on chromosome 8qA2 (Figure S1B-C). An alternative splicing, which splices out the fourth exon (159 bp), was detected in all major organs tested (Figure S1D), leading to the in-frame translation of a variant protein without a TSP1 domain (Figure S1B). The human orthologue of the splicing variant is also deposited in the EST database (GenBank accession number: BC063842).
Characterization of mouse Tmtsp protein
By using the fusion protein of the Tmtsp extracellular region and human IgG1 Fc portion as an immunogen, anti–mouse Tmtsp mAbs were generated. To verify the specificity of the Abs, cell surface proteins of BW5147 cells expressing mouse Tmtsp FLAG-tagged at the N-terminus were biotinylated and the total cell lysates were used for immunoprecipitation with anti-Tmtsp mAb, followed by detection with streptavidin-HRP (Figure 2A). Specific bands were detected only in BW5147 cells expressing Tmtsp. In addition to the main band at the size of 95 kDa, several smaller bands were detected, indicative of various forms of proteolytic processing. The specificity of the mAb was also confirmed by the flow cytometric analysis (Figure 2B).
In view of structural similarity with Unc5h proteins, the binding of Tmtsp with netrin-1, the ligand for Unc5h proteins, was assessed. DCC, another receptor for netrin-1, was used as a positive control. Netrin-1 bound to the surface of COS7 cells expressing DCC, but not Tmtsp, indicating that netrin-1 is not the ligand for Tmtsp (Figure 2C).
Tmtsp is highly expressed on hematopoietic stem and progenitor cells
Quantitative RT-PCR analysis confirmed the highest expression of Tmtsp in CD34–KSL HSCs (relative copy number: 210 copy), and relatively high expression in CD34+KSL (173 copy) and Lin– (46.4 copy) progenitor cells. Additionally, the expression of Tmtsp was detectable in thymic CD4–CD8– (double-negative [DN]) cells and gradually decreased during T-cell differentiation (relative copy numbers: DN cells, 10.1 copy; DP cells, 5.74 copy; CD4SP cells, 1.82 copy; CD8SP cells, 0.72 copy). Consistent with RT-PCR data, FACS analysis demonstrated that Tmtsp protein is highly expressed on CD34–KSL HSCs and on CD34+KSL progenitor cells in the BM (Figure 3A), but not the lineage marker–positive cells, except for DN cells in the thymus (Figure 3B).
Localization of Tmtsp protein on endothelial cells
Distribution of Tmtsp mRNA in major organs was next investigated by Northern blotting analysis (Figure 4A). Signals were detected in all organs tested and the highest expression was observed in the lung, where numerous blood vessels exist. Immunohistochemical analysis using anti-Tmtsp mAb revealed that its expression is tightly restricted to endothelial cells in all the tissues analyzed. For example, Tmtsp-specific signals were detected only at the blood vessel like–structure in the forebrain (Figure 4B). Double staining of E15.5 fetal liver sections demonstrated that Tmtsp protein is expressed on VE-cadherin–positive endothelial cells26 with spindle nuclei surrounding the luminal duct filled with enucleated erythrocytes and some DAPI-positive blood cells (Figure 4C). Of interest, Tmtsp protein was also detected on some hematopoietic cell–like round cells outside the ducts and was exactly colocalized with VE-cadherin. These cells supposedly represent recently described VE-cadherin+ fetal liver HSCs.27 Flow cytometric analysis also demonstrated Tmtsp expression on bEND.3 cells, a mouse brain–derived endothelioma cell line, and on CD45–CD31+ endothelial cells, but not on CD45–CD31– nonendothelial cells prepared from adult mouse liver (Figure 4D). To further characterize the expression of Tmtsp protein on endothelial cells, we next used the differentiation model of mouse ES cells. ES cells were first allowed to differentiate by formation of EBs, aggregates containing 3 germ layers. After 4 days of differentiation, dissociated cells were cultured on a layer of OP-9 stromal cells in the presence of VEGF for 4 days to generate an endothelial network. Immunostaining demonstrated that VE-cadherin+ endothelial cells express Tmtsp protein (Figure 4E). All of these data strongly indicate that Tmtsp expression is specific to endothelial cells in addition to hematopoietic cells.
Gene targeting and visualization of transcription at the Tmtsp locus
For visualization of gene expression profiles at the Tmtsp locus, we designed a targeting vector to replace 2 putative translation start codons of Tmtsp gene by Venus, a variant of yellow fluorescence protein,28 and neomycin-resistant gene (Figure 5A). Germ-line transmission of the targeted homologous recombinant was verified by Southern blotting (Figure 5B). To confirm the correlation between Venus expression from mutated allele and authentic gene expression from wild-type allele, targeted ES cells were differentiated by EB formation followed by coculture with OP-9 cells, and then analyzed by flow cytometry and immunostaining. During EB formation, the Venus signal became detectable from day 4 under fluorescence microscopy (Figure 5C). Flow cytometric analysis of EBs (day 7) with the anti-Tmtsp mAb demonstrated a good correlation of Venus expression and authentic Tmtsp expression (Figure 5D). In immunostaining analyses of endothelial cells (7 days of differentiation) on OP-9 cells, Venus+ cells coexpressed Tmtsp protein, as well as CD31/PECAM-1 (pan-endothelial cell marker). Detailed analyses also demonstrated that Venus expression is detectable both in VE-cadherin+ vascular endothelial cells and LYVE-1+ lymphatic endothelial cells21 (Figure 5E). In addition, Venus was expressed in the CD31+ vascular network of the E10.5 embryo body (Figure 5F). These data indicate that Venus fluorescence faithfully reflects Tmtsp gene expression and marks endothelial cells in the embryo.
Expression of Tmtsp/Venus during embryogenesis
To investigate the expression of Tmtsp at the early embryonic stage, EBs derived from knock-in ES cells (TmtspVenus/+) were used as a developmental model. Venus was detectable within the Flk-1–positive mesodermal precursor population at day 3 by flow cytometry. CD41, an early marker molecule for primitive and definitive hematopoiesis protein,29-31 was clearly expressed from day 4 to day 6, and a part of CD41+ cells expressed Venus (Figure 6A). These findings allow us to hypothesize that Tmtsp is expressed in embryonic hematopoietic progenitors. To verify this idea, we examined whether colony-forming cells on OP-9 stromal cells are enriched in the Venus-positive population in TmtspVenus/+ embryos. Expression of Venus in the yolk sac (YS) was scarcely detected before embryonic day 8.5 (E8.5). After E8.5, Venus+ cells rapidly increased in number (Figure 6B-C). Cells from YS were fractionated by the expression of CD41 and Venus, and then cultured on OP-9 cells to determine the frequencies of hematopoietic progenitors. Of note was that among 4 fractions, hematopoietic colony-forming cells in E8.5 and E9.5 YSs were highly enriched in the CD41+Venus+ fraction (Figure 6B-C). Similarly, cells from the E10.5 AGM region, where HSCs with adult hematopoietic reconstitution capacity first appear,32 were fractionated and cultured on OP-9 cells. Further fractionation of the CD41+Venus+ fraction by the levels of Venus and CD41 expression promoted enrichment, and, strikingly, 31.5% of CD41lowVenushigh cells retained a colony-forming capacity (Figure 6B). Even in the ES cell differentiation system, EB (day 6)–derived hematopoietic colony-forming cells were also enriched in the same CD41+Venus+ population (data not shown).
Expression of Tmtsp marks hematopoietic progenitors in the developing embryo. (A) Expression kinetics of Venus, Flk-1, and CD41 in EBs (days 3-6). (B-D) Hematopoietic colony-forming cells were highly enriched in CD41lowVenushigh fraction. Yolk sac cells from E8.5 (B) and E9.5 (C) TmtspVenus/+ embryos and AGM cells from E10.5 TmtspVenus/+ embryo (D) were divided into 4 or 6 populations according to the expression levels of CD41 and Venus. Sorted cells were cultured on OP-9 stromal cells in the presence of SCF, TPO, IL-3, and EPO for 3 days. Sorting gates (left) and number of hematopoietic colony-forming cells per 1000 plating cells (right) are indicated. The results are shown as the mean value ± SD of triplicate cultures.
Expression of Tmtsp marks hematopoietic progenitors in the developing embryo. (A) Expression kinetics of Venus, Flk-1, and CD41 in EBs (days 3-6). (B-D) Hematopoietic colony-forming cells were highly enriched in CD41lowVenushigh fraction. Yolk sac cells from E8.5 (B) and E9.5 (C) TmtspVenus/+ embryos and AGM cells from E10.5 TmtspVenus/+ embryo (D) were divided into 4 or 6 populations according to the expression levels of CD41 and Venus. Sorted cells were cultured on OP-9 stromal cells in the presence of SCF, TPO, IL-3, and EPO for 3 days. Sorting gates (left) and number of hematopoietic colony-forming cells per 1000 plating cells (right) are indicated. The results are shown as the mean value ± SD of triplicate cultures.
Expression level of Tmtsp/Venus correlates hematopoietic activity
Tmtsp expression in adult BM hematopoietic stem and progenitor cells was investigated using Venus knock-in (TmtspVenus/+) mice. Consistent with the Tmtsp expression presented in Figure 4A, Venus was highly expressed in both CD34–KSL HSCs and CD34+KSL multipotential progenitor cells (Figure 7A). Tmtsp expression was next determined in the committed myeloid progenitor cells: common myeloid progenitors (CMPs), granulocyte/macrophage progenitors (GMPs), and megakaryocyte/erythrocyte lineage-restricted progenitors (MEPs).33 Venus expression gradually declined along with myeloid differentiation from CMPs to GMPs and MEPs (Figure 7B). In contrast, common lymphoid progenitors (CLPs), the committed lymphoid progenitor cells in the BM,34 displayed comparable Venus expression with CD34–KSL HSCs and CD34+KSL cells (Figure 7B). Venus expression was again gradually decreased along with the maturation of B- and T-cell lineages in the BM and thymus, respectively (Figure S2). Thus, the intensity of Venus is an indicator of differentiation in both myeloid and lymphoid lineages. To functionally confirm this idea, Lin– cells were fractionated by the level of Venus, and the content of colony-forming units and the hematopoietic reconstitution capacity in lethally irradiated mice were determined for each fraction. HPP-CFCs, which represent primitive progenitors, were highly enriched in the Lin–Venushigh fraction. About 20% of HPP-CFCs were multipotential in terms of myeloid differentiation and formed so-called mixed colonies (data not shown). LPP-CFCs, which represent committed progenitors, were also enriched in the Venushigh fraction. In contrast, the Lin–Venuslow fraction preferentially gave rise to small colonies, and the Lin–Venus– fraction generated no colony (Figure 7C and Table 1). In the competitive repopulation assay, only Lin–Venushigh cells exhibited the ability to repopulate hematopoiesis in lethally irradiated mice after transplantation (Table 2). These data functionally demonstrated that the expression of the Tmtsp gene marks both long-term and short-term repopulating HSCs, and its expression level correlates with the proliferative and differentiation potential both in vitro and in vivo.
Methylcellulose colony assay of fractionated Lin– cells from adult TmtspVenus/+ mice.
. | No. cells plated . | Colony number/dish, ± SD . | . | |
|---|---|---|---|---|
| Cell type . | . | Large . | Small . | |
| Total Lin- | 500 | 3.33 ± 1.15 | 20.3 ± 6.03 | |
| Venus- | 400 | ND | ND | |
| Venuslow | 1600 | 4.67 ± 0.58 | 42.7 ± 4.16 | |
| Venushigh | 200 | 16.7 ± 1.15 | 15.0 ± 3.61 | |
. | No. cells plated . | Colony number/dish, ± SD . | . | |
|---|---|---|---|---|
| Cell type . | . | Large . | Small . | |
| Total Lin- | 500 | 3.33 ± 1.15 | 20.3 ± 6.03 | |
| Venus- | 400 | ND | ND | |
| Venuslow | 1600 | 4.67 ± 0.58 | 42.7 ± 4.16 | |
| Venushigh | 200 | 16.7 ± 1.15 | 15.0 ± 3.61 | |
Competitive hematopoietic repopulation capacity of Lin–Venushigh cells
. | . | No. positive mice/total mice . | . | Chimerism, % . | . | ||
|---|---|---|---|---|---|---|---|
| Cell population . | No. test cells/mouse . | 4 wk . | 12 wk . | 4 wk . | 12 wk . | ||
| Lin-Venushigh | 1 674 | 7/7 | 6/7 | 7.30 ± 1.22 | 17.8 ± 19.6 | ||
| Lin-Venuslow | 17 555 | 0/8 | 0/7 | — | — | ||
| Lin-Venus- | 2 712 | 0/4 | 0/4 | — | — | ||
. | . | No. positive mice/total mice . | . | Chimerism, % . | . | ||
|---|---|---|---|---|---|---|---|
| Cell population . | No. test cells/mouse . | 4 wk . | 12 wk . | 4 wk . | 12 wk . | ||
| Lin-Venushigh | 1 674 | 7/7 | 6/7 | 7.30 ± 1.22 | 17.8 ± 19.6 | ||
| Lin-Venuslow | 17 555 | 0/8 | 0/7 | — | — | ||
| Lin-Venus- | 2 712 | 0/4 | 0/4 | — | — | ||
Lin- cells fractionated by the level of Venus expression (Ly5.1+C57BL/6; test cells) were transplanted into lethally irradiated Ly5.2 C57BL/6 mice with 2 × 105 total BM cells (Ly5.2+ C57BL/6; competitor cells). Peripheral blood cells of recipient mice were analyzed 4 and 12 weeks after transplantation. Percent chimerism (± SD) indicates the percentage of test cells (donor; Ly5.1+) per total Ly5+ (Ly5.1+ + Ly5.2+) cells.
— indicates not engrafted.
Expression level of Tmtsp/Venus correlates to hematopoietic activity. (A) Expression of Venus in hematopoietic stem and progenitor cells. (B) Expression of Venus in hematopoietic progenitors. Bone marrow–derived IL-7R–Lin– Sca-1–c-Kit+ cells were subdivided into FcγRlowCD34+ (CMP), FcγRhighCD34+ (GMP), and FcγRlowCD34– (MEP) fractions (top left), and the expression of Venus is shown in the histograms (top right). Venus signal in IL-7R+Lin–c-KitlowSca-1low cells (CLP) (bottom panel). (C) Colony-forming cells were highly enriched in the Lin–Venushigh fraction. Numbers of HPP-CFCs and LPP-CFCs are presented as the mean value ± SD of triplicate cultures. Numbers of HPP- and LPP-CFCs in the Lin–Venushigh fraction were significantly different from the Lin–Venuslow fraction (Student t test, ***P < .001, *P < .05).
Expression level of Tmtsp/Venus correlates to hematopoietic activity. (A) Expression of Venus in hematopoietic stem and progenitor cells. (B) Expression of Venus in hematopoietic progenitors. Bone marrow–derived IL-7R–Lin– Sca-1–c-Kit+ cells were subdivided into FcγRlowCD34+ (CMP), FcγRhighCD34+ (GMP), and FcγRlowCD34– (MEP) fractions (top left), and the expression of Venus is shown in the histograms (top right). Venus signal in IL-7R+Lin–c-KitlowSca-1low cells (CLP) (bottom panel). (C) Colony-forming cells were highly enriched in the Lin–Venushigh fraction. Numbers of HPP-CFCs and LPP-CFCs are presented as the mean value ± SD of triplicate cultures. Numbers of HPP- and LPP-CFCs in the Lin–Venushigh fraction were significantly different from the Lin–Venuslow fraction (Student t test, ***P < .001, *P < .05).
Discussion
In this study, we successfully identified a novel gene encoding transmembrane protein with TSP1 domain expressed on hematopoietic stem/progenitor cells as well as endothelial cells. A wide variety of molecules, including Tie-2 (receptor tyrosine kinase),5 Endoglin (TGF-β receptor),11-13 Scl/Tal-1 (a helix-loop-helix transcription factor),35 and Gata-2 (transcription factor with zinc finger motif),36,37 is commonly expressed in immature hematopoietic and endothelial cells and considered to be important for the development and functions of these cell lineages. Among a list of molecules shared by HSCs and endothelial cells, Tmtsp is the first one that possesses the TSP1 domain, which is implicated in cell adhesion and migration.
Tmtsp mRNA was expressed preferentially in hematopoietic stem and progenitor cells (Figure 1A). Corresponding to this observation, Tmtsp transcript has been included on the list of HSC-enriched genes.38 Using monoclonal antibodies against Tmtsp, we confirmed that the expression profile of the Tmtsp protein correlates well to its mRNA expression in the lymphohematopoietic system (Figure 3), and that its expression is restricted to hematopoietic and endothelial cells (Figures 3 and 4, respectively). The specific expression of Tmtsp prompted us to visualize its expression with fluorescent protein by introducing Venus cDNA into the Tmtsp gene locus. Genetic marking of Tmtsp was successful and Venus expression faithfully recapitulated the authentic Tmtsp expression in ES cells, as well as in mice (Figures 5, 6, 7). Detailed analyses of TmtspVenus/+ mice clearly demonstrated that the level of Venus correlates well to the differentiation stage of hematopoietic cells. Starting from HSCs, Venus expression gradually declined as differentiation proceeded, and the cells with the highest Venus expression were enriched for primitive hematopoietic cells with a higher hematopoietic activity (Figure 7). Venus expression also traced the lymphoid differentiation, starting from the highest expression in CLPs down to the T- and B-cell progenies. Of interest, among the DN cells in thymus, DN2 exhibited a high level of Venus expression, indicating the functional involvement of Tmtsp in T-cell differentiation at this specified stage.
Two waves of hematopoiesis occur in the mouse embryo. The first transient wave of primitive hematopoiesis is detected in the YS as early as E7.0. At E8.25, just prior to the onset of circulation, definitive hematopoiesis takes place in the YS. Over the subsequent 24 hours, definitive hematopoietic progenitors increase in numbers within the yolk sac and also enter the newly formed bloodstream.39 On the other hand, definitive hematopoiesis also arises in the E10.5 intraembryonic AGM.40-42 Until now, only few molecules have been reported as markers for early hematopoietic progenitors during development. Although CD41 and c-Kit are efficient and practical markers for developing hematopoietic stem and progenitor cells in the YS and AGM,29,30,41 the enrichment of definitive hematopoietic progenitors is hampered by the limited number of available markers. Notably, expression of the Venus reporter coincided with the emergence of definitive hematopoietic progenitors in the YS, and Venus expression in combination with CD41 promoted the enrichment of developing hematopoietic progenitors in both YS and AGM (Figure 6B-D). Strikingly, in E10.5 AGM region, the CD41+Venushigh fraction exhibited the highest seeding efficiency, at 31.5% (Figure 6D). Among CD41+ cells, the level of Venus expression correlated well to the hematopoietic activity, as observed in the adult BM. In contrast, the CD41–Venus+ fraction that gave rise to no hematopoietic colonies included endothelial cells (data not shown). These findings define Tmtsp to be a novel marker for developing definitive hematopoietic stem and progenitor cells in the embryo. As demonstrated in Figure 4C, Tmtsp protein is expressed on a subset of VE-cadherin+ fetal liver hematopoietic cells, which supposedly represent recently described VE-cadherin+ fetal liver HSCs.27 Thus, it would be intriguing to analyze fetal liver HSCs and their niche by using Tmtsp/Venus as markers. Several mouse models have been generated for the analysis of HSC ontogeny. These include the LacZ knock-in mouse in the Scl locus,43 LacZ knock-in mouse in the Runx1 locus,44 and Ly-6A (Sca-1) promoter-driven GFP transgenic mouse.40 These mouse models facilitated the temporal and anatomic identification of HSC ontogeny. Taken together with the specific expression of Tmtsp in endothelial cells, TmtspVenus/+ mice would serve as a useful mouse model for the analysis of embryonic and adult hematopoiesis, and also for the analysis of vasculogenesis.
In the human genome, 41 genes encode the TSP1 domain.45 Some of them are grouped under subfamilies, such as thrombospondins, ADAMTSs, and Unc5hs.24 Tmtsp does not belong to those subfamilies, but its protein motif composition in the extracellular region is close to that of Unc5h proteins. Proteins with a TSP1 domain have a variety of functions, including interaction with the ECM, axonal guidance, cell adhesion, and the control of migration. The Unc5h family is known to be related to the formation of axon guidance by its interaction with netrin-1, the laminin-related secreted protein.46 Recent reports have also shown that Unc5B controls the morphogenesis of the vascular system as a receptor of netrin-1.47 Moreover, the secretory protein, thrombospondin-1, binds to CD36 and controls vasculogenesis and angiogenesis.48 In this context, Tmtsp might be also implicated in the regulation of vasculogenesis and/or angiogenesis through interaction with its specific ligand. Identification of the specific ligand is one of the key steps to clarify the function of the Tmtsp protein.
Expression profiles also indicate that Tmtsp functions in primitive hematopoietic cells in the BM niche. Osteoblasts, one of the components of the HSC niche, express several adhesion molecules, including N-cadherin7 and integrins. For instance, ICAM-1 on osteoblasts interacts with LFA-1 on HSCs and mediates signals essential for the maintenance of stemness.8 Tmtsp possesses the TSP1 domain, which has been implicated in cell migration, and 3 Ig-like domains. Likewise, through these domains, Tmtsp might play a role in HSC homing to the niche and following cell-to-cell contact to maintain HSC quiescence.
To elucidate the biologic role of Tmtsp, analysis of the Tmtsp-deficient mouse is crucial and is now under investigation. Characterization of Tmtsp function and identification of its ligand would give new insights into hematopoiesis and vascular biology.
Prepublished online as Blood First Edition Paper, February 2, 2006; DOI 10.1182/blood-2005-09-3747.
Supported in part by grants from the Ministry of Education, Culture, Sport, Science, and Technology, Japan; the Naito Foundation; and the Terumo Lifescience Foundation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs A. Miyawaki (RIKEN, Saitama, Japan), T. Kitamura (IMSUT, Tokyo, Japan), M. Ogawa (Kumamoto University Kumamoto, Japan), S. Takahashi (Tsukuba University, Ibaraki, Japan), and M. Tessier-Lavigne (Stanford University, CA) for providing Venus cDNA; pMX retrovirus vector; APC-conjugated anti–VE-cadherin antibody; a knock-in construct containing splicing acceptor, splicing donor, and SV40-derived polyadenylation signal; and chick netrin-1 (VI · V)-Fc, respectively. We also thank all laboratory members and especially Drs M. Osawa, Yuji Yamazaki, Chie Furuta, Azusa Matsubara, Jun Seita, Tohru Morisada, and Reiko Sakamoto for technical assistance and helpful discussion.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal