Abstract
The therapeutic potential of hematopoietic stem cell (HSC) gene therapy can be fully exploited only by reaching efficient gene transfer into HSCs without compromising their biologic properties. Although HSCs can be transduced by HIV-derived lentiviral vectors (LVs) in short ex vivo culture, they display low permissivity to the vector, requiring cytokine stimulation to reach high-frequency transduction. Using stringent assays of competitive xenograft repopulation, we show that early-acting cytokines synergistically enhanced human HSC gene transfer by LVs without impairing engraftment and repopulation capacity. Using S-phase suicide assays, we show that transduction enhancement by cytokines was not dependent on cell cycle progression and that LVs can transduce quiescent HSCs. Pharmacologic inhibition of the proteasome during transduction dramatically enhanced HSC gene transfer, allowing the reach of very high levels of vector integration in their progeny in vivo. Thus, LVs are effectively restricted at a postentry step by the activity of this proteolytic complex. Unexpectedly, cytokine stimulation rapidly and substantially down-regulated proteasome activity in hematopoietic progenitors, highlighting one mechanism by which cytokines may enhance permissiveness to LV gene transfer. These findings demonstrate that antiviral responses ultimately mediated by proteasomes strongly limit the efficiency of HSC transduction by LVs and establish improved conditions for HSC-based gene therapy.
Introduction
Hematopoietic stem cells (HSCs) are attractive targets for the cell and gene therapy of several inherited and acquired diseases. Because of self-renewing ability, multipotency, and clonogenic potential, they can reconstitute all hematopoietic lineages in a host undergoing transplantation. Upon ex vivo gene transfer, they may generate a progeny of gene-corrected cells potentially for a life span. For gene therapy to be efficacious, however, effective gene transfer into HSCs must be reached without inducing detrimental effects on their biologic properties.
The vectors most often used for HSC gene transfer are gamma-retroviral vectors (RVs). More recently, HIV-derived lentiviral vectors (LVs) have been proposed as improved tools for this task. Both RVs and LVs integrate into the genome of host cells but, while RV integration is dependent on target cell mitosis, LVs transduce both proliferating and nonproliferating cells.1 Because HSCs are considered to be mostly quiescent cells, they require stimulation with cytokines that trigger them into the cell cycle in order to be transduced by RVs. Consequently, all the protocols developed for RV-mediated HSC gene transfer involve prolonged ex vivo manipulation and/or induction of cell cycle entry and proliferation,2,6 conditions that in most cases were shown to decrease the frequency of HSCs in the culture, and compromise their long-term repopulating ability.6,9 Nevertheless, recent clinical trials have demonstrated that RVs can be successfully used for HSC-based severe combined immunodeficiency (SCID) gene therapy.10,12 However, the in vivo selective growth advantage of transduced cells, which enabled amplification of a small input of transduced HSCs, was a key factor in the success of these trials. To broaden significantly the scope of HSC-based gene therapy, preservation and transduction of most of the HSCs harvested for transplantation would be required.
We and others have shown efficient transduction of cord blood (CB) nonobese diabetic (NOD)/SCID mouse–repopulating cells (SRCs), considered to be closely related to HSCs, after a short incubation with LVs, in the absence of cytokine stimulation.13,15 Surprisingly, and in apparent contrast with the lack of LV requirement for target cell proliferation, we later found a significant enhancement in gene transfer in the presence of a combination of early-acting cytokines (interleukin-6 [IL-6], stem cell factor [SCF], thrombopoietin [TPO], and Flt3 ligand [Flt3L]),16 previously shown to induce moderate ex vivo SRC expansion.17 It remains to be established whether the gain in gene transfer could be offset, even for a short cytokine exposure, by a decreased engraftment or long-term repopulation capacity of the transduced cells. To address these issues in the SRC model, stringent experimental approaches are needed.
LVs have been shown to be restricted in quiescent T lymphocytes, which must exit from G0 and presumably progress to the G1b phase of the cell cycle in order to be infected by HIV-1 or transduced by LVs.18,21 The transduction block in quiescent cells may be due to the lack of required cellular cofactors and/or the expression of restrictive factors.22 Several recent works have revealed that mammalian cells have developed defense mechanisms against retroviral infection (reviewed by Goff23 ). Some of these factors may affect viral uncoating and reverse transcription and/or target the internalized viral particles to degradation by the ubiquitin-proteasome pathway, whose antiviral activity was observed in earlier studies of HIV-infected lymphocytes.24
In this work, we assessed the impact of early-acting cytokines on HSCs in stringent models of NOD/SCID mouse repopulation, investigated factors limiting the efficiency of LV gene transfer in HSCs, and developed new strategies that may effectively overcome them.
Materials and methods
Cells
HeLa, 293T, and K562 cells were maintained in IMDM, U937 cells in RPMI 1640 (Sigma, St Louis, MO), both containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). CD34+ cells were obtained from human CB, as described,16 after informed consent according to the Declaration of Helsinki and the San Raffaele Hospital Bioethical Committee and used fresh or after freezing. Approval for these studies were obtained from the San Raffaele Hospital institutional review board.
Transduction
Vector stocks were prepared, concentrated, and titered as described.25,26 Transfer vectors were pRRLsin.cPPT.hPGK.eGFP.Wpre, pRRLsin.cPPT. hPGK.YFP.Wpre, and pCCLsin.cPPT.hPGK.eGFP.Wpre; packaging plasmids were pCMVΔR8.74 or pMDLg/pRRE, pRSV-Rev, and pMD2.VSV-G.26 From 5 × 105 to 1 × 106 CD34+ cells per milliliter were incubated with 108 (unless otherwise indicated) LV transducing units per milliliter (TU/mL) for 18 to 24 hours in serumfree medium (StemSpan; StemCell Technologies, Vancouver, BC, Canada) with or without recombinant human (rh) IL-6 (20 ng/mL), SCF (100 ng/mL), Flt3L (100 ng/mL), and TPO (20 ng/mL) (Peprotech, Rocky Hill, NJ), tested individually or in combination. A total of 105 HeLa or 293T cells per milliliter, 106 U937 cells per milliliter, and 2 × 105 K562 cells per milliliter were transduced for 12 hours with 5 × 104, 2 × 105, and 5 × 105 LV TU/mL, respectively. Proteasome inhibitors MG132 (Calbiochem, San Diego, CA) and PS-341 (Millennium, Cambridge, MA) were added to the transduction medium at the indicated concentration. Transduced cells were analyzed by a fluorescence-activated cell sorter (FACS) (FACSCalibur, Becton Dickinson, San Jose, CA) 6 to 14 days after transduction and by quantitative real-time polymerase chain reaction (qPCR) after 3 weeks; qPCR primers and probe complementary to the GFP or ψ vector sequences and conditions were as described17,27 ; primers and probe complementary to the human TERT gene28 were used to normalize DNA content. Colony-forming cell (CFC) assays were performed as described16 and colonies scored by light and fluorescence microscopy at days 10 to 14.
NOD/SCID transplantation
NOD/LtSz scid/scid (NOD/SCID) mice29 obtained from the British Columbia Cancer Research Center (Vancourver, BC, Canada) or Charles River (Calco, Italy) were handled as described16 according to protocols approved by the San Raffaele Hospital Animal Care and Use Committee (IACUC 244) and communicated to the Ministry of Health and local authorities. Each noncompetitively repopulated (reference) mouse received 4 × 105 CD34+ cells transduced with or without cytokines with either LV-GFP or LV-YFP; each competitively repopulated mouse received 2 × 105 CD34+ cells transduced with LV-GFP (or LV-YFP) in the presence of cytokines and 2 × 105 CD34+ cells transduced with LV-YFP (or LV-GFP) without cytokines. In S-phase suicide assays, transduced CD34+ cells were injected together with 5 × 105 CD34– accessory cells. Six to 10 weeks after transplantation, bone marrow (BM) cells were retrieved and plated in CFC assays, as described.16 For FACS analysis, the following human-specific Cy5-phycoerythrin–(for the competitive repopulation assays) or phycoerythrin–(for other assays) conjugated antibodies were used: anti-hCD45, anti-hCD34, anti-hCD19, anti-hCD13, and IgG1 isotype control (Dako, Glostrup, Denmark). Cells were stained with 2 μg/mL PI, and the percentage of antibody-positive/transgene-positive cells was determined after excluding PI-positive, nonviable cells. PI was not used in combination with Cy5-phycoerythrin–conjugated antibodies and YFP because of emission wavelength overlapping. Mice were considered engrafted if hCD45+ cells exceeded 1% of total BM cells. For qPCR, genomic DNA was extracted from BM-derived cells and analyzed using primers and probe complementary to the vector RRE sequence (fw 5′-TGAGGGCTATTGAGGCGC3-′ 600 nM; rv 5′-TGCCTGGAGCTGCTTGATG-3′ 600 nM; probe 5′-6-FAM-TTGCAACTCACAGTCTG-MGB-3′ 200 nM), to the human TERT gene28 (fw 200 nM; rv 600 nM; probe 200 nM), and to the murine β-actin gene (fw: 5′-AGAGGGAAATCGTGCGTGAC-3′ 300 nM; rv 5′-CAATAGTGATGACCTGGCCGT-3′ 750 nM; probe 5′-VIC-CACTGCCGCATCCTCTTCCTCCC-MGB-3′ 200 nM); qPCR conditions were as described.17,27 LV copy numbers were normalized to human DNA content. Percentage of human engraftment was calculated as the ratio between human and total (human + murine) DNA. Samples were analyzed using Abi Prism 7900 HT (Applied Biosystems, Freiburg, Germany).
S-phase suicide assay
After transduction with or without cytokines, 5 × 105 CD34+ cells per milliliter were exposed to 2 × 10–6 M cytosine 1-β-d-arabinofuranoside (Ara-C; SIGMA) for 12 hours or left untreated and then washed twice and plated in liquid culture and CFC assays. Killing efficiency was assessed by scoring CFC numbers.
Cell cycle analysis
CD34+ cells were incubated in DNA staining buffer (PBS 1% saponin with 300 Kuntz units [KU] RNAse and 50 μg/mL PI) for 10 minutes in ice and then analyzed by FACS. Cell aggregates and dead cells were eliminated from further analysis by gating individual cells in the FL3-area versus FL3-width plot.
Proteasome assays
Cellular extracts were prepared as described30 with minor modifications. Briefly, cells were sonicated in ice-cold buffer (50 mM Tris-HCl [pH 7.5], 1 mM DTT, 0.25 M sucrose, 5 mM MgCl2, 0.5 mM EDTA, 2 mM ATP), and extracts were prepared by centrifugation. Peptidase activities were assayed by monitoring production of 7-amino-4-methylcoumarin (amc) from fluorogenic peptides, as described.31 Suc-LLVY-amc (for chymotrypsin-like activity), Boc-LRR-amc (for trypsin-like activity), and Ac-YVAD-amc (for caspase-like activity) were used at 100 μM in 20 mM Tris-HCl (pH 7.5), 1 mM ATP, 2 mM MgCl2, 0.1% BSA. Fluorescence of released amc (excitation, 380 nm; emission, 460 nm) was monitored continuously at 37°C, following addition of cellular extracts, by a Carry Eclipse spectrofluorimeter. Background activity was determined in the presence of proteasome inhibitors MG132 (for chymotrypsin- and caspase-like activities; 10 μM) and β-lactone (for trypsin-like activity; 20 μM). Assays were calibrated using standard solutions of free fluorophores. Substrate consumption at the end of the reaction never exceeded 1%. Concentration of dimethyl sulfoxide never exceeded 0.7%.32
Statistical analysis
The Wilcoxon signed rank test was used for comparison between paired samples; the Wilcoxon Mann-Whitney rank-sum test for comparison between 2 independent samples; and the Kruskal-Wallis test, a nonparametric analog of the analysis of variance (ANOVA) test, for comparison between more than 2 independent samples. Calculations were done using the function wilcox.test, native to the free statistical software R (http://www.R-project.org). For the data in Figure 3E, to make the differences in percent GFP comparable across different doses, a logarithmic transformation was applied. A standard sign test was used for the data in Figure 5. For the analysis of the competitive repopulation assay, see Table S1 and accompanying text (at the Blood website, see the Supplemental Materials link at the top of the online article).
Results
Cytokine stimulation preserved NOD/SCID repopulating ability of human HSCs
To stringently assess repopulation capacity of CB SRCs after a short ex vivo culture with and without a combination of early-acting cytokines (SCF, TPO, IL-6, Flt3L), we performed competitive NOD/SCID repopulation assays. We transduced CB CD34+ cells in both culture conditions with either of 2 self-inactivating LVs26 differing only for the transgene (GFP or YFP). We injected half cells transduced with cytokines by LV-YFP and half cells transduced without cytokines by LV-GFP into the same NOD/SCID mouse (YFP + cyt/GFP – cyt group, n = 12). To avoid any transgene-specific effect, we swapped transgene and culture condition in a parallel group of mice (YFP – cyt/GFP + cyt group, n = 8). In parallel, mice received transplants with the same total number of the same cells transduced in only one culture condition and with either of the 2 vectors (reference, noncompetitively repopulated groups: GFP + cyt, GFP – cyt, YFP + cyt, YFP – cyt; n = 4, 6, 5, and 5, respectively).
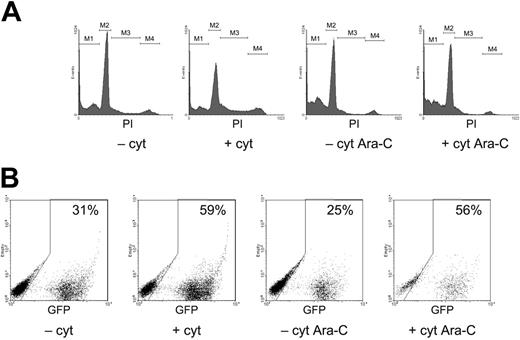
Aliquots of the transduced cells were kept in culture and plated in colony-forming cell (CFC) assays for assessing transduction. Analysis of suspension cultures and CFC assay showed a higher transduction level when cytokines were used for both vectors (data not shown). Ten weeks after transplantation, we analyzed NOD/SCID BM. Representative FACS plots of the 3-channel analysis are shown in Figure 1. Transgene expression was assessed both in the total human CD45+ cell population and, to verify transduction of multipotent HSCs, in the progenitor, lymphoid, and myeloid fractions (Figure 1B-C). All repopulated mice (40 of 44 assayed; numbers of mice per group refer to the successfully repopulated mice) showed multilineage engraftment by transduced SRCs.
As shown in Table 1, human cells engrafted mice at variable levels, as expected for the NOD/SCID model. The engraftment level was slightly lower in competitively than in noncompetitively repopulated mice (21% versus 35% CD45+ cells on average, respectively; n = 20 and 20).
Competitive repopulation assay: SRC analysis
Mouse . | %hCD45* . | %GFP† . | %YFP† . |
|---|---|---|---|
| Reference mice | |||
| GFP + cyt | |||
| Mouse no. 1 | 45 | 71 | — |
| Mouse no. 2 | 41 | 80 | — |
| Mouse no. 3 | 64 | 83 | — |
| Mouse no. 4 | 16 | 69 | — |
| Average | 41 | 76 | — |
| GFP - cyt | |||
| Mouse no. 1 | 66 | 60 | — |
| Mouse no. 2 | 53 | 58 | — |
| Mouse no. 3 | 17 | 23 | — |
| Mouse no. 4 | 18 | 33 | — |
| Mouse no. 5 | 20 | 35 | — |
| Mouse no. 6 | 59 | 37 | — |
| Average | 39 | 41 | — |
| YFP + cyt | |||
| Mouse no. 1 | 40 | — | 85 |
| Mouse no. 2 | 24 | — | 71 |
| Mouse no. 3 | 50 | — | 78 |
| Mouse no. 4 | 2 | — | 35 |
| Mouse no. 5 | 50 | — | 44 |
| Average | 33 | — | 63 |
| YFP - cyt | |||
| Mouse no. 1 | 45 | — | 82 |
| Mouse no. 2 | 15 | — | 53 |
| Mouse no. 3 | 60 | — | 63 |
| Mouse no. 4 | 19 | — | 18 |
| Mouse no. 5 | 11 | — | 18 |
| Average | 30 | — | 47 |
| Competitively repopulated mice | |||
| YFP + cyt/GFP - cyt | |||
| Mouse no. 1 | 18 | 32 | 27 |
| Mouse no. 2 | 10 | 40 | 17 |
| Mouse no. 3 | 19 | 6 | 73 |
| Mouse no. 4 | 15 | 7 | 65 |
| Mouse no. 5 | 6 | 9 | 82 |
| Mouse no. 6 | 40 | 1 | 85 |
| Mouse no. 7 | 3 | 26 | 31 |
| Mouse no. 8 | 4 | 47 | 17 |
| Mouse no. 9 | 25 | 24 | 20 |
| Mouse no. 10 | 15 | 20 | 16 |
| Mouse no. 11 | 45 | 68 | 22 |
| Mouse no. 12 | 9 | 18 | 11 |
| Average | 17 | 25 | 39 |
| YFP - cyt/GFP + cyt | |||
| Mouse no. 1 | 58 | 30 | 48 |
| Mouse no. 2 | 15 | 33 | 33 |
| Mouse no. 3 | 5 | 44 | 22 |
| Mouse no. 4 | 7 | 6 | 85 |
| Mouse no. 5 | 40 | 45 | 15 |
| Mouse no. 6 | 10 | 50 | 8 |
| Mouse no. 7 | 30 | 67 | 20 |
| Mouse no. 8 | 43 | 51 | 2 |
| Average | 26 | 41 | 29 |
Mouse . | %hCD45* . | %GFP† . | %YFP† . |
|---|---|---|---|
| Reference mice | |||
| GFP + cyt | |||
| Mouse no. 1 | 45 | 71 | — |
| Mouse no. 2 | 41 | 80 | — |
| Mouse no. 3 | 64 | 83 | — |
| Mouse no. 4 | 16 | 69 | — |
| Average | 41 | 76 | — |
| GFP - cyt | |||
| Mouse no. 1 | 66 | 60 | — |
| Mouse no. 2 | 53 | 58 | — |
| Mouse no. 3 | 17 | 23 | — |
| Mouse no. 4 | 18 | 33 | — |
| Mouse no. 5 | 20 | 35 | — |
| Mouse no. 6 | 59 | 37 | — |
| Average | 39 | 41 | — |
| YFP + cyt | |||
| Mouse no. 1 | 40 | — | 85 |
| Mouse no. 2 | 24 | — | 71 |
| Mouse no. 3 | 50 | — | 78 |
| Mouse no. 4 | 2 | — | 35 |
| Mouse no. 5 | 50 | — | 44 |
| Average | 33 | — | 63 |
| YFP - cyt | |||
| Mouse no. 1 | 45 | — | 82 |
| Mouse no. 2 | 15 | — | 53 |
| Mouse no. 3 | 60 | — | 63 |
| Mouse no. 4 | 19 | — | 18 |
| Mouse no. 5 | 11 | — | 18 |
| Average | 30 | — | 47 |
| Competitively repopulated mice | |||
| YFP + cyt/GFP - cyt | |||
| Mouse no. 1 | 18 | 32 | 27 |
| Mouse no. 2 | 10 | 40 | 17 |
| Mouse no. 3 | 19 | 6 | 73 |
| Mouse no. 4 | 15 | 7 | 65 |
| Mouse no. 5 | 6 | 9 | 82 |
| Mouse no. 6 | 40 | 1 | 85 |
| Mouse no. 7 | 3 | 26 | 31 |
| Mouse no. 8 | 4 | 47 | 17 |
| Mouse no. 9 | 25 | 24 | 20 |
| Mouse no. 10 | 15 | 20 | 16 |
| Mouse no. 11 | 45 | 68 | 22 |
| Mouse no. 12 | 9 | 18 | 11 |
| Average | 17 | 25 | 39 |
| YFP - cyt/GFP + cyt | |||
| Mouse no. 1 | 58 | 30 | 48 |
| Mouse no. 2 | 15 | 33 | 33 |
| Mouse no. 3 | 5 | 44 | 22 |
| Mouse no. 4 | 7 | 6 | 85 |
| Mouse no. 5 | 40 | 45 | 15 |
| Mouse no. 6 | 10 | 50 | 8 |
| Mouse no. 7 | 30 | 67 | 20 |
| Mouse no. 8 | 43 | 51 | 2 |
| Average | 26 | 41 | 29 |
— indicates not applicable.
Percentage of human cells in the mouse BM.
Percentage of human cells expressing the indicated transgene.
The frequency of transgene-expressing cells was on average higher for SRCs transduced with cytokines and in all permutations of vector type and experimental condition tested.
To assess whether cytokine stimulation had an impact on the repopulating capacity of the transduced SRCs, we developed a statistical model (Table S1). We confirmed that cytokines had a significant effect (P = .011) on the frequency of transgene-positive cells, with an estimated positive effect in the logodd scale equal to +1.044. This means that if the mean frequency of transduction is, for example, 30% for unstimulated cells (–cyt), the estimated mean frequency increases to 55% when stimulated (+cyt).
Because the interaction between the parameters “cytokines” and “xenotransplantation model” (competitive or not) was not significant (P = .77), we concluded that cytokine stimulation had a positive significant impact on SRC transduction without changing significantly its extent and direction between reference and competitively repopulated mice, thus showing that cytokine-stimulated SRCs had an engraftment capacity similar to that of unstimulated cells. We confirmed our findings at the vector DNA level, performing PCR analysis on DNA extracted from human colonies derived from engrafted mouse BM (Figure S1).
Representative FACS analysis of competitively repopulated NOD/SCID mice. Three-channel FACS analysis of mouse BM 10 weeks after transplantation of human CD34+ cells transduced with either LV-GFP or LV-YFP. (A) Frequency of transgene expression in the human cell (hCD45+) graft. FACS plots from a competitively repopulated mouse, showing both GFP- and YFP-positive cells (top panels), and 2 reference mice, showing either YFP- or GFP-positive cells (middle and bottom panels), as indicated. Percentages of transgene-positive cells detected in each channel (FL-2: YFP; FL-1: GFP + YFP, FL-3: human CD45 [hCD45]) are given in the respective quadrant. (B-C) Multilineage engraftment by transduced SRCs. Percentages shown represent the fraction of transgene-expressing cells among the indicated lineage marker-positive cells (scored in the FL-3 channel). Because both GFP and YFP signals emit in the FL-1 channel, we calculated the frequency of GFP-expressing cells by subtracting the fraction of YFP-expressing cells, scored in FL-2, from the percentage of cells scored positive in FL-1. Mouse numbers are as in Table 1.
Representative FACS analysis of competitively repopulated NOD/SCID mice. Three-channel FACS analysis of mouse BM 10 weeks after transplantation of human CD34+ cells transduced with either LV-GFP or LV-YFP. (A) Frequency of transgene expression in the human cell (hCD45+) graft. FACS plots from a competitively repopulated mouse, showing both GFP- and YFP-positive cells (top panels), and 2 reference mice, showing either YFP- or GFP-positive cells (middle and bottom panels), as indicated. Percentages of transgene-positive cells detected in each channel (FL-2: YFP; FL-1: GFP + YFP, FL-3: human CD45 [hCD45]) are given in the respective quadrant. (B-C) Multilineage engraftment by transduced SRCs. Percentages shown represent the fraction of transgene-expressing cells among the indicated lineage marker-positive cells (scored in the FL-3 channel). Because both GFP and YFP signals emit in the FL-1 channel, we calculated the frequency of GFP-expressing cells by subtracting the fraction of YFP-expressing cells, scored in FL-2, from the percentage of cells scored positive in FL-1. Mouse numbers are as in Table 1.
Overall, these results showed that cytokine treatment, while increasing LV transduction of human HSCs, did not affect their engraftment and repopulating ability in a stringent xenotransplantation model.
Cytokine enhancement of SRC gene transfer does not require cell cycle progression
A possible explanation for the cytokine enhancement of LV transduction was that only the SRCs in the G1 phase of the cell cycle were susceptible to transduction and that cytokines enhanced transduction triggering the exit of SRCs from G0. Progression into late G1, however, commits the cells to DNA replication and mitosis and may thus impair the long-term repopulating ability of HSCs, a difficult feature to assess in the NOD/SCID model. Because we could not measure the cell cycle status of the few SRCs contained in the CD34+ population, we performed an S-phase suicide assay on the transduced cells.
We stimulated or not CB-derived CD34+ cells with cytokines and then exposed the cells or not to 1-β-d-arabinofuranoside (Ara-C) to kill cycling cells. Ara-C was previously reported to effectively kill hematopoietic stem/progenitor cells (HSPCs) passing through S phase.33,36 To verify the cell cycle status of the cells after treatment, we stained them with propidium iodide and analyzed by FACS (Figure 2A; Table 2). Cytokines increased the fraction of cells in S phase (17% for +cyt versus 9% for –cyt), and this difference was completely abrogated when the cells were exposed to Ara-C.
S-phase suicide assay: HSPC cell-cycle analysis
Phase . | -cyt, % . | +cyt, % . | -cyt Ara-C, % . | +cyt Ara-C, % . |
|---|---|---|---|---|
| Apoptotic/necrotic cells; M1 | 32 | 28 | 42 | 42 |
| G0/G1; M2 | 52 | 43 | 48 | 49 |
| S; M3 | 9 | 17 | 6 | 5 |
| G2/M; M4 | 7 | 12 | 4 | 4 |
Phase . | -cyt, % . | +cyt, % . | -cyt Ara-C, % . | +cyt Ara-C, % . |
|---|---|---|---|---|
| Apoptotic/necrotic cells; M1 | 32 | 28 | 42 | 42 |
| G0/G1; M2 | 52 | 43 | 48 | 49 |
| S; M3 | 9 | 17 | 6 | 5 |
| G2/M; M4 | 7 | 12 | 4 | 4 |
Human CD34+ were stimulated (+cyt) or not (-cyt) with cytokines for 18 hours, then exposed or not to Ara-C for another 12 hours, as indicated, and analyzed immediately. Distribution of the analyzed cell populations is shown in the indicated cell-cycle phases, marked as M1 to M4 in Figure 2A. Average values of 2 experiments are shown.
S-phase suicide assay of HSPCs assayed in vitro. Human CD34+ cells were stimulated (+cyt) or not (–cyt) with cytokines for 18 hours, then exposed or not to Ara-C for another 12 hours, as indicated, and analyzed immediately (A) or after 1 week (B) of culture. For B, the cells were also transduced with LV-GFP during the first 18 hours of incubation. (A) Cell-cycle analysis: representative FACS histograms of propidium iodide (PI) staining; distribution of the analyzed cell populations in the indicated cell-cycle phases is shown in Table 2. M1 indicates apoptotic/necrotic cells; M2, G0/G1 phases; M3, S phase; and M4, G2/M phases. (B) Representative FACS analysis of GFP expression in suspension culture. Percentages of GFP-positive cells are given.
S-phase suicide assay of HSPCs assayed in vitro. Human CD34+ cells were stimulated (+cyt) or not (–cyt) with cytokines for 18 hours, then exposed or not to Ara-C for another 12 hours, as indicated, and analyzed immediately (A) or after 1 week (B) of culture. For B, the cells were also transduced with LV-GFP during the first 18 hours of incubation. (A) Cell-cycle analysis: representative FACS histograms of propidium iodide (PI) staining; distribution of the analyzed cell populations in the indicated cell-cycle phases is shown in Table 2. M1 indicates apoptotic/necrotic cells; M2, G0/G1 phases; M3, S phase; and M4, G2/M phases. (B) Representative FACS analysis of GFP expression in suspension culture. Percentages of GFP-positive cells are given.
We then transduced CD34+ cells with LV-GFP, exposed them or not to Ara-C, performed in vitro assays, and injected the cells into NOD/SCID mice. CFC assay showed that Ara-C killed most of the cytokine-stimulated cells and a much smaller fraction of nonstimulated cells, indicating that cytokines triggered committed progenitors into S phase, as expected. Nevertheless, when scoring transduction frequency in the surviving cultures, both suspension cultures and CFCs, we did not observe a decrease in transduction levels in the cells exposed to Ara-C (Figure 2B; Table 3). We obtained similar results using 3H-thymidine as killing agent (not shown).
S-phase suicide assay: CFC analysis
. | -cyt . | +cyt . | -cyt Ara-C . | +cyt Ara-C . |
|---|---|---|---|---|
| % GFP | 39 | 65 | 34 | 55 |
| CFCs | 80 | 83 | 40 | 16 |
| % killing | — | — | 50 | 80 |
. | -cyt . | +cyt . | -cyt Ara-C . | +cyt Ara-C . |
|---|---|---|---|---|
| % GFP | 39 | 65 | 34 | 55 |
| CFCs | 80 | 83 | 40 | 16 |
| % killing | — | — | 50 | 80 |
Human CD34+ were transduced in the presence (+cyt) or absence (-cyt) of cytokines for 18 hours, then exposed or not to Ara-C for another 12 hours, as indicated, plated in methylcellulose, and analyzed after 2 weeks. Average values of 2 experiments are shown.
% GFP indicates percentage of GFP+ colonies scored by fluorescence microscopy; CFCs, total number expressed as number of colonies per 103 CD34+ cells plated; % killing, percentage of killing calculated as the ratio between the number of colonies arising form the Ara-C—treated and —untreated populations.
For transplantation, transduced cells were matched for the number of viable cells prior to Ara-C treatment and injected with CD34– accessory cells. NOD/SCID BM cells were harvested 6 to 8 weeks after transplantation, analyzed by FACS, and plated for CFC assay (Table 4). FACS analysis showed, for the cells not exposed to Ara-C, a higher SRC transduction level when cytokines were added (91% versus 54% for +cyt versus –cyt). Interestingly, we observed a similarly significant difference in the cells exposed to Ara-C (61% versus 29% for +cyt Ara-C and –cyt Ara-C, respectively). CFC assay confirmed these data.
S-phase suicide assay: SRC analysis
. | FACS analysis . | . | . | CFC assay % GFP§ . | ||
|---|---|---|---|---|---|---|
. | %hCD45* . | % GFP† . | P‡ . | . | ||
| -cyt | 28 ± 14 | 54 ± 20 | .029∥ | 39 ± 14 | ||
| +cyt | 42 ± 32 | 91 ± 5 | 68 ± 15 | |||
| -cyt Ara-C | 23 ± 22 | 29 ± 8 | .028¶ | 18 ± 13 | ||
| +cyt Ara-C | 7 ± 6 | 61 ± 22 | 45 ± 22 | |||
. | FACS analysis . | . | . | CFC assay % GFP§ . | ||
|---|---|---|---|---|---|---|
. | %hCD45* . | % GFP† . | P‡ . | . | ||
| -cyt | 28 ± 14 | 54 ± 20 | .029∥ | 39 ± 14 | ||
| +cyt | 42 ± 32 | 91 ± 5 | 68 ± 15 | |||
| -cyt Ara-C | 23 ± 22 | 29 ± 8 | .028¶ | 18 ± 13 | ||
| +cyt Ara-C | 7 ± 6 | 61 ± 22 | 45 ± 22 | |||
All values are expressed as mean ± SD of 2 experiments performed. For -cyt group, n = 4; for +cyt group, n = 4; for -cyt Ara-C group, n = 6; and for +cyt Ara-C group, n = 5.
n indicates number of mice per group.
Percentage of human cells in the mouse BM.
Percentage of human GFP-expressing cells.
P value calculated by rank sum 2-sided test.
Percentage of human GFP-expressing cells, scored by fluorescence microscopy.
-cyt versus +cyt.
-cyt Ara-C versus +cyt Ara-C.
Because cytokine enhancement of SRC transduction was not abrogated by killing proliferating cells, it did not require progression into the S phase of the cell cycle. Ara-C treatment decreased engraftment and transduction levels in both stimulated and nonstimulated SRCs, indicating that some SRCs were sensitive to Ara-C, most likely because they were induced to proliferate early after transplantation. One can thus postulate that the quiescent SRC fraction was mainly responsible for repopulating the mice receiving transplants with Ara-C–exposed cells; because these cells were transduced to substantial levels, these data strongly suggest that LVs are able to transduce quiescent SRCs ex vivo.
Lentiviral gene transfer is restricted by proteasome in HSPCs
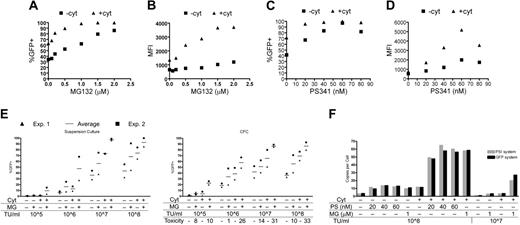
We showed that cytokine enhancement of LV gene transfer was not dependent on the envelope pseudotype (Figure S2), indicating that cytokine stimulation did not affect a specific entry pathway. Thus, we evaluated factors influencing the postentry steps of viral infection. Because of its emerging effector role in many cellular response pathways, we assessed the influence of proteasome on LV transduction of HSPCs. In a first set of experiments we transduced CD34+ cells with the standard dose of LV-GFP, with or without cytokine stimulation, and added different doses of the reversible peptide-aldehyde proteasome inhibitor MG13237 during the transduction period. After transduction, cells were washed to remove the inhibitor and kept in suspension culture. After 2 weeks, FACS analysis revealed a substantial MG132 dose-dependent increase in the frequency of GFP-expressing cells and in their mean fluorescence intensity (MFI), for both the stimulated and nonstimulated cells (Figure 3A-B). Remarkably, at high concentrations of MG132, we were able to transduce virtually all the stimulated cells, and to very high MFI, and up to 90% of the nonstimulated cells. We were not able to further increase the transduction level in nonstimulated cells without reaching toxic doses of MG132. To verify that the observed enhancement of transduction was specifically dependent on proteasome inhibition, we repeated these experiments using the more powerful peptide-boronate inhibitor PS-341 and obtained similar results (Figure 3C-D).
In a second set of experiments we evaluated the effect of a constant concentration of MG132 (1 μM) on CD34+ cells transduced with or without cytokines and different LV amounts. The same number of cells was transduced with or without MG132, cultured, and plated in CFC assay to test the toxicity of MG132 on HSPCs. Proteasome inhibitor substantially enhanced transduction at each vector dose, for both cytokine-stimulated and nonstimulated cells, whether assayed in suspension cultures or by CFCs (Figure 3E) (P = .008 and P = .016 for MG132-treated versus untreated cells, in +cyt and –cyt conditions, respectively, both for suspension culture cells and CFCs). Stimulated cells showed higher toxicity in response to the inhibitor than nonstimulated cells. This could be due, however, to the extremely high GFP expression level and vector integration load, as indicated by the increasing percentage of CFC death with increasing LV dose (Figure 3E and data not shown).
To confirm at the molecular level the enhancement of transduction by proteasome inhibitor, we performed quantitative real-time PCR (qPCR) analysis on the transduced progenitors' DNA. We used 2 primer-probe systems to amplify different vector sequences and found that both proteasome inhibitors (PS-341 and MG132) increased from 3- to 7-fold the average vector copy number per cell in both nonstimulated and stimulated cells The enhancement was dependent on the inhibitor dose, reaching an average of 14 and 61 copies per cell in nonstimulated and stimulated cells, respectively, at the highest doses of vector and inhibitor (Figure 3F).
Overall, these data indicated that the proteasome effectively restricted LV gene transfer in HSPCs.
Cytokines down-regulate the proteasome activity of HSPCs
We assessed the contribution of individual cytokines to the observed enhancement of LV transduction by taking advantage of the response amplification mediated by MG132. We transduced CD34+ cells with or without 1 μM MG132 and in the absence or presence of IL-6, SCF, TPO, and Flt3L, tested separately or in combination (Figure 4). Each cytokine induced only a modest increase in gene transfer as compared with the unstimulated cells, both when scored in liquid culture and in CFC assay. However, addition of MG132 during transduction revealed a higher response of the cells stimulated with a single cytokine than that seen in nonstimulated cells. When comparing cells stimulated with each individual cytokine with cells stimulated by their combination, the effect of cytokines on gene transfer appeared to be synergistic, as indicated by the frequency of transduction, which reached saturation at 100%, and by the transduced cells' MFI.
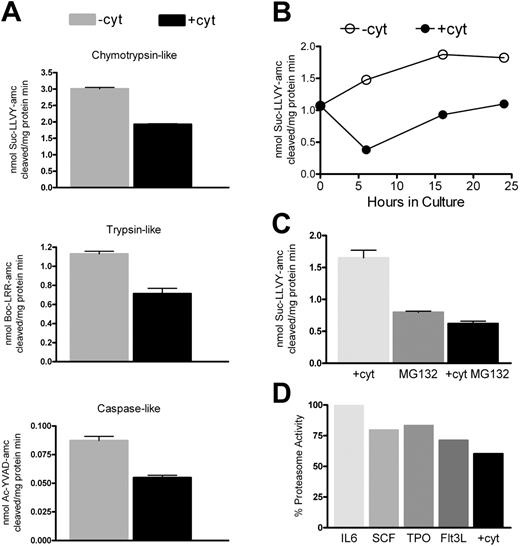
We then assayed the proteasomal chymotryptic-, tryptic-, and caspase-like activities in CD34+ cells stimulated or not with cytokines. We found that, for all activities tested, cytokine-stimulated CD34+ cells displayed a significantly lower specific activity—on average 60%—compared with nonstimulated cells (global P = 2 × 2–18 ) (Figure 5A). We then performed a time-course analysis of the chymotryptic-like proteasome activity (which accounts for 90% of the total activity of this complex) of CD34+ cells stimulated or not with cytokines (Figure 5B). Whereas the proteasome activity progressively increased in cultured unstimulated CD34+ cells, it was rapidly and strongly down-regulated by exposure to cytokines. Indeed, the difference in specific activity between stimulated and nonstimulated cells was higher at 6 hours (earliest time analyzed; 30% of unstimulated cells) than at 24 hours.
Effect of proteasome inhibition on HSPC transduction by LVs. (A,C) Percentage of GFP-positive cells and (B,D) mean fluorescence intensity (MFI) of CD34+ cells transduced with 108 TU/mL LV-GFP with (▴) or without (▪) cytokine stimulation in the presence of increasing doses of the proteasome inhibitors MG132 (A-B) and PS-341 (C-D) and analyzed 2 weeks after transduction. Each panel represents the average of 2 experiments. (E) Percentages of GFP-positive cells in suspension culture (left panel) and GFP-positive colonies from CFC assays (right panel) from CD34+ cells transduced with or without cytokine (Cyt) stimulation with increasing doses of LV-GFP (TU/mL) and in the absence or presence of 1μM MG132 (MG), as indicated, and analyzed 2 weeks after. Toxicity was calculated from the ratio between the number of colonies arising from MG132-exposed and not exposed cells and shown as percentage of CFCs killed. Average (—) and individual values (▴, ▪) from 2 experiments. Statistical analysis is reported in “Results.” (F) Quantitative real-time PCR analysis of DNA extracted after 3 weeks of suspension culture from HSPCs stimulated or not with cytokines (Cyt), transduced with different doses of LV-GFP (TU/mL), and treated or not with different doses of PS-341 (PS) or MG132 (MG), as indicated. Vector sequences were amplified with a system annealing to the ψ of the vector (PSI system) and a system annealing to the GFP sequence (GFP system). For each condition, an average of 3 detections is shown.
Effect of proteasome inhibition on HSPC transduction by LVs. (A,C) Percentage of GFP-positive cells and (B,D) mean fluorescence intensity (MFI) of CD34+ cells transduced with 108 TU/mL LV-GFP with (▴) or without (▪) cytokine stimulation in the presence of increasing doses of the proteasome inhibitors MG132 (A-B) and PS-341 (C-D) and analyzed 2 weeks after transduction. Each panel represents the average of 2 experiments. (E) Percentages of GFP-positive cells in suspension culture (left panel) and GFP-positive colonies from CFC assays (right panel) from CD34+ cells transduced with or without cytokine (Cyt) stimulation with increasing doses of LV-GFP (TU/mL) and in the absence or presence of 1μM MG132 (MG), as indicated, and analyzed 2 weeks after. Toxicity was calculated from the ratio between the number of colonies arising from MG132-exposed and not exposed cells and shown as percentage of CFCs killed. Average (—) and individual values (▴, ▪) from 2 experiments. Statistical analysis is reported in “Results.” (F) Quantitative real-time PCR analysis of DNA extracted after 3 weeks of suspension culture from HSPCs stimulated or not with cytokines (Cyt), transduced with different doses of LV-GFP (TU/mL), and treated or not with different doses of PS-341 (PS) or MG132 (MG), as indicated. Vector sequences were amplified with a system annealing to the ψ of the vector (PSI system) and a system annealing to the GFP sequence (GFP system). For each condition, an average of 3 detections is shown.
Because we observed an additive effect of cytokines and proteasome inhibitor in enhancing transduction (Figures 3, 4), we measured the proteasome activity 24 hours after either treatment or their combination (Figure 5C). MG132 down-regulated the proteasome activity more effectively than the cytokines, and the combination was slightly but not significantly more effective than the drug alone.
Finally, we assessed the contribution of each individual cytokine (Figure 5D). We observed a moderate down-regulation of proteasome activity when CD34+ cells were exposed to SCF, TPO, or Flt3L, while no effect was seen with IL-6. No cytokine tested alone reached the effect of the cocktail, indicating an additive effect of 3 cytokines.
Contribution of individual cytokines to enhanced HSPC transduction by LVs. (A) Percentage of GFP-positive cells in suspension culture and colonies (CFC) of CD34+ cells transduced with 108 TU/mL LV-GFP without (–cyt) or with the indicated cytokine, added separately (IL-6, SCF, TPO, Flt3L) or all together (+cyt), and with or without (NT [not treated]) 1μM MG132. (B) Mean fluorescence intensity (MFI) of suspension culture cells shown in panel A. One experiment of 2 performed with similar results is shown.
Contribution of individual cytokines to enhanced HSPC transduction by LVs. (A) Percentage of GFP-positive cells in suspension culture and colonies (CFC) of CD34+ cells transduced with 108 TU/mL LV-GFP without (–cyt) or with the indicated cytokine, added separately (IL-6, SCF, TPO, Flt3L) or all together (+cyt), and with or without (NT [not treated]) 1μM MG132. (B) Mean fluorescence intensity (MFI) of suspension culture cells shown in panel A. One experiment of 2 performed with similar results is shown.
Overall, the rapid onset and the substantial extent of proteasome activity down-regulation induced by cytokines in HSPCs indicate a likely role of this response in mediating the enhancement of LV transduction. However, because the addition of cytokines to cells exposed to proteasome inhibitor further enhanced transduction without likely inducing a comparatively stronger proteasome activity down-regulation, it is conceivable that other pathways activated by cytokines beside their effect on the proteasome contribute to the increased permissiveness to LV transduction.
Proteasome activities in HSPC cells. CD34+ cells were treated as indicated, lysed, and tested for proteasome peptidase activities. Peptidase activities were assayed by monitoring the production of 7-amino-4-methylcoumarin (amc) from fluorogenic peptides: Suc-LLVY-amc for the chymotrypsin-like activity, Boc-LRR-amc for the trypsin-like activity, and Ac-YVAD-amc for the caspase-like activity. (A) Peptidase activities of cells exposed to cytokines (+cyt) or not (–cyt) for 24 hours. (B-D) Chymotrypsin-like activity of cells kept in culture for the indicated times with (+cyt) or without (–cyt) cytokines (B); exposed to cytokines (+cyt), to MG132 (MG132), or to both treatments (+cyt MG132) for 24 hours (C); and exposed to the indicated cytokine for 24 hours (D). Values in panels A-C are mean ± SD of 3 measurements from 1 of 2 experiments with similar results, performed with different cell preparations. Values in panel D are the average of 2 experiments and are expressed as percentage of the proteasome activity of unstimulated cells.
Proteasome activities in HSPC cells. CD34+ cells were treated as indicated, lysed, and tested for proteasome peptidase activities. Peptidase activities were assayed by monitoring the production of 7-amino-4-methylcoumarin (amc) from fluorogenic peptides: Suc-LLVY-amc for the chymotrypsin-like activity, Boc-LRR-amc for the trypsin-like activity, and Ac-YVAD-amc for the caspase-like activity. (A) Peptidase activities of cells exposed to cytokines (+cyt) or not (–cyt) for 24 hours. (B-D) Chymotrypsin-like activity of cells kept in culture for the indicated times with (+cyt) or without (–cyt) cytokines (B); exposed to cytokines (+cyt), to MG132 (MG132), or to both treatments (+cyt MG132) for 24 hours (C); and exposed to the indicated cytokine for 24 hours (D). Values in panels A-C are mean ± SD of 3 measurements from 1 of 2 experiments with similar results, performed with different cell preparations. Values in panel D are the average of 2 experiments and are expressed as percentage of the proteasome activity of unstimulated cells.
Proteasome restriction of LVs appears to be a specific feature of HSPCs
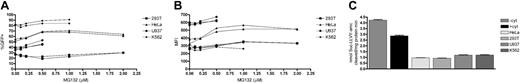
To verify if proteasome restriction of LVs was common to different cell types, we evaluated the effect of MG132 on the transduction of different cell lines, either of hematopoietic origin (K562 and U937) or not (293T and HeLa). Cells were transduced with a constant LV-GFP dose and with increasing MG132 doses, washed, and analyzed 2 weeks later by FACS. The effect of MG132 on transduction was much lower for all the cell lines tested than that observed in HSPCs (Figure 6A-B; compare with Figure 3A-B). To exclude the possibility that proteasome inhibitor was not effective because the level of transduction for some cell lines had already reached saturation without the drug, we repeated the experiments using lower doses of vector and obtained similar results (data not shown). We then assayed the proteasome chymotryptic-like activity in all cell types tested. The activity in all cell line extracts was about one third to one fourth of that found in HSPCs, stimulated or not with cytokines, respectively (Figure 6C).
These results suggest that proteasome restriction of LVs is a specific feature of HSPCs and that it could be due to the high proteasome activity observed in these cells.
Proteasome inhibition strongly enhances SRC gene transfer by LVs
We then verified the effect of proteasome inhibition on SRC transduction. We transduced CD34+ cells stimulated or not with cytokines with 2 × 107 TU/mL LV-GFP and with or without 0.7 μM MG132, performed in vitro assays, and transplanted them into NOD/SCID mice. We chose a lower vector dose than that used in previous NOD/SCID experiments to better detect drug-dependent enhancement of transduction. We analyzed NOD/SCID BM by FACS and qPCR. FACS analysis showed increasingly higher transduction levels when comparing unstimulated cells with “unstimulated plus MG132 cells” with stimulated cells (+cyt > –cyt MG > –cyt). However, we reached such a high frequency of transduction in the latter condition that any possible further enhancement by MG132 addition could not be detected (Figure 7A). Thus, we used qPCR specific for LV sequence and for human DNA to evaluate the number of integrated vector copies per human cell and the level of human engraftment per mouse (Figure 7B-C). Proteasome inhibitor increased the average vector copy number by 2-fold in unstimulated cells and 1.5-fold in stimulated cells, reaching the remarkable number of 34 copies per human cell in the graft (highest score) and an average of 23 in SRCs transduced with cytokines and inhibitor. Importantly, consistently with the results shown before, cytokine treatment alone increased 3-fold the vector copy number per cell. The differences between the 4 groups were statistically significant (P = .002).
Effect of proteasome inhibition on LV transduction of different cell lines. (A) Frequency of GFP-positive cells and (B) MFI of 293T, HeLa, U937, and K562 cells transduced with LV-GFP in the presence of increasing concentrations (0.1 to 2 μM) of MG132 for 12 hours. Missing points at the highest doses in K562 and U937 curves are due to cell death because of MG132 toxicity. Two to 3 experiments are shown for each cell line. (C) CD34+ cells exposed (+cyt) or not (–cyt) to cytokines for 20 to 24 hours; HeLa, 293T, U937 and K562 cells were lysed and tested for proteasome chymotrypsin-like activity. Chymotrypsin-like activity was assayed by monitoring the production of 7-amino-4-methylcoumarin (amc) from fluorogenic peptide Suc-LLVY-amc. Values are mean ± SD of 3 measurements from 1 of 2 experiments with similar results.
Effect of proteasome inhibition on LV transduction of different cell lines. (A) Frequency of GFP-positive cells and (B) MFI of 293T, HeLa, U937, and K562 cells transduced with LV-GFP in the presence of increasing concentrations (0.1 to 2 μM) of MG132 for 12 hours. Missing points at the highest doses in K562 and U937 curves are due to cell death because of MG132 toxicity. Two to 3 experiments are shown for each cell line. (C) CD34+ cells exposed (+cyt) or not (–cyt) to cytokines for 20 to 24 hours; HeLa, 293T, U937 and K562 cells were lysed and tested for proteasome chymotrypsin-like activity. Chymotrypsin-like activity was assayed by monitoring the production of 7-amino-4-methylcoumarin (amc) from fluorogenic peptide Suc-LLVY-amc. Values are mean ± SD of 3 measurements from 1 of 2 experiments with similar results.
Effect of proteasome inhibition on SRC transduction. CD34+ cells were transduced with LV-GFP without any stimulation (–cyt), in the presence of 0.7 μM MG132 (–cyt MG), in the presence of cytokines (+cyt), or in the presence of both cytokines and 0.7 μM MG132 (+cyt MG) and transplanted into NOD/SCID mice. Six weeks after transplantation, mouse BM was analyzed by FACS (A) or by quantitative real-time PCR (B-C). (A) Percentage of GFP-positive cells in the human graft. (B) Number of LV integrated copies per human cell. (C) Percentage of human engraftment, calculated as the ratio between human and total (human + murine) DNA. Each dot represents a mouse; for each group, the average (—) is shown. (D) FACS analysis of a mouse undergoing transplantation with cells transduced in the presence of cytokines and MG132 and carrying 34 LV copies per cell; multilineage repopulation is shown by a similar percentage of GFP-positive cells in all the indicated lineages. Percentages were calculated within the gated populations. Human engraftment level was 15%.
Effect of proteasome inhibition on SRC transduction. CD34+ cells were transduced with LV-GFP without any stimulation (–cyt), in the presence of 0.7 μM MG132 (–cyt MG), in the presence of cytokines (+cyt), or in the presence of both cytokines and 0.7 μM MG132 (+cyt MG) and transplanted into NOD/SCID mice. Six weeks after transplantation, mouse BM was analyzed by FACS (A) or by quantitative real-time PCR (B-C). (A) Percentage of GFP-positive cells in the human graft. (B) Number of LV integrated copies per human cell. (C) Percentage of human engraftment, calculated as the ratio between human and total (human + murine) DNA. Each dot represents a mouse; for each group, the average (—) is shown. (D) FACS analysis of a mouse undergoing transplantation with cells transduced in the presence of cytokines and MG132 and carrying 34 LV copies per cell; multilineage repopulation is shown by a similar percentage of GFP-positive cells in all the indicated lineages. Percentages were calculated within the gated populations. Human engraftment level was 15%.
Importantly, we did not observe a detrimental effect of MG132 on the SRC repopulating ability. In fact, we obtained similar engraftment levels in all groups tested (P = .46). Furthermore, when we analyzed the mouse BM by FACS, we observed multilineage engraftment by transduced cells in all the groups (Figure 7D).
These results prove that proteasome effectively restricts LVs in SRCs and highlight a new strategy to overcome the low permissiveness of human HSCs to LV gene transfer.
Discussion
Using a stringent assay of competitive xenograft repopulation, we show that human CB HSCs transduced to high efficiency by a short exposure to LVs in the presence of SCF, TPO, IL-6, and Flt3L maintain comparable repopulation capacity as cells transduced in the absence of stimulation. Both types of cells contributed similarly to the multilineage human graft and yielded similar numbers of progenitors. The enhancement of LV gene transfer was dependent on the synergy of all 4 cytokines.
Our marking studies provide rigorous, and until now lacking, proof that early-acting cytokines, which are currently investigated for ex vivo HSC expansion, do not impair stem cell properties in the conditions tested. Previous studies reported a decreased homing capacity of CB CFCs after exposure to some of the cytokines used in this study.38 Although our results do not suggest such impairment, we cannot exclude that cytokine-stimulated cells may compensate a decreased homing capacity with a higher in vivo self-renewal and/or clonogenicity. Clonal analysis of the human graft in the repopulated mice will help clarify these issues. On the other hand, because the SRC assay does not provide extended long-term follow-up and does not support T lymphopoiesis, we cannot exclude detrimental effects of the cytokines on these more-difficult-to-assess stem cell features. Further studies in new xenograft models39,42 or, more likely, the follow-up of patients treated with LV-mediated HSC gene therapy will address these issues.
Our studies indicate that, according to the absence or presence of cytokines, HSC gene transfer can be tuned to limit the average level of vector integration and reduce the risk of insertional mutagenesis or, instead, to maximize transduction and transgene expression levels. Because LV integration preferentially targets active genes, cytokine stimulation may also affect the integration sites spectrum, making less or more likely that cytokine-responsive, growth-related genes are targeted.
Cytokine enhancement of SRC transduction was not linked to cell progression into S phase, because it was not abrogated by treating the cells with the S-phase poison Ara-C after transduction. Ara-C–treated cells, both stimulated with cytokines and unstimulated, yielded lower, although not statistically significant, levels of human engraftment and transduction in mice receiving transplants. This suggests that a fraction of SRCs may become sensitive to Ara-C in vivo and that cells in this fraction are more permissive to LVs. These more permissive cells may be those exiting from quiescence; because of an earlier S-phase induction upon transplantation, they may become sensitive to the delayed effect of Ara-C in vivo. The cells less susceptible to LVs may be the quiescent cells; because of a delayed S-phase induction upon transplantation, they may be resistant to Ara-C. Because the SRCs successfully engrafting after Ara-C treatment were transduced to substantial levels, these findings strongly suggest that LVs are able to transduce quiescent SRCs. In this respect, HSCs may behave differently than lymphocytes that restrict LV transduction, and HIV-1 infection, in the G0 phase of the cell cycle.
In recent studies, restriction of retroviral and lentiviral infection has been ascribed to different families of cytosolic proteins that target viral uncoating or edit the viral genome and inhibit viral replication (reviewed by Goff23 ). Factors interfering with viral uncoating, such as TRIM5α, are typically identified by saturating their activity with increasing viral input. We could not overcome the relative resistance of unstimulated, as compared with cytokine-stimulated, cells by increasing the vector input. Rather than being dependent on cell cycle progression or the elimination of a cytosolic restriction factor, the enhancement of LV transduction by cytokines in HSPCs may involve, at least in part, down-regulation of the proteasome activity. By directly testing the proteasome activities in stimulated and unstimulated HSPCs, we showed that they were significantly down-regulated by exposure to the cytokines. Importantly, this cytokine response was rapid, with a maximal effect at early time points corresponding to the period in which the first steps of LV transduction take place. Although it is difficult to estimate the actual level of inhibition reached in cells treated with MG132, the activity recovered after 24 hours of exposure was similar to that recovered from cells exposed for 6 hours to cytokines, suggesting that the extent of down-regulation induced by the latter was substantial. However, because cytokine treatment further enhanced transduction in drug-treated cells, it is likely that other mechanisms are also involved in increasing cell permissiveness to LV.
The down-regulation of proteasome activity in HSPCs in response to cytokine stimulation is a novel and intriguing finding, because it likely affects several cellular functions. Interestingly, SCF, TPO, and Flt3L all had an additive effect on the proteasome activity. Further studies will identify the signaling pathway from the cytokine receptors to the proteasome.
Direct measurement of integrated vector copy numbers in CD34+ cells and in SRCs transduced in the presence and absence of proteasome inhibitors showed a dramatic increase in transduction of cells exposed to the drugs. This indicates that a large excess of vector particles is uptaken but does not become integrated and that the proteasome effectively restricts LV transduction in HSPCs. The mechanism of restriction still needs to be elucidated. The proteasome could act directly and target the uncoating vector core, most likely after it has been labeled for degradation by specific ubiquitin-conjugation systems, possibly recruited by restrictive factors; the proteasome could also act indirectly by targeting factors required for the early steps of transduction.
Because we obtained a very high vector copy number in SRCs transduced with cytokines and proteasome inhibitor, further studies are needed to assess the impact on the target cell genome and evaluate the possible consequences of the increased insertional mutagenesis. However, as shown in Figure 3, using proteasome inhibitors in the absence of cytokine stimulation allows the reach of high-frequency transduction without necessarily increasing the average vector copy number to extremely high levels, thus providing a possible new strategy to optimize gene transfer. It is conceivable that both minimal and maximal thresholds of vector copy numbers providing the best risk-benefit ratio will have to be set according to each proposed gene therapy application.
Whereas proteasome inhibition increased LV transduction to some extent in several cell lines, effective proteasome restriction appeared to be a specific feature of HSPCs (this study) and lymphocytes.24 We correlated effective restriction with the high proteasome activity observed in HSPCs. Two recent works43,44 reported increased HIV infection of some cell lines by proteasome inhibition, although the cells were scored 2 days after infection, thus without ruling out transient effects of the treatment on reporter expression.
In conclusion, we report a substantial increase in SRC gene transfer in the presence of proteasome inhibitor and, importantly, no apparent adverse effect on the cell engraftment capacity. Further studies will determine the safety and efficacy of this approach to improve HSC gene therapy.
Prepublished online as Blood First Edition Paper, February 9, 2006; DOI 10.1182/blood-2005-10-4047.
Supported by grants from Telethon (TIGET) (L.N.), National Institutes of Health (NIH) (2 P01 HL053750-11 CFDA no. 93.839) (L.N.), European Union (QLK3-1999-00859 and Concerted Safety and Efficiency Evaluation of Retroviral Transgenesis for Gene Therapy of Inherited Diseases [CONSERT]) (L.N.), and the Italian Ministry of Scientific Research (L.N., M.G.).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to S. Cenci, R. Sitia, M. De Palma, A. Thrasher, and C. Di Serio for fruitful advice; F. Benedicenti and L. Sergi Sergi for technical help; A. Palini for help with FACS analysis; A. Ferrari and the San Raffaele Obstetric Department for CB collection; and F. L. Cosset, A. Stornaiuolo, and B. Piovani for reagents.

![Figure 1. Representative FACS analysis of competitively repopulated NOD/SCID mice. Three-channel FACS analysis of mouse BM 10 weeks after transplantation of human CD34+ cells transduced with either LV-GFP or LV-YFP. (A) Frequency of transgene expression in the human cell (hCD45+) graft. FACS plots from a competitively repopulated mouse, showing both GFP- and YFP-positive cells (top panels), and 2 reference mice, showing either YFP- or GFP-positive cells (middle and bottom panels), as indicated. Percentages of transgene-positive cells detected in each channel (FL-2: YFP; FL-1: GFP + YFP, FL-3: human CD45 [hCD45]) are given in the respective quadrant. (B-C) Multilineage engraftment by transduced SRCs. Percentages shown represent the fraction of transgene-expressing cells among the indicated lineage marker-positive cells (scored in the FL-3 channel). Because both GFP and YFP signals emit in the FL-1 channel, we calculated the frequency of GFP-expressing cells by subtracting the fraction of YFP-expressing cells, scored in FL-2, from the percentage of cells scored positive in FL-1. Mouse numbers are as in Table 1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/11/10.1182_blood-2005-10-4047/4/m_zh80110696420001.jpeg?Expires=1767736799&Signature=1VzKQ6149igpk6dd2ndW3HOgPR8P67h5HBSmx3bJKgxoN0UCONF52f4SIuksnlOd8wjpBvA-OyBUXHMsEHYwEYnwnv7kXeKtdASIO-tYrDzh4KkmljjOQl9bQrKcw6pB9xYdsADgPffoqfmTD51to3m172SB4EXzj9C7ZURq5batYh-n9matDaPBXUgvW5BZwZMJOkka5HW~Wb4tCRGgMhLmHb1MCoBdQu8O9UKwjpOtcsII1xnaZ83bl5hy-SoepL2APBigC-fzO3FqLir0zJ97HCFhq6AAYKUV08CK-wx7CSdC~kDirttG-6qaQDtnN3AhGxj~IOX~VLll~dVNKQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Contribution of individual cytokines to enhanced HSPC transduction by LVs. (A) Percentage of GFP-positive cells in suspension culture and colonies (CFC) of CD34+ cells transduced with 108 TU/mL LV-GFP without (–cyt) or with the indicated cytokine, added separately (IL-6, SCF, TPO, Flt3L) or all together (+cyt), and with or without (NT [not treated]) 1μM MG132. (B) Mean fluorescence intensity (MFI) of suspension culture cells shown in panel A. One experiment of 2 performed with similar results is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/11/10.1182_blood-2005-10-4047/4/m_zh80110696420004.jpeg?Expires=1767736799&Signature=MxubDdFBhaj9dXWzHSp1R7JHWy9HXpuX30RV3CdpHodKh4u0DFoyWwIgrCWnDrUsuS4X7dTiHYwZyxW4xu0UaYSDfMRzNzDcTaa77yYMEie~tLHigso7BMLREF3m9318cOykN8SVqJFndlvmDSw3~-3kckk4-GI0LFkCio4vDmL2b6R4-VluD1bNJH6YZH0P9jLXfok1tP5OdrCg22cFor7bG1lLIaGjQBmhl8-3LgbGpl5xtybDpobOsP417fkUyplhiP6JdGA93-cVNiHs2pUb5mUn~3Ws7npCeHzv-P3J3BbyYNfXeYXY4gZqEf0a3XFjZG~vLcNMYEjXWuV2jA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal