Abstract
JAK2, a member of the Janus kinase (JAK) family of protein tyrosine kinases (PTKs), is an important intracellular mediator of cytokine signaling. Mutations of the JAK2 gene are associated with hematologic cancers, and aberrant JAK activity is also associated with a number of immune diseases, including rheumatoid arthritis. Accordingly, the development of JAK2-specific inhibitors has tremendous clinical relevance. Critical to the function of JAK2 is its PTK domain. We report the 2.0 Å crystal structure of the active conformation of the JAK2 PTK domain in complex with a high-affinity, pan-JAK inhibitor that appears to bind via an induced fit mechanism. This inhibitor, the tetracyclic pyridone 2-tert-butyl-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinoline-7-1, was buried deep within a constricted ATP-binding site, in which extensive interactions, including residues that are unique to JAK2 and the JAK family, are made with the inhibitor. We present a structural basis of high-affinity JAK-specific inhibition that will undoubtedly provide an invaluable tool for the further design of novel, potent, and specific therapeutics against the JAK family.
Introduction
The Janus kinases (JAKs) are an important family of intracellular protein tyrosine kinases (PTKs), with 4 mammalian members, JAK1, JAK2, JAK3, and TYK2,1-5 as well as homologs in chicken,6 fish,7 and Drosophila.8 The JAKs play critical roles in several important intracellular signaling pathways, including the eponymous JAK/STAT pathway,9 central to the mediation of cytokine signaling.10,11 It is this pivotal role in cytokine signaling that underpins the notion that specific JAK inhibitors may be therapeutically deployed in situations where cytokine activity results in disease. Important examples of this include autoimmune diseases such as rheumatoid arthritis and psoriasis,12,13 myeloproliferative syndromes such as polycythemia vera,14-17 leukemias,18-20 lymphomas,21 and cardiovascular disease22,23 inter alia.
Members of the JAK family each share a characteristic domain structure,2 with a C-terminal PTK domain (known as the JAK homology-1 [JH1] domain), immediately adjacent to a kinase-like domain (JH2), and 5 additional JAK homology domains (JH3-JH7). While the JH2 domain appears to possess an important regulatory role on the PTK activity of the JH1 domain,24-29 the precise mechanism by which this control is exerted is currently poorly understood. The role of a putative SH2-like domain (JH3/JH4)2,30 is also unknown at present, whereas the function of a well-defined band F ezrin-radixin-moesin homology (FERM) domain (JH7)31,32 appears to be critical for interaction of the JAKs with their cognate receptors and regulatory proteins.
The JAKs coordinate specifically to different receptors, for example, JAK3 appears to be associated with cytokine receptors that include the γc chain of the interleukin-2 (IL-2) receptor (eg, IL-4, IL-7, etc), whereas JAK2 is associated with a wide range of cytokine receptors, including those activated by growth hormone,33-35 erythropoietin,36 prolactin,37 granulocyte colony-stimulating factor (G-CSF),38 and IL-3,39 as well as some G-protein-coupled receptors. It is therefore unsurprising that murine JAK2 knockouts are embryonically lethal, highlighting JAKs' essential physiologic role.40,41
With their anticipated role in pathologies, the JAKs represent attractive therapeutic targets, yet also represent a considerable challenge in the design of small-molecule JAK-specific inhibitors. Specifically, the JAK2 enzyme represents an important new drug discovery target, with potential therapeutic benefit for JAK2 inhibitors in the treatment of polycythemia vera14-17 and cancer.18-20 The potential for toxic side effects in the therapeutic use of a JAK2 inhibitor, as a consequence of the important role the enzyme plays in embryonic development,40,41 and in the ongoing provision of erythrocytes and thrombocytes via its requirement for signaling by the EPO and TPO receptors, would be abrogated in a clinical situation through regular blood transfusions.
In this study we have determined the 2.0 Å resolution crystal structure of JAK2 kinase domain in complex with the JAK-specific inhibitor 2-tert-butyl-9-fluoro-3,6-dihydro-7H-benzo[h]imidazo-[4,5-f]isoquinoline-7-one.42 The structure provides not only critical insights into how this molecule exerts its JAK specificity but also an important first step in trying to understand the mechanism by which JAK kinases are regulated.
Materials and methods
Protein expression and purification
The kinase domain of human JAK2 (residues 835-1132 [PubMed NM 004972]) was cloned into pFastBac, which allows the protein to be expressed fused to a GST cleavable tag. Recombinant bacmid DNA containing the JAK2 insert was isolated and transfected to Spodoptera frugiperda (Sf9) insect cells. Baculovirus obtained from the transfection was used to infect Sf9 cells grown in suspension to a density of 2 × 106 cells per mL at a multiplicity of infection greater than 10 and harvested 48 hours after infection. Cells were resuspended into a buffer consisting of 20 mM Tris HCl, pH 8.5, 250 mM NaCl, 0.5% thesit, 5% glycerol, and 1 mM DTT supplemented with complete protease inhibitors mixture (Roche Diagnostics, Mannheim, Germany), lysed by sonication, and centrifuged at 45 000g for 1 hour. The supernatant was filtered and recirculated onto a GST resin (Scientifix, Victoria, Australia). After extensive washes, the fusion protein was eluted, and fractions containing GST-JAK2 were pooled and concentrated to 2 mL and incubated with α-thrombin (Sigma, St Louis, MO) overnight at 4°C. The kinase was demonstrated to be catalytically active. The protein was then incubated with 3 × molar ratio of inhibitor (2-tert-butyl-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinoline-7-1) (termed CMP6)42 before being loaded onto Superdex 75 gel filtration column (HiLoad 16/60) equilibrated in 20 mM Tris pH 8.5, 250 mM NaCl, and 1 mM DTT. JAK2-inhibitor complex containing fractions were pooled and concentrated to 10 mg/mL for crystallization trials.
Crystallization
Crystals were grown at 20°C using the hanging drop vapor-diffusion method. Purified JAK2-CMP6 complex was mixed with an equal volume of a reservoir solution containing 28% polyethylene glycol 4000, 0.2 M ammonium acetate, and 0.1 M citrate pH 6.0. Crystals formed after 1 to 3 days.
X-ray data collection, structure determination, and refinement
The crystals were flash frozen prior to data collection using 5% glycerol as a cryoprotectant. A 2.0 Å data set was merged and processed with an HKL software package (HKL Research, Charlottesville, VA). The crystals, with unit cell dimensions a = b = 111.2 Å and c = 70.5 Å belong to space group P41, with 2 monomers in the asymmetric unit. The structure was determined by the molecular replacement method using the program AmoRe in the CCP4 suite. EGFRK was used as a search probe (Protein Data Bank code 1M14). Subsequent runs of manual fitting and crystallographic refinement was performed in CNS43 and O.44 Further refinement was carried out using the molecular refinement program REFMAC.45 The final model comprises 2 monomers (residues 843-1132), 395 water molecules and 2 inhibitor molecules. See Table 1 for a summary of data collection and refinement statistics. The loop connecting β4 and β5 (residues 919-924) is poorly ordered and as such is not included in the final model of the JAK2 structure. The coordinates have been deposited in the Protein Data Bank (2B7A).
Data collection and refinement statistics
Statistic . | Measurement . |
|---|---|
| Data collection statistics | |
| Temperature | 100K |
| Space group | P41 |
| Cell dimensions, Ȧ; a, b, c | 111.2, 111.2, 70.5 |
| Resolution, Ȧ, range* | 100-2.00 (2.07-2.00) |
| Total no. observations | 146053 |
| No. unique observations | 56521 |
| Multiplicity | 2.58 |
| Data completeness, %* | 96.8 (97.1) |
| No. data greater than 2σ1* | 73 (39.8) |
| I/σ1* | 15.95 (2.12) |
| Rmerge1, %* | 6.5 (55.5) |
| Refinement statistics | |
| Nonhydrogen atoms, no. | |
| Protein | 4766 |
| Ligand | 46 |
| Water | 395 |
| Resolution, Ȧ, range | 100-2.00 |
| Rfactor†, % | 20.8 |
| Rfree‡, % | 24.9 |
| Rms deviations from ideality, Ȧ | |
| Bond lengths | 0.005 |
| Bond angles | 1.075 |
| Impropers | 0.667 |
| Dihedrals | 21.66 |
| Ramachandran plot, most favored and allowed region, % | 88 (11.6) |
| B factors, Ȧ2 | |
| Average main chain | 39.7 |
| Average side chain | 43.0 |
| Average water molecule | 44.1 |
| Ligand | 26.0 |
| Rms deviation bonded Bs | 2.0 |
Statistic . | Measurement . |
|---|---|
| Data collection statistics | |
| Temperature | 100K |
| Space group | P41 |
| Cell dimensions, Ȧ; a, b, c | 111.2, 111.2, 70.5 |
| Resolution, Ȧ, range* | 100-2.00 (2.07-2.00) |
| Total no. observations | 146053 |
| No. unique observations | 56521 |
| Multiplicity | 2.58 |
| Data completeness, %* | 96.8 (97.1) |
| No. data greater than 2σ1* | 73 (39.8) |
| I/σ1* | 15.95 (2.12) |
| Rmerge1, %* | 6.5 (55.5) |
| Refinement statistics | |
| Nonhydrogen atoms, no. | |
| Protein | 4766 |
| Ligand | 46 |
| Water | 395 |
| Resolution, Ȧ, range | 100-2.00 |
| Rfactor†, % | 20.8 |
| Rfree‡, % | 24.9 |
| Rms deviations from ideality, Ȧ | |
| Bond lengths | 0.005 |
| Bond angles | 1.075 |
| Impropers | 0.667 |
| Dihedrals | 21.66 |
| Ramachandran plot, most favored and allowed region, % | 88 (11.6) |
| B factors, Ȧ2 | |
| Average main chain | 39.7 |
| Average side chain | 43.0 |
| Average water molecule | 44.1 |
| Ligand | 26.0 |
| Rms deviation bonded Bs | 2.0 |
The value in parentheses is for the highest resolution bin (approximate interval, 0.1A)
Rmerge = Σ|/hkl–</hkl>|/Σ/hkl
Rfactor = Σ/hkl||Fo–|Fc|||/Σ/hkl|Fo| for all data except for 4%, which was used for the Rfree calculation
Results
JAK2 crystal structure
To determine the precise atomic architecture of the JAK2 PTK domain and the mechanism by which tetracyclic pyridones achieve high specificity toward the JAKs, we determined the 2.0 Å resolution crystal structure of human JAK2 PTK domain (843-1132) in complex with 2-tert-butyl-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinoline-7-one (CMP6) (Figure 1A). JAK2 crystallizes as 2 molecules per asymmetric unit, although there is nothing to suggest a higher-order oligomeric JAK2 assembly within the unit cell. The root mean square (rms) deviation between the 2 monomers in the asymmetric unit is 0.56 Å; accordingly, unless explicitly stated, structural analysis will be confined to one monomer.
JAK2 PTK domain exhibits an architecture typical of previously reported protein kinases, namely a small and large N-terminal and C-terminal lobe, respectively. The N-terminal lobe comprises a curled β sheet of 5 antiparallel β-strands (β1 to β5) and one α-helix (αC). The COOH-terminal lobe is mainly α-helical with 8 α-helices (αD-αK) and 3 3/10 helices (3/10B, C, D), and 3 pairs of antiparallel β-strands (β7-β8, β6-β9, and β10-β11).
While the JAK2 kinase domain appears to conform in most respects to the structures of other PTKs, a loop structure located between amino acids 1056 and 1078, termed the JAK2 kinase insertion loop, does not resemble a feature observed in any other kinase. Nonetheless, this loop is a conserved feature of the JAK family (Figure 1B) and most likely plays an important functional role within the family. This loop packs loosely against the base of the C-terminal lobe and is relatively mobile and solvent accessible. Notably, the serine located at position 1056 (Figure 1B) is conserved in all known JAK family kinases, with the exception of the Drosophila JAK family member hopscotch. Moreover, the exposed nature of this serine suggests that it may be involved in a phosphorylation-dependent regulatory role in JAK function.
Overview of the crystal structure of JAK2 kinase domain and sequence alignment of the members of the JAK family around the lip region. (A) Ribbon representation of the crystal structure of JAK2 PTK domain in complex with CMP6 (2-tert-butyl-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinoline-7-one). The N-terminal lobe (residues 840-931) shown in gray comprises a 5-stranded antiparallel β-sheet (β1 to β5) and one α-helix (αC). The COOH-terminal lobe (residues 932-1132) shown in green comprises 8 α-helices (αD-αK), and 3 3/10 helices (3/10B, 3/10C, 3/10D), and 3 pairs of antiparallel β-strands (β7-β8, β6-β9, and β10-β11). The JAK2 lip contains one 3/10 helix (3/10C) and one α-helix (αH) connected by a short linker. The glycine loop is colored in orange, the hinge region between the 2 lobes in yellow, the catalytic loop in blue, the activation loop in red, and the JAK2 lip in dark blue. The bound CMP6 is presented in a ball-and-stick representation with carbon atoms in gray, oxygen atoms in red, nitrogen atoms in blue, and fluoride atom in pink. (B) Amino acid sequence alignment of human JAK2 PTK domain with the other members of the JAK family TYK2, JAK3, and JAK1 and the kinase domain of FAK and LCK around the Lip region. The secondary structure of JAK2 is illustrated directly above the sequence alignment. Cylinders delineate α-helices. Residues that are conserved among the JAK kinases sequence are highlighted in red. Dark gray boxes indicate residues conserved to at least 75% within the JAK kinase family. Light gray boxes indicate conservatively substituted residues. GenBank accession codes for JAK2, TYK2, JAK3, JAK1 are NP_004963, AAS37680, NP_000206, and NP_002218, respectively.
Overview of the crystal structure of JAK2 kinase domain and sequence alignment of the members of the JAK family around the lip region. (A) Ribbon representation of the crystal structure of JAK2 PTK domain in complex with CMP6 (2-tert-butyl-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinoline-7-one). The N-terminal lobe (residues 840-931) shown in gray comprises a 5-stranded antiparallel β-sheet (β1 to β5) and one α-helix (αC). The COOH-terminal lobe (residues 932-1132) shown in green comprises 8 α-helices (αD-αK), and 3 3/10 helices (3/10B, 3/10C, 3/10D), and 3 pairs of antiparallel β-strands (β7-β8, β6-β9, and β10-β11). The JAK2 lip contains one 3/10 helix (3/10C) and one α-helix (αH) connected by a short linker. The glycine loop is colored in orange, the hinge region between the 2 lobes in yellow, the catalytic loop in blue, the activation loop in red, and the JAK2 lip in dark blue. The bound CMP6 is presented in a ball-and-stick representation with carbon atoms in gray, oxygen atoms in red, nitrogen atoms in blue, and fluoride atom in pink. (B) Amino acid sequence alignment of human JAK2 PTK domain with the other members of the JAK family TYK2, JAK3, and JAK1 and the kinase domain of FAK and LCK around the Lip region. The secondary structure of JAK2 is illustrated directly above the sequence alignment. Cylinders delineate α-helices. Residues that are conserved among the JAK kinases sequence are highlighted in red. Dark gray boxes indicate residues conserved to at least 75% within the JAK kinase family. Light gray boxes indicate conservatively substituted residues. GenBank accession codes for JAK2, TYK2, JAK3, JAK1 are NP_004963, AAS37680, NP_000206, and NP_002218, respectively.
Activation loop and implications for regulation
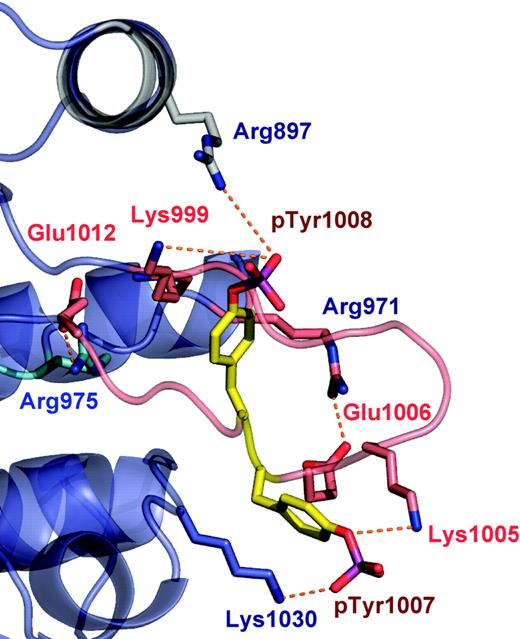
PTKs exist in either a catalytically inactive state or catalytically active state.46 These conformational states are largely governed by the phosphorylation of tandem tyrosine residues within the activation loop that results in the expulsion of the activation loop from the active site. In addition, this conformational switch repositions the highly conserved Asp-Phe-Gly motif (residues 994-996 in JAK2) in the proximity of the active site, allowing a shift in the position of the αC helix. The functional role of these tyrosine residues varies between the JAKs, and in JAK2, phosphorylation of the Tyr 1007 is critical for activity.47 We have crystalized the JAK2 PTK domain in an active conformation, in which the activation loop is expelled fully from the ATP-binding pocket, phosphorylated at positions Tyr 1007 and Tyr 1008 (Figure 2). The 2 Fo - Fc and Fo - Fc electron density maps showed clearly that Tyr 1007 and Tyr 1008 were phosphorylated (data not shown). Moreover, mass spectrometry analysis on tryptic fragments of the JAK2 PTK domain also was consistent with these residues being phosphorylated (data not shown). In addition, a salt bridge observed between Lys882 (β3) and Glu898 (αC helix) also represents a characteristic feature of active PTKs.
The well-ordered conformation of the JAK2 activation loop (residues 994-1023) is stabilized by a large number of interactions, including 2 antiparallel (β9/β6 and β10/β11) and 2 arginine residues, Arg 971 and Arg 975, which interact with the base and the tip of the activation loop, respectively (Figure 2). In addition, a number of lysine residues stabilize the conformation of pTyr 1007 (Lys 1005, Lys 1009, and Lys 1030) and pTyr 1008 (Lys 999). The conformation of the JAK2 activation loop is similar to that of other PTKs, providing a docking site for protein substrates, ATP analogs, and other regulatory proteins. The high degree of solvent exposure of pTyr1007 is consistent with its critical role in binding to the JAK2 regulatory proteins such as SOCS-1 and PTP1B.48-51
Comparative analyses
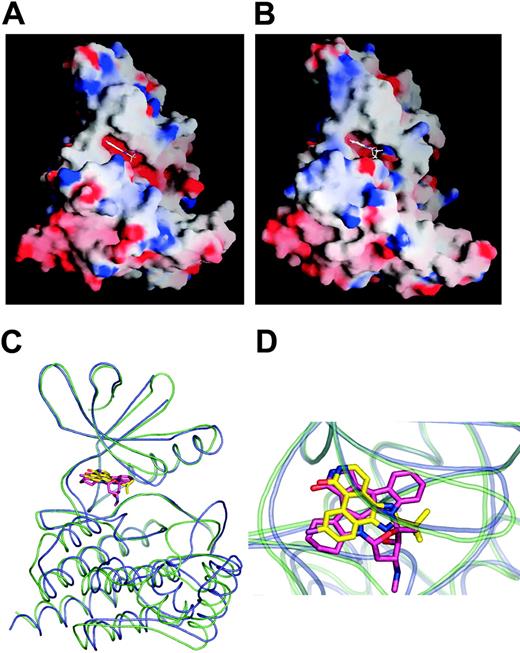
The individual N- and C-terminal lobes superpose well with previously determined PTK structures. For example, the r.m.s.d and sequence similarity of the N-terminal lobe of JAK2 PTK domain and other active PTK N-terminal lobes are IRK (1.45 Å over 63 Cα atoms, 26% identity); EGFR (1.38 Å over 69 Cα atoms, 23% identity); LCK (1.19 Å over 65 Cα atoms, 38% identity); FAK (1.46 Å over 69 Cα atoms, 27% identity); and ZAP 70 (1.25 Å over 66 Cα atoms, 35% identity). The root mean square deviation (rmsd) and sequence similarity of the C-terminal lobe of JAK2 PTK domain and other active PTK C-terminal lobes are IRK (1.17 Å over 151 Cα atoms, 39% identity); EGFR (0.90 Å over 163 Cα atoms, 43% identity); LCK (1.04 Å over 155 Cα atoms, 39% identity); FAK (0.97 Å over 155 Cα atoms, 40% identity); and ZAP 70 (1.19 Å over 166 Cα atoms, 36% identity). However, in comparison to the other PTKs, the juxtapositioning of the respective lobes is significantly different. For example, after superposing the N-terminal lobe of JAK2 PTK onto the N-terminal lobes of IRK, EGF, LCK, and FAK kinases, a 13.9°, 13.6°, 10.3°, and 18.6° rotation, respectively, is required to superpose the corresponding C-terminal lobes. As a consequence, the opening angle of the active JAK2 PTK structure is significantly more “closed” than any other active PTK structure determined in presence of the nucleotides or analogs (Figure 3A,B). The swing of the N-terminal lobe toward the C-terminal lobe markedly narrows the JAK2 ATP binding site, which is constricted further via the conformations of the activation loop and the glycine loop; the glycine loop (consensus sequence G-xG-x-Φ-G, where Φ is either mainly Phe or Tyr), known to be important in substrate and nucleotide binding, is oriented toward and makes contacts with the activation loop and catalytic loop.
The activation loop of JAK2 PTK domain. The ribbon representation of the polypeptide backbone of the activation loop is shown in pink. Tyr1007 and Tyr 1008 are the sites of phosphorylation within the activation loop. Hydrogen bonds interactions are shown as dotted lines.
The activation loop of JAK2 PTK domain. The ribbon representation of the polypeptide backbone of the activation loop is shown in pink. Tyr1007 and Tyr 1008 are the sites of phosphorylation within the activation loop. Hydrogen bonds interactions are shown as dotted lines.
Comparison of JAK2 and JAK3 structures. (A) Molecular surface representation of JAK2 PTK domain in complex with CMP6. (B) Molecular surface representation of JAK3 PTK domain in complex with AFN941. The Graphical Representation and Analysis of Surface Properties program (GRASP)52 was used to color code surfaces by electrostatic potential. (C) Superposition of JAK2 kinase domain bound to CMP6 and JAK3 kinase domain bound to AFN941. JAK2 is colored in violet, and JAK3 is colored in green. (D) Close-up of the catalytic cleft of JAK2 and JAK3 kinase domain. CMP6 is colored in yellow and AFN941 in pink.
Comparison of JAK2 and JAK3 structures. (A) Molecular surface representation of JAK2 PTK domain in complex with CMP6. (B) Molecular surface representation of JAK3 PTK domain in complex with AFN941. The Graphical Representation and Analysis of Surface Properties program (GRASP)52 was used to color code surfaces by electrostatic potential. (C) Superposition of JAK2 kinase domain bound to CMP6 and JAK3 kinase domain bound to AFN941. JAK2 is colored in violet, and JAK3 is colored in green. (D) Close-up of the catalytic cleft of JAK2 and JAK3 kinase domain. CMP6 is colored in yellow and AFN941 in pink.
Superposition of JAK2/CMP6 complex with the recently reported JAK3/AFN94153 complex exemplifies the “closed” conformation that JAK2 adopts when bound to CMP6 (Figure 3C,D). Although the N-terminal domain of JAK2 superposes well with JAK3 (0.86 Å over 83 Cα atoms, 70% identity), a 5-degree rotation is required to superpose the corresponding C-terminal lobes. Significant differences also are seen in the conformations of the hinge region, the glycine loop, the activation loop, and the JAK insertion loop. In the reported JAK3 structure, the presence of a DTT molecule in the phosphate-binding region, in addition to the bulky staurosporine analog, appears to push the glycine loop away from the active site, resulting in a loss of contacts with the activation and catalytic loop and a more open site than the constricted site seen in JAK2. Furthermore, the activation loops differ markedly just before the APE motif, while by contrast the phosphorylated tyrosines adopt a similar conformation in both structures.
Mode of inhibitor binding
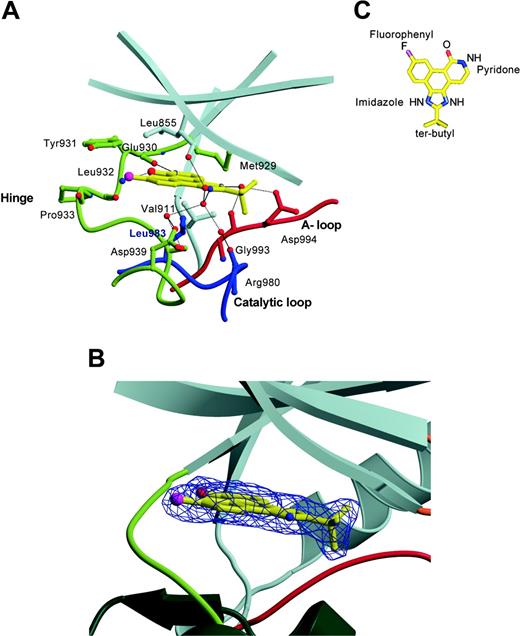
The JAK-specific inhibitor sits snugly within the constricted ATP-binding site that lies deep between the 2 lobes, occupying a site where the adenine base resides. The inhibitor is well ordered; moreover, the mode of binding of the inhibitor within the JAK2 PTK domain structure is unambiguous, as evidenced by the electron density maps. The inhibitor is orientated such that the fluorophenyl moiety points toward the bulk solvent, the pyridone moiety is orientated toward the gatekeeper residue (Met 929), and the tert-butyl group points toward the tip of the glycine loop (Figure 4). There is high shape complementarity between the planar ring system of the inhibitor and the JAK2 PTK, in which the inhibitor buries 225 Å2 of its available 516 Å2 surface area, thereby making numerous contacts with the residues lining the active site.
CMP6 is predominantly hydrophobic and, accordingly, forms a large number of van der Waals interactions with JAK2 PTK domain (Figure 4 and Table 2). The planar ring system of the inhibitor is sandwiched between the hydrophobic residues of the N-terminal lobe (Leu 855, Val 863, Ala 880, Val 911), the C-terminal lobe (Leu 983 and Gly 935), and the hinge (Met 929, Tyr 931). In addition, the pyridone ring forms 2 direct hydrogen bonds, with the hinge region between the N- and C-lobes of JAK2 PTK (pyridoneN2 and pyridoneO0 interacts with Glu 930O and Leu 932N, respectively) that mimic those observed between the adenine group of ATP and other PTKs. The imidazole moiety participates in a network of water-mediated hydrogen bonds, the imidazoleN1 group interacting with Asp 939OD1, Ser 936N,OG, Leu855O, and Arg 980O; the imidazoleNO interacting with Gly 993O and Asp 994OD1. Interestingly, the carbonyl group of Gly 993 points toward the ATP-binding pocket, whereas in all the PTK structures examined (including JAK3), the corresponding carbonyl group points toward the core of the C-terminal lobe. The hydrophobic t-butyl group of the inhibitor is not well accommodated in the JAK2 active site, being located within and adjacent to a polar pocket that includes Asp 994, Arg 980, Asn 981, Asn 859, and Lys 882, a pocket that typically coordinates Mg2+ ions. The glycine loop was observed not to participate in inhibitor contacts in the JAK2 PTK domain, with the Phe 860 residue pointing away from the active site. Instead, the glycine loop collapses over and restricts the active site, with Asn 859 making a water-mediated hydrogen bond to the conserved Asp 994 and a hydrogen bond to the conserved catalytic residue Asp 976 of the C-terminal lobe. Previously, induced-fit binding of inhibitors to PTKs have been described, and a similar conformation of this loop has been observed in both the fibroblast growth factor receptor tyrosine kinase (FGFR1) in complex with a high-affinity inhibitor,54 and the Abl kinase in complex with Glivec.55 Neither ATP nor the pan-kinase inhibitor staurosporine can be accommodated within the observed active site of JAK2 PTK without having to invoke conformational changes in either the glycine loop or Asp 994, which is indicative of JAK2 PTK being able to exhibit a degree of malleability in accommodating substrates and inhibitors.
Contacts between inhibitor and JAK2 PTK domain
Inhibitor . | JAK2 PTK . | Nature of interaction . |
|---|---|---|
| Imidazole moiety | ||
| C0 | Leu 983CD2 | VDW |
| C1 | Leu 983CD2 | VDW |
| N0 | Val 863CG2, Asp 994OD1, Gly 993O | VDW; Water-mediated H-BOND |
| N1 | Asp 939OD1, Ser 936N,OG, Arg 980O, Leu 855O | Water-mediated H-BOND |
| C4 | Leu 855O | VDW |
| C5 | Leu 855CD2, Gly 935CA | VDW |
| C6 | Leu 855CD2, Leu 932CD2, Gly 935CA | VDW |
| C7 | Leu 855CD2, Leu 932O, Tyr 931CE1 | VDW |
| C8 | Leu 855CD1, Leu 983CD2 | VDW |
| F | Leu 855CD2, Leu 932O, Gly 935CA,N, Tyr 931CE1,OH | VDW |
| Pyridone moiety | ||
| C9 | Leu 983CD1,CD2 | VDW |
| C10 | Leu 983CD1,CD2 | VDW |
| C11 | Leu 983CD1, Ala 880CB, Gly 930O, Leu 932N | VDW |
| N2 | Leu 983CD1, Ala 880CB | VDW |
| N2 | Glu 930O | H-BOND |
| C12 | Leu 983CD1, Met 929SD, Ala 880CB, Glu 930O, Val 911CG2 | VDW |
| C13 | Gly 993O, Leu 983CD1, Met 929CE | VDW |
| O0 | Leu 932CA,CB,O, Ala 880CB, Glu 930O, Tyr 931CD1,CA,C, | VDW |
| O0 | Leu 932N | H-BOND |
| t-butyl moiety | ||
| C15 | Asn 981OD1, Arg 980O | VDW |
| C16 | Asp 994OD1, Val 863CG2 | VDW |
| C17 | Lys 857N,C,O Gly 856CA,C | VDW |
Inhibitor . | JAK2 PTK . | Nature of interaction . |
|---|---|---|
| Imidazole moiety | ||
| C0 | Leu 983CD2 | VDW |
| C1 | Leu 983CD2 | VDW |
| N0 | Val 863CG2, Asp 994OD1, Gly 993O | VDW; Water-mediated H-BOND |
| N1 | Asp 939OD1, Ser 936N,OG, Arg 980O, Leu 855O | Water-mediated H-BOND |
| C4 | Leu 855O | VDW |
| C5 | Leu 855CD2, Gly 935CA | VDW |
| C6 | Leu 855CD2, Leu 932CD2, Gly 935CA | VDW |
| C7 | Leu 855CD2, Leu 932O, Tyr 931CE1 | VDW |
| C8 | Leu 855CD1, Leu 983CD2 | VDW |
| F | Leu 855CD2, Leu 932O, Gly 935CA,N, Tyr 931CE1,OH | VDW |
| Pyridone moiety | ||
| C9 | Leu 983CD1,CD2 | VDW |
| C10 | Leu 983CD1,CD2 | VDW |
| C11 | Leu 983CD1, Ala 880CB, Gly 930O, Leu 932N | VDW |
| N2 | Leu 983CD1, Ala 880CB | VDW |
| N2 | Glu 930O | H-BOND |
| C12 | Leu 983CD1, Met 929SD, Ala 880CB, Glu 930O, Val 911CG2 | VDW |
| C13 | Gly 993O, Leu 983CD1, Met 929CE | VDW |
| O0 | Leu 932CA,CB,O, Ala 880CB, Glu 930O, Tyr 931CD1,CA,C, | VDW |
| O0 | Leu 932N | H-BOND |
| t-butyl moiety | ||
| C15 | Asn 981OD1, Arg 980O | VDW |
| C16 | Asp 994OD1, Val 863CG2 | VDW |
| C17 | Lys 857N,C,O Gly 856CA,C | VDW |
Mode of binding of CMP6 to JAK2 kinase domain. (A) Interactions between CMP6 and JAK2 kinase domain. The side chains of residues that interact with the inhibitor are shown, as are main-chain atoms and water molecules participating in hydrogen bonds. The N-terminal lobe is colored in gray, the hinge region in yellow-green, the activation loop in red, and the catalytic loop in blue. Selected hydrogen bonds are shown as black lines. (B) View of the catalytic cleft of JAK2 bound to CMP6 presented in a ball-and-stick representation and covered with the final 2Fo-Fc electron density map contoured at 1σ. The coloring of the backbone representation is the same as in panel A. (C) Structural formula of CMP6 presented in a ball-and-stick representation.
Mode of binding of CMP6 to JAK2 kinase domain. (A) Interactions between CMP6 and JAK2 kinase domain. The side chains of residues that interact with the inhibitor are shown, as are main-chain atoms and water molecules participating in hydrogen bonds. The N-terminal lobe is colored in gray, the hinge region in yellow-green, the activation loop in red, and the catalytic loop in blue. Selected hydrogen bonds are shown as black lines. (B) View of the catalytic cleft of JAK2 bound to CMP6 presented in a ball-and-stick representation and covered with the final 2Fo-Fc electron density map contoured at 1σ. The coloring of the backbone representation is the same as in panel A. (C) Structural formula of CMP6 presented in a ball-and-stick representation.
Discussion
With many kinase inhibitors in clinical development, an important new class of therapeutic drugs is beginning to emerge. Spearheaded by drugs such as imatinib (Glivec),56 gefitinib (Iressa), and erlotinib (Tarceva),57 the PTKs offer fertile ground for drug discovery.58
While members of the broader JAK family each present themselves as excellent drug discovery targets for diseases of the immune system, JAK2, by dint of gene rearrangement, appears to be an important therapeutic target for a number of hematologic cancers such as atypical chronic myelogenous leukemia18-20 and T-cell acute lymphoblastic leukemia.21 More recently, the observation that activating JAK2 mutations seem to underpin the etiology of a spectrum of myeloproliferative diseases, such as PV, ET, and chronic IMF,14-17 suggests that JAK2 inhibitors might be therapeutic in a range of hematologic malignancies. To date, however, it has proven challenging to generate JAK-specific kinase inhibitors. Moreover, the high degree of identity within the kinase domains of all members of the JAK family has made it exceedingly difficult to design inhibitors specific for one or other members of the family. Although Boggon et al53 have recently reported the 2.55 Å crystal structure of JAK3 kinase domain, the JAK3 structure described is complex with a nonspecific staurosporine-based inhibitor and therefore does not adequately provide a structural basis for the design of JAK-specific inhibitors.
The published literature is replete with references to the use of AG490, a purported JAK2-specific inhibitor, in the elucidation of JAK2-dependent pathways. In our hands and others, this molecule is equipotent on at least 5 other PTKs.59 WHI-P131, a JAK3-specific inhibitor,60 is similarly not as specific for JAK3 as it was originally asserted to be (data not shown). To date, the most potent and specific JAK inhibitors reported in the literature are the JAK3 inhibitor CP690550 developed by Pfizer that prevents organ transplant rejection61 and the JAK2/TYK2/JAK3-selective JAK inhibitor developed by Merck Research Laboratories.42 This latter inhibitor (CMP6), based on a tetracyclic pyridone scaffold, has been reported to potently inhibit the JAK kinase family with a IC50 of 1 nM for JAK2 and TYK2, 5 nM for JAK3, and 15 nM for JAK1, while displaying significantly weaker affinities (130 nM to more than 10 μM) for other PTKs. Our own in-house data confirm these relative IC50 values (data not shown). To better understand the prospects of generating specific JAK family member-specific inhibitors, we have focused upon the generation of co-crystal structures of members of this family with potent and specific JAK inhibitors.
What are the unique features of the JAK2 PTK active site that confer specificity toward CMP6? Due to the high degree of conservation of the ATP-binding site within the PTKs, specificity toward tetracyclic pyridones is likely to be achieved by a unique architecture and constellation of residues within the ATP-binding pocket. First, in JAK2, the unique constricted nature of the active site permits extensive interactions to be made with the inhibitor (the inhibitor is akin to a penny in a slot), whereas other PTK family members have a more accessible active site and consequently will not exhibit a high degree of shape complementarity with the JAK-specific inhibitor (Figure 2B). The planarity of the compound could therefore be an important factor in determining selectivity. The related trisubstituted imidazole, compound 5 (Supplemental Document S1 and Figure S1, available on the Blood website; click on the Supplemental Materials link at the top of the online article), a nonplanar precursor of CMP6,42 displays only μM affinity toward JAK2 and JAK3 in our hands (data not shown), suggesting that this planarity is a key feature of the preference of CMP6 for JAK family members. Comparison of our JAK2/CMP6 complex structure with the recently published JAK3/AFN941 complex structure exemplifies how the size and shape of the ligand can impact binding characteristics and potency (Figure 3). We anticipate that the presence of the indolocarbazole ring system of staurosporine would displace the side chain of Asp 994 and disrupt the interaction between Asp 994 and Lys 882. In addition, the presence of the tetrahydroindole ring and the inherent rigidity of staurosporine is likely to relate to the outward movement of the hinge region observed in the JAK3 structure, which results in a more open structure, a reduction in shape complementarity, and therefore a decrease in potency for the pan-kinase inhibitor.
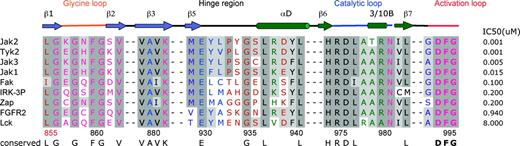
Second, a number of side chains (Met 929, Tyr 931, Leu 932) within the hinge of JAK2 PTK domain, a hypervariable region across the PTK family, interact with CMP6. However, this hinge region is well conserved within the JAK family and appears to be a key region that determines selectivity toward the tetracyclic pyridone (Figure 5). Moreover, the presence of a conserved Pro (Pro 933 in JAK2) among the JAK family is likely to introduce rigidity into the hinge region, which may represent an important factor in selectivity. Together, these features could account for the relative JAK specificity of CMP6 compared to other members of the PTK family.
In addition, the Met 929 gatekeeper residue appears to be a reasonable indicator of potent binding to CMP6. For example, PTKs that possess a methionine at an equivalent position (FAK, IRK, and ZAP 70) display an IC50 for CMP6 around the 200-nM range, whereas other PTKs that possess either a Thr or Val display an IC50 in the μM range (Figure 5). The gatekeeper residue is known to determine the shape and size of the so-called back pocket, which is also defined by the invariant Glu 898 and Leu 902, Val 911, Leu 927, Gly 993, and Asp 994. In JAK2, Met 929 is orientated toward the center of the pocket, sterically hindering the close contact of CMP6 with Leu 902 and Leu 927 of the back pocket. The presence of Met 929 also precludes a reverse orientation of the tetracyclic pyridine, where the hydrophobic fluorophenyl group would position itself in the back pocket. Such a binding mode is favored by the structurally related first-generation p38 MAP kinase inhibitors, such as SB-203580 (which also possesses a fluorophenyl moiety), when bound to the serine/threonine kinases p38 MAPK and ERK.62-65 Consequently, Met 929 simultaneously constricts the active site and maximizes its shape complementarity to and sterically constrains the pyridone group. However, Met 929 is not the sole diagnostic for selectivity toward that JAK-specific inhibitor: the combination Met 929, Tyr/Phe 931, and Leu/Val 932 within the JAK family appears to represent a prerequisite for tetracyclic pyridone specificity and a unique characteristic of the JAK kinases (Figure 5). Although the hinge region is well conserved between members of the JAK family, subtle yet significant differences could be exploited for the design of selective JAK inhibitors.
Amino acid sequence alignment of the ATP-binding site region of human JAK2 PTK domain with the other members of the JAK family TYK2, JAK3, and JAK1 and the kinase domain of FAK, IRK, ZAP-70, FGFR2, and LCK. The secondary structure of JAK2 is illustrated directly above the sequence alignment. Arrows delineate β-strands, and cylinders delineate α-helices. Dark gray boxes indicate conserved residues; light gray boxes, conservatively substituted residues. Residues highlighted in blue are located in the adenine-binding region; in green, in the sugar pocket; and in pink, in the phosphate-binding region. Residues accessible to solvent are colored in brown and buried residues are colored in purple. IC50s of CMP6 for each kinase are indicated on the right.
Amino acid sequence alignment of the ATP-binding site region of human JAK2 PTK domain with the other members of the JAK family TYK2, JAK3, and JAK1 and the kinase domain of FAK, IRK, ZAP-70, FGFR2, and LCK. The secondary structure of JAK2 is illustrated directly above the sequence alignment. Arrows delineate β-strands, and cylinders delineate α-helices. Dark gray boxes indicate conserved residues; light gray boxes, conservatively substituted residues. Residues highlighted in blue are located in the adenine-binding region; in green, in the sugar pocket; and in pink, in the phosphate-binding region. Residues accessible to solvent are colored in brown and buried residues are colored in purple. IC50s of CMP6 for each kinase are indicated on the right.
It is intriguing to note the 15-fold difference in the relative potency of this inhibitor for JAK2 versus JAK1. Examination of the contacts between the inhibitor and the JAK2 PTK active site (Table 1), together with the residual alignment of the JAK PTK active site (Figure 5), highlights a constellation of residues (853, 857, 859, 862, 865, 931, 934, 938, 979, and 982) that may account for the observed differences in the binding affinity of CMP6 to JAK1 and JAK2. Of these, 857 and 931 are the sole locations that make contact with the inhibitor in the JAK2 crystal structure. Lys 857, located in the phosphate-binding region of JAK2, makes a number of van der Waals contacts with the inhibitor. In JAK1 and TYK2, lysine is replaced by a glutamic acid residue, which may alter the strength and nature of the enzyme contacts with the tert-butyl moiety. Position 931, located in the adenine binding region of the hinge, is the only unique residual difference between JAK1 and the rest of the JAK family that makes direct contact with CMP6. In JAK2, JAK3, and TYK2, this residue is a tyrosine, while in JAK1 it is a phenylalanine. The presence of an intermolecular halogen bond between the fluorophenyl moiety of the inhibitor and the hydroxyl of Y931 may contribute to the observed difference in activity. Halogen bond interactions are primarily electrostatic in nature, with involvement from charge transfer, polarization, and dispersion contributing to estimated bond strengths ranging from about half to slightly greater than that of a hydrogen bond, depending on the environment.66
If we broaden our outlook and examine the nearest neighbors of those residues within the JAK2 active site that interact with CMP6, we can see a number of changes that may serve to indirectly impact the binding affinity of the inhibitor to JAK2 and JAK1. Asn859His, Ser862Lys of the glycine loop; Gln853Arg, Met865Leu, and Tyr934Ser of the solvent accessible region; Arg938Lys, Thr979Ala of the sugar region; and Ile982Val of the adenine binding region may all have the capacity to alter the environment of their adjacent residues that interact with the various moieties of the tetracyclic pyridone. We feel it is most likely the sum of these subtle changes that may account for the differences in the binding affinity of CMP6 to JAK1 and the other members of the JAK family.
The determination of the crystal structure of JAK2 PTK domain in complex with a pan-JAK inhibitor provides not only insights into the mechanism by which tetracyclic pyridones achieve specificity but also a greater understanding of the JAK-ATP binding pocket that will enable more selective drug targets to be developed against a myriad of diseases where the JAKs play a pivotal role.
Prepublished online as Blood First Edition Paper, September 20, 2005; DOI 10.1182/blood-2005-06-2413.
Supported in part by the Australian Research Council, a National Health and Medical Research Council Research Fellowship (I.L.), and a Wellcome Trust Senior Research Fellowship in Biomedical Science in Australia (J.R.)
Several authors (E.F., M.S., C.J.B., H.T., A.F.W.) have declared a financial interest in a company whose product was studied in the present work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ian Phillips and Claire Mitchell for assistance with cloning.

![Figure 1. Overview of the crystal structure of JAK2 kinase domain and sequence alignment of the members of the JAK family around the lip region. (A) Ribbon representation of the crystal structure of JAK2 PTK domain in complex with CMP6 (2-tert-butyl-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinoline-7-one). The N-terminal lobe (residues 840-931) shown in gray comprises a 5-stranded antiparallel β-sheet (β1 to β5) and one α-helix (αC). The COOH-terminal lobe (residues 932-1132) shown in green comprises 8 α-helices (αD-αK), and 3 3/10 helices (3/10B, 3/10C, 3/10D), and 3 pairs of antiparallel β-strands (β7-β8, β6-β9, and β10-β11). The JAK2 lip contains one 3/10 helix (3/10C) and one α-helix (αH) connected by a short linker. The glycine loop is colored in orange, the hinge region between the 2 lobes in yellow, the catalytic loop in blue, the activation loop in red, and the JAK2 lip in dark blue. The bound CMP6 is presented in a ball-and-stick representation with carbon atoms in gray, oxygen atoms in red, nitrogen atoms in blue, and fluoride atom in pink. (B) Amino acid sequence alignment of human JAK2 PTK domain with the other members of the JAK family TYK2, JAK3, and JAK1 and the kinase domain of FAK and LCK around the Lip region. The secondary structure of JAK2 is illustrated directly above the sequence alignment. Cylinders delineate α-helices. Residues that are conserved among the JAK kinases sequence are highlighted in red. Dark gray boxes indicate residues conserved to at least 75% within the JAK kinase family. Light gray boxes indicate conservatively substituted residues. GenBank accession codes for JAK2, TYK2, JAK3, JAK1 are NP_004963, AAS37680, NP_000206, and NP_002218, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/1/10.1182_blood-2005-06-2413/4/m_zh80010688830001.jpeg?Expires=1769296270&Signature=3YwmfREgsab5q4kyiiaWyY-TsEonf-iFh7J04BR8nrSHO7GXeYOTAfOg-ybV2G6I9vMPoF8UP0K5N84f3k15RuT~M-FKcpOBD2bprgIs8UeC3k1mlSHmziT7UGkuA4vHpca4NhnW8p7iOvR46m0NTTfCLakP2BJdA89iMFuCpOcZFdGPWVG9ZUx11z9jv3Dh6LewkDs4QeIU37JhghTSZGWAvQ~vhbxk0fxsWzSY7TtCjRkKWD6DjiUy00ToIaFEuirjAU~aCdX~40ie7hBKZf7alPUidsgzY~knGkYLmPFNloQlTlxDz3beB7BH5~h9~eedG7~L9jRghsOI-n4DIg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal