Abstract
Gene-expression profiling has identified 3 major subgroups of diffuse large B-cell lymphoma (DLBCL): germinal center B-cell-like (GCB), activated B-cell-like (ABC), and primary mediastinal DLBCL (PMBCL). Using comparative genomic hybridization (CGH), we investigated the genetic alterations of 224 cases of untreated DLBCL (87 GCB-DLBCL, 77 ABC-DLBCL, 19 PMBCL, and 41 unclassified DLBCL) previously characterized by gene-expression profiling. The DLBCL subgroups differed significantly in the frequency of particular chromosomal aberrations. ABC-DLBCL had frequent trisomy 3, gains of 3q and 18q21-q22, and losses of 6q21-q22, whereas GCB-DLBCL had frequent gains of 12q12, and PMBCL had gains of 9p21-pter and 2p14-p16. Parallel analysis of CGH alterations, locus-specific gene-expression profiles, and global gene-expression signatures revealed that DNA amplifications and gains had a substantial impact on the expression of genes in the involved chromosomal regions, and some genes were overexpressed in a DLBCL subgroup-specific fashion. Unexpectedly, specific chromosomal alterations were associated with significant changes in gene-expression signatures that reflect various aspects of lymphoma cell biology as well as the host response to the lymphoma. In addition, gains involving the chromosomal region 3p11-p12 provided prognostic information that was statistically independent of the previously defined gene-expression-based survival model, thereby improving its predictive power.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most frequent lymphoma in adults worldwide, accounting for 30% to 40% of lymphoid neoplasms.1 The diversity in clinical presentation and outcome, as well as the pathologic and biologic heterogeneity, suggest that DLBCL comprises several disease entities that may require different therapeutic approaches. Gene-expression profiling has identified 3 major subgroups of DLBCL, termed germinal center B-cell-like DLBCL (GCB-DLBCL), activated B-cell-like DLBCL (ABC-DLBCL), and primary mediastinal DLBCL (PM-BCL).2-5 These 3 subgroups of DLBCL are associated with a widely disparate clinical outcome with 5-year survival rates of 59%, 30%, and 64% in patients with GCB-DLBCL, ABC-DLBCL, and PMBCL, respectively.2-4 In addition, GCB-DLBCL is characterized by frequent REL amplifications, BCL2 translocations,3,6 and ongoing somatic hypermutation of the immunoglobulin genes.7 In contrast, ABC-DLBCL8,9 and PMBCL4,5,9 have a constitutive activation of the nuclear factor κB (NF-κB) pathway that they require for survival, which is not a feature of GCB-DLBCL.
Malignant lymphomas are genetically characterized by distinctive recurrent primary chromosomal translocations such as the t(11;14) or t(14;18) in mantle-cell and follicular lymphoma, respectively. By identifying genomic imbalances, comparative genomic hybridization (CGH) has the potential to detect less-well-characterized chromosomal aberrations in lymphomas that may play an important role in the development and progression of the disease. In DLBCL, previous cytogenetic studies have identified a plethora of clonal chromosomal aberrations, some of which are associated with particular morphologic or clinical manifestations.10-18 For example, PMBCL is associated with recurrent gains and amplifications of chromosomes 9p and 2p.19-21 The identification of GCB-DLBCL and ABC-DLBCL by gene-expression profiling now provides a new framework to evaluate the genetic alterations in DLBCL.
In the present study, we used CGH to address 3 specific questions regarding the relationship between chromosomal aberrations and gene-expression profiles in DLBCL. First, we investigated whether the 3 DLBCL subgroups defined by gene expression are characterized by distinct sets of chromosomal alterations. Second, we examined the influence that individual chromosomal aberrations have on gene-expression profiles. Finally, we tested whether specific genetic alterations add prognostic information to the gene-expression-based survival predictor for DLBCL.3
Patients, materials, and methods
Samples and patients
We studied 224 untreated de novo DLBCL samples previously characterized by gene-expression profiling using Lymphochip cDNA microarrays.3 Tumors were selected for the study on the sole basis of availability of genomic DNA obtained simultaneously with mRNA extraction from the same frozen tissue used for gene-expression profiling. There were 87 tumors classified as GCB-DLBCL, 77 as ABC-DLBCL, 19 as PMBCL, and 41 as unclassified DLBCL.22 Clinical data had been obtained from all patients according to a protocol approved by the National Cancer Institute Institutional Review Board.3 All patients had received anthracycline-based chemotherapy. Median follow-up was 2.7 years and 58% of patients died during this period. The median age of the patients was 60 years and 54% were men. Sixteen percent of patients had Ann Arbor stage I disease and 30%, 19%, and 35% had stage II, III, and IV, respectively. Thirty-eight percent of patients with DLBCL (78 cases) with available data were in the low-risk International Prognostic Index (IPI) group (IPI 0-1), 48% (99 cases) were in the intermediate-risk IPI group (IPI 2-3), and 14% (30 cases) were in the high-risk IPI group (IPI 4-5).
Comparative genomic hybridization
Comparative genomic hybridization (CGH) was performed using a commercially available CGH kit provided by Vysis (Downers Grove, IL). Hybridizations and digital image acquisition, processing, and evaluation were performed on a Cytovision Ultra workstation (Applied Imaging, Sunderland, United Kingdom) as described previously.23 Signal ratios greater than 1.25 or less than 0.75 were considered as chromosomal gains or losses, respectively. Ratios exceeding 1.5 and/or strong focal signals with the ration profile showing overrepresentation were considered as genomic amplifications. All CGH data are available at http://www.ncbi.nlm.nih.gov/sky.
Molecular analysis
To evaluate genomic gains and amplifications of potential target genes, we performed real-time quantitative polymerase chain reaction (RQ-PCR) using the ABI Prism 7700 Sequence Detector System (Applied Biosystems, Foster City, CA). The primers and probes used are listed in Supplemental Table S1, available at the Blood website; click on the Supplemental Materials link at the top of the online article. For controls, β2-microglobulin (β2M) was used in all cases and albumin (ALB) in a subset of cases. Each assay was analyzed by the comparative cycle threshold (CT) method, using the arithmetic formula provided by the manufacturer. To determine the cut-off values for a genomic gain/amplification in each probe set, 8 DNA samples from peripheral blood or placenta from healthy individuals were studied. The cut-off ratio for a genomic gain was determined as the mean ratio plus 3 standard deviation units (approximately 1.3 for each gene). In the lymphoma specimens, a ratio between the cut-off value and 2 was considered a gain, and a ratio greater than 2 was considered an amplification. A subset of the cases studied (31/109, 30%) was also investigated using a second reference locus (ALB). The results between the 2 reference genes were totally concordant in 87% of the cases and partially concordant (gain vs amplification) in 13% of the cases. In 4 cases in which the β2M locus (15q21.1) was altered by CGH (lost in 3 cases and gained in one case), ALB was used as the sole reference gene.
Statistical analysis
CGH alterations in individual cytobands were treated as categoric variables and their associations with DLBCL subgroups or gene-expression signatures were analyzed as follows. Preliminary analyses did not reveal significant differences in the effects of gains and amplifications, so we treated them as equivalent chromosomal abnormalities. Since a large number of individual chromosomal abnormalities were analyzed, there was a danger that some of the abnormalities would appear to be significant purely by chance. To avoid such false positives, we used a stepwise permutation test,24,25 which generated nominal P values that accounted for multiple hypothesis testing. This test takes into account the correlation between the different chromosomal abnormalities. Differences in abnormality frequency between subtypes were detected using a chi-squared test. Differences in gene-expression signature measures affected by genomic imbalances were detected using a t test. To further reduce the effects of multiple comparisons, we analyzed only those chromosomal abnormalities that were present in a substantial portion of our data. For the subgroup analysis, we considered only those alterations that had a frequency of greater than 20% in one or more of the DLBCL subgroups (GCB-DLBCL, ABC-DLBCL, or PMBCL). For correlation with gene-expression signatures, chromosomal abnormalities were only considered if they occurred in at least 5% of all DLBCL samples.
P values for the associations between gene-expression levels (as a continuous variable) and genomic imbalances (amplification vs gain vs normal copy number) were calculated using an analysis of variance (ANOVA) test. P values of less than .01 were considered significant to account for multiple comparisons. Overall survival was modeled using a Cox proportional hazards approach and visualized using the Kaplan-Meier method. The P values were adjusted for multiple comparisons, with the follow-up time and status at follow-up being permuted and once an abnormality was found to be significant univariately, a likelihood ratio test was used to determine whether this variable added significantly to the survival model based on gene expression.
Results
Diffuse large B-cell lymphoma subgroups identified by expression profiling are genetically distinct
CGH analysis was performed on 224 DLBCL tumors that had previously been analyzed by gene-expression profiling3 (Figure 1A-C; Table 1). Chromosomal alterations were observed in 164 of the 224 patients (73%). The number of chromosomal imbalances did not differ statistically between GCB-DLBCL (3.1 ± 3.7, n = 87), ABC-DLBCL (4.5 ± 4.5, n = 77), PM-BCL (3.3 ± 2.7, n = 19), and unclassified DLBCL (1.7 ± 2.2, n = 41). Among cases with chromosomal imbalances, most (81%) had more than one abnormality. In those cases with only one abnormality, the most frequent alteration was loss of 6q (8 cases), with 2 minimally lost regions in 6q21-q22 and 6q25-qter; these chromosomal deletions may represent early events in the development of these lymphomas.
Commonly altered chromosomal regions in different subgroups of DLBCL
. | Whole series, n (%); N = 224 . | ABC, n (%); n = 77 . | GCB, n (%); n = 87 . | PMBCL, n (%); n = 19 . | Unclassified, n (%); n = 41 . |
|---|---|---|---|---|---|
| Altered cases | 164 (73) | 63 (81) | 63 (72) | 16 (84) | 22 (54) |
| Mean no. of alterations | 3.3 | 4.5 | 3.1 | 3.3 | 1.7 |
| Mean no. of gains | 1.9 | 2.5 | 1.6 | 2.1 | 1.0 |
| Mean no. of amplifications | 0.3 | 0.4 | 0.3 | 0.4 | 0.0 |
| Mean no. of losses | 1.2 | 1.6 | 1.1 | 0.8 | 0.6 |
| Gains | |||||
| Xp | 27 (12) | 12 (16) | 12 (14) | 3 (16) | 0 (0) |
| 1q25-q32 | 26 (12) | 9 (12) | 9 (10) | 1 (5) | 7 (17) |
| 2p14-p16* | 39 (17) | 12 (15) | 15 (17) | 9 (47) | 3 (7) |
| Trisomy 3* | 14 (6) | 12 (15) | 0 (0) | 1 (5) | 1 (2) |
| 3p* | 28 (12) | 24 (31) | 1 (1) | 1 (5) | 2 (5) |
| 3q† | 22 (10) | 20 (26) | 0 (0) | 1 (5) | 1 (2) |
| 3q27-qter† | 35 (16) | 26 (33) | 4 (5) | 3 (16) | 2 (5) |
| 6p | 30 (13) | 13 (17) | 11 (13) | 1 (5) | 5 (12) |
| 7p | 22 (10) | 8 (10) | 13 (15) | 1 (5) | 0 (0) |
| 7q | 25 (11) | 10 (13) | 13 (15) | 1 (5) | 1 (2) |
| 8q23-qter | 23 (10) | 8 (10) | 10 (11) | 2 (11) | 3 (7) |
| 9p† | 14 (6) | 5 (6) | 0 (0) | 7 (37) | 2 (5) |
| 12p | 19 (8) | 4 (5) | 14 (16) | 1 (5) | 0 (0) |
| 12q12‡ | 24 (11) | 4 (5) | 18 (21) | 1 (5) | 1 (2) |
| 12q22-qter | 22 (10) | 7 (9) | 13 (15) | 1 (5) | 1 (2) |
| 18q21-q22* | 42 (19) | 26 (34) | 9 (10) | 3 (16) | 4 (10) |
| Losses | |||||
| 6q16 | 50 (22) | 26 (34) | 19 (22) | 0 (0) | 5 (12) |
| 6q21-c22* | 55 (25) | 31 (40) | 19 (22) | 0 (0) | 5 (12) |
| 8p22-pter | 19 (8) | 8 (10) | 3 (3) | 3 (16) | 5 (12) |
| 17p | 22 (10) | 14 (18) | 7 (8) | 0 (0) | 1 (2) |
. | Whole series, n (%); N = 224 . | ABC, n (%); n = 77 . | GCB, n (%); n = 87 . | PMBCL, n (%); n = 19 . | Unclassified, n (%); n = 41 . |
|---|---|---|---|---|---|
| Altered cases | 164 (73) | 63 (81) | 63 (72) | 16 (84) | 22 (54) |
| Mean no. of alterations | 3.3 | 4.5 | 3.1 | 3.3 | 1.7 |
| Mean no. of gains | 1.9 | 2.5 | 1.6 | 2.1 | 1.0 |
| Mean no. of amplifications | 0.3 | 0.4 | 0.3 | 0.4 | 0.0 |
| Mean no. of losses | 1.2 | 1.6 | 1.1 | 0.8 | 0.6 |
| Gains | |||||
| Xp | 27 (12) | 12 (16) | 12 (14) | 3 (16) | 0 (0) |
| 1q25-q32 | 26 (12) | 9 (12) | 9 (10) | 1 (5) | 7 (17) |
| 2p14-p16* | 39 (17) | 12 (15) | 15 (17) | 9 (47) | 3 (7) |
| Trisomy 3* | 14 (6) | 12 (15) | 0 (0) | 1 (5) | 1 (2) |
| 3p* | 28 (12) | 24 (31) | 1 (1) | 1 (5) | 2 (5) |
| 3q† | 22 (10) | 20 (26) | 0 (0) | 1 (5) | 1 (2) |
| 3q27-qter† | 35 (16) | 26 (33) | 4 (5) | 3 (16) | 2 (5) |
| 6p | 30 (13) | 13 (17) | 11 (13) | 1 (5) | 5 (12) |
| 7p | 22 (10) | 8 (10) | 13 (15) | 1 (5) | 0 (0) |
| 7q | 25 (11) | 10 (13) | 13 (15) | 1 (5) | 1 (2) |
| 8q23-qter | 23 (10) | 8 (10) | 10 (11) | 2 (11) | 3 (7) |
| 9p† | 14 (6) | 5 (6) | 0 (0) | 7 (37) | 2 (5) |
| 12p | 19 (8) | 4 (5) | 14 (16) | 1 (5) | 0 (0) |
| 12q12‡ | 24 (11) | 4 (5) | 18 (21) | 1 (5) | 1 (2) |
| 12q22-qter | 22 (10) | 7 (9) | 13 (15) | 1 (5) | 1 (2) |
| 18q21-q22* | 42 (19) | 26 (34) | 9 (10) | 3 (16) | 4 (10) |
| Losses | |||||
| 6q16 | 50 (22) | 26 (34) | 19 (22) | 0 (0) | 5 (12) |
| 6q21-c22* | 55 (25) | 31 (40) | 19 (22) | 0 (0) | 5 (12) |
| 8p22-pter | 19 (8) | 8 (10) | 3 (3) | 3 (16) | 5 (12) |
| 17p | 22 (10) | 14 (18) | 7 (8) | 0 (0) | 1 (2) |
The group of unclassified tumors was not included in the statistical analysis.
P < .05, after adjustment for multiple variable comparisons
P < .001, after adjustment for multiple variable comparisons
P = .059, after adjustment for multiple variable comparisons
Summary of chromosomal imbalances detected in 224 cases of untreated de novo DLBCL classified by gene-expression profiling. Red bars on the left side of the ideogram indicate losses of chromosomal material; green bars on the right side indicate gains of chromosomal material; thick green bars indicate chromosomal gains exceeding the cut-off value of 1.5 in a large chromosomal region; solid dots indicate high-level DNA amplifications. Each bar represents a chromosomal region gained or lost in a single sample. (A) GCB-DLBCL (n = 87); (B) ABC-DLBCL (n = 77); (C) PMBCL (n = 19); and (D) bar diagram indicating the frequencies of chromosomal imbalances that distinguish between ABC-DLBCL, GCB-DLBCL, and PMBCL (for statistical details see “Patients, materials, and methods”). All differences were statistically significant at P < .05, with the exception of 12q12 gains (P = .059).
Summary of chromosomal imbalances detected in 224 cases of untreated de novo DLBCL classified by gene-expression profiling. Red bars on the left side of the ideogram indicate losses of chromosomal material; green bars on the right side indicate gains of chromosomal material; thick green bars indicate chromosomal gains exceeding the cut-off value of 1.5 in a large chromosomal region; solid dots indicate high-level DNA amplifications. Each bar represents a chromosomal region gained or lost in a single sample. (A) GCB-DLBCL (n = 87); (B) ABC-DLBCL (n = 77); (C) PMBCL (n = 19); and (D) bar diagram indicating the frequencies of chromosomal imbalances that distinguish between ABC-DLBCL, GCB-DLBCL, and PMBCL (for statistical details see “Patients, materials, and methods”). All differences were statistically significant at P < .05, with the exception of 12q12 gains (P = .059).
Irrespective of the DLBCL subgroup, the most frequent imbalances in DLBCL were loss of 6q21-q22 (25%), loss of 6q16 (22%), gain of 18q21-q22 (19%), gain of 2p14-p16 (17%), gain of 3q27-qter (16%), gain of 6p (13%), and gain of Xp, 1q25-q32, and 3p (12% each). DNA amplifications were identified in 33 different chromosomal regions, most frequently in 2p14-p16 and 18q21-q22 (11 cases and 20 cases, respectively). Some chromosomal abnormalities occurred frequently in the same tumors, suggesting that they may be part of a recurrent pathway of lymphomagenesis. For example, among ABC-DLBCL, 17 of 26 cases (65%) with 3q27-qter gains also had 18q21-q22 gains (P < .001; odds ratio: 9.23; 95% confidence interval [CI]: 3.14-27.2).
Notably, several chromosomal alterations were differentially distributed among the DLBCL subgroups (Figure 1D). ABC-DLBCL showed characteristic and recurrent gains of chromosome 3, gains and amplification of 18q21-q22, and loss of 6q21-q22. Frequently, ABC-DLBCL had either gains of the whole 3q arm (26%) or trisomy 3 (15%), but these events were never observed in GCB-DLBCL and in only one case of PMBCL. Thirty-four percent of ABC-DLBCLs were characterized by recurrent gains of 18q21-q22 (compared with 10% in GCB-DLBCL and 16% in PMBCL; P < .05). Amplification of chromosomal region 18q21, which contains the BCL2 gene, was also more frequent in ABC-DLBCL (18%) compared with GCB-DLBCL (5%) and PMBCL (5%). Previously, we used PCR-based and fluorescent in situ hybridization (FISH) methods to detect the t(14;18) translocation involving the BCL2 gene in a subset of the DLBCL cases studied here, and found that this translocation occurs in GCB-DLBCL (46% and 53% of cases, respectively), but never in ABC-DLBCL.6,26 Interestingly, 3 of the 4 GCB-DLBCLs with an amplification of 18q21 also had the t(14;18), and the remaining case was not analyzed for the translocation. High expression of the BCL2 gene is a characteristic feature of all ABC-DLBCL, but only occurs in GCB-DLBCLs that have the t(14;18).2,3,6 Together, these data suggest that amplification of the 18q21 region occurs preferentially in lymphomas that have the ability to transcribe the BCL2 gene.
GCB-DLBCL was characterized by more frequent gains of 12q12 as compared with ABC-DLBCL and PMBCL, although this did not reach statistical significance (21% vs 5% and 5%, respectively, P = .059). Compared with GCB-DLBCL and ABC-DLBCL, PMBCL was characterized by frequent gains of 9p21-pter (37% vs 0% in GCB-DLBCL and 6% in ABC-DLBCL; P < .001), and gains of 2p14-p16 (47% vs 17% in GCB-DLBCL and 11% in ABC-DLBCL; P < .02). Taken together, these CGH data demonstrate that GCB-DLBCL, ABC-DLBCL, and PMBCL are genetically distinct.
To confirm some of the more frequent CGH abnormalities, we used real-time quantitative PCR (RQ-PCR) to quantify the copy number of the following genes: REL, BCL11A (2p14-16); SAS, CDK4, MDM2 (12q13-q14); RFC4, BCL6 (3q27); and MADH4, MALT1, BCL2 (18q21) (Supplemental Figure S1). REL was amplified in virtually all GCB-DLBCL cases with high-level DNA amplifications in 2p14-16 detected by CGH. BCL11A gene copy number was increased in all but one of these cases, albeit usually at lower levels than REL. Although CGH demonstrated high-level DNA amplifications in 2p14-16 in 2 cases of ABC-DLBCL, REL showed only a gain by RQ-PCR in these cases, whereas BCL11A was amplified in one case and gained in the other (Supplemental Figure S1A). This confirms our previous observation that REL may not be the primary target of amplification in ABC-DLBCL.3 CDK4 and SAS, mapping to chromosomal bands 12q13-q14, were frequently gained/amplified in GCB-DLBCL showing 12q gains by CGH, whereas MDM2 was less commonly altered. In contrast, all 3 cases of ABC-DLBCL with chromosome 12q13-q14 gains showed gains of CDK4, but less frequently SAS or MDM2 gains (Supplemental Figure S1B). RFC4 and BCL6 located in 3q27 were gained/amplified in all ABC-DLBCL and GCB-DLBCL with CGH gains/amplifications of 3q27-qter (Supplemental Figure S1C). MALT1 and BCL2 genes were commonly gained/amplified in ABC-DLBCL and GCB-DLBCL with chromosomal gains of 18q21, whereas MADH4 was less frequently altered (Supplemental Figure S1D). In summary, RQ-PCR confirmed many of the CGH findings and further demonstrated that in some instances, chromosomal alterations affect different genes in different DLBCL subgroups.
Chromosomal alterations influence locus-specific gene expression
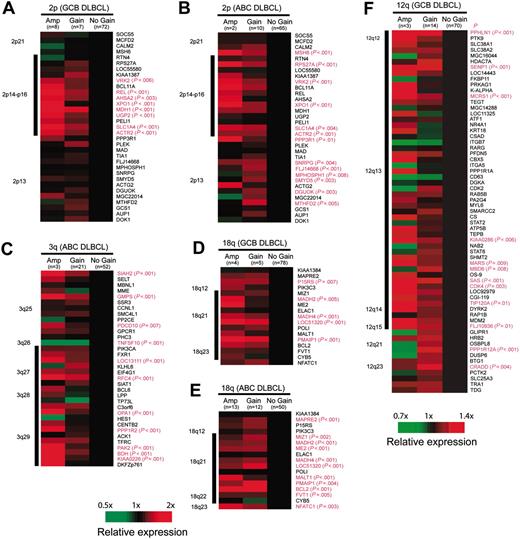
To determine the influence of chromosomal imbalances on locus-specific gene expression, mRNA levels of genes located in 4 recurrently gained/amplified chromosomal regions (2p14-p16, 3q27-qter, 12q12-q15, and 18q21-q22) were correlated with chromosomal copy number changes. Interestingly, we found that these chromosomal abnormalities were associated with higher expression of a subset of genes within the involved regions, but the particular genes that were overexpressed differed between the DLBCL subgroups. Fourteen genes were mapped to the chromosomal region 2p14-p16, and GCB-DLBCL and ABC-DLBCL with increased genomic copy number in this region showed significant overexpression of 8 (57%) and 5 (36%) of these 14 genes, respectively. Four genes (VRK2, XPO1, SLC14A, and ACTR2) were significantly overexpressed in both DLBCL subgroups. In contrast, REL, ASHA2, MDH1, and UGP2 were only overexpressed in GCB-DLBCL with 2p14-p16 gains (Figure 2A-B). In GCB-DLBCL and ABC-DLBCL with gains/amplifications of 12q12-q15, 10 (19%) and 12 (23%) of the 52 genes represented on the Lymphochip microarray were significantly overexpressed, but only 5 genes were overexpressed in both subgroups (SENP1, MCRS1, MARS, SAS, and CDK4; Figure 2F). Most of the overexpressed genes clustered to the chromosomal region 12q13. Similarly, 7 (33%) of the 21 genes mapping to chromosome 3q27-qter were significantly overexpressed in ABC-DLBCL (Figure 2C), in contrast to only 2 (13%) genes in GCB-DLBCL with overrepresentation of this region. Nine (75%) of the 12 genes mapping to chromosome 18q21-q22 were significantly overexpressed in ABC-DLBCL (Figure 2E), whereas only 4 (33%) genes were overexpressed in GCB-DLBCL with similar chromosomal alterations (Figure 2D). All 4 genes overexpressed in GCB-DLBCL (MADH2, MADH4, LOC51320, and PMAIP1) were also overexpressed in ABC-DLBCL, whereas overexpression of MIZ1, ME2, MALT1, BCL2, and FVT1 was restricted to ABC-DLBCL. We conclude that genomic copy number gains in 2p14-p16, 12q12-q15, 3q27-qter, and 18q21-q22 lead to a significant and DLBCL subgroup-specific up-regulation of genes located in the involved chromosomal regions.
Chromosomal alterations influence gene-expression signatures
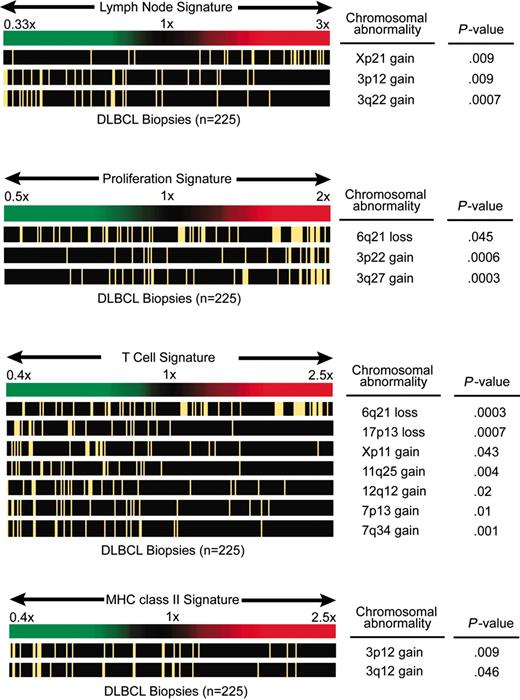
Previous studies have shown that certain gene-expression signatures reflect variable biologic features of DLBCL tumors, some of which are associated with survival.2,3,27,28 Gene-expression signatures that reflect variable biologic features of the malignant cells include the germinal center B-cell signature, the proliferation signature, and the major histocompatibility complex (MHC) class II signature. Other gene-expression signatures reflect properties of the nonmalignant cells in DLBCL, among these the T-cell signature2 and the “lymph node” signature, which was associated with favorable outcome in DLBCL and encompasses genes that are predominantly expressed in tumor-infiltrating immune cells and stromal cells.3 To determine whether genetic alterations influence previously defined gene-expression signatures, we created a gene-expression signature average for each DLBCL, which was then evaluated within tumors with specific chromosomal imbalances. Statistically significant associations were observed between several chromosomal alterations and the proliferation, lymph node, T-cell, and MHC class II signatures. In particular, gains of various cytobands of chromosome 3 as well as losses in 6q21 were associated with increased expression of the proliferation signature (Figure 3). Copy number gains of the chromosomal regions 3p12 and 3q12 were associated with decreased expression of the MHC class II gene-expression signature. The expression of the T-cell signature was negatively influenced by genetic losses of 6q21 and other cytobands of chromosome 6 as well as by losses of 17p13, gains of Xp11, gains of 11q24-q25, gains of 12q12, and gains of several cytobands in 7p and 7q. Finally, gains of Xp21 were associated with increased expression of the lymph node signature, whereas this signature was found to be decreased in DLBCL cases that harbored gains of 3q22 or several additional cytobands in 3p and 3q. In summary, certain genomic aberrations in DLBCL appear to be correlated with gene-expression signatures derived from nonmalignant tumor-infiltrating cells (lymph node and T-cell gene-expression signatures) or from the malignant cells (proliferation and MHC class II signatures).
Influence of chromosomal gains and amplifications on locus-specific gene-expression levels. Changes in gene-expression levels are depicted for each gene (averaged in each cohort) with regard to the locus-specific genetic status (wild-type vs gain vs amplification). Genes are ordered according to their chromosomal position shown on the right. Gene-locus information was obtained from the website for Genes On Sequence Map (Homo sapiens built 33). For genes with more than one microarray element on the Lymphochip, the average expression from different clones was calculated. The black bar on the left indicates the minimally gained region in all cases. The comparison of the expression levels was performed using the ANOVA test. Genes with significant differences (P < .01) are highlighted in red.
Influence of chromosomal gains and amplifications on locus-specific gene-expression levels. Changes in gene-expression levels are depicted for each gene (averaged in each cohort) with regard to the locus-specific genetic status (wild-type vs gain vs amplification). Genes are ordered according to their chromosomal position shown on the right. Gene-locus information was obtained from the website for Genes On Sequence Map (Homo sapiens built 33). For genes with more than one microarray element on the Lymphochip, the average expression from different clones was calculated. The black bar on the left indicates the minimally gained region in all cases. The comparison of the expression levels was performed using the ANOVA test. Genes with significant differences (P < .01) are highlighted in red.
Prognostic significance of chromosomal alterations
The prognostic value of CGH alterations was analyzed both in the entire set of DLBCL cases and within each DLBCL subgroup separately. Although several chromosomal alterations were individually associated with a significantly inferior or superior overall survival, only gains of different regions of chromosome 3 were significantly associated with shorter overall survival after adjustment for multiple comparisons in the whole series of patients. Thus, gains of 3p11-p12 (P < .001; relative risk [RR]: 3.07), 3q11-q13 (P = .002; RR: 2.7), 3q21-q24 (P < .01; RR: 2.4), and 3q25-q27 (P < .05; RR: 2.1) were associated with a shorter survival of the patients.
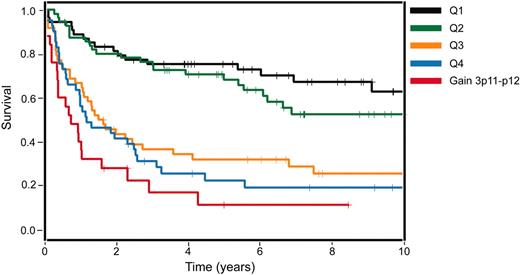
Previously, we had developed a gene-expression-based survival model for DLBCL that combines the prognostic influence of 4 gene-expression signatures.3 This model could divide patients with DLBCL into 4 quartile groups with 5-year survival rates of 73%, 71%, 34%, and 15%. To determine whether gains of various regions of chromosome 3 identified by CGH could improve the survival prediction for patients with DLBCL based on their gene expression,3 we performed a multivariate analysis. In this analysis, chromosome 3 gains involving the 3p11-p12 region had an independent prognostic value and improved the survival prediction based on the gene-expression-based model alone (Figure 4). The cases with gains of 3p11-p12 were primarily included in the least favorable quartile survival group, as defined by gene expression alone. However, some cases with 3p11-p12 gains were present in the more favorable quartile survival groups and, altogether, patients whose tumors had 3p11-p12 gains had a significantly worse clinical prognosis than predicted by gene expression alone (P = .029, likelihood ratio test; Figure 4).
Discussion
Using CGH, we uncovered 3 important relationships between chromosomal imbalances in DLBCL and their biologic and clinical attributes. First, DLBCL subgroups defined by gene-expression profiling had distinct patterns of genomic alterations. Second, particular chromosomal alterations affected discrete biologic features of the tumors as defined by gene-expression signatures. Third, several chromosomal abnormalities were associated with clinical outcome, and one abnormality added prognostic value to an optimal gene-expression-based survival model for DLBCL.
Chromosomal imbalances influence the lymph node, proliferation, T-cell, and MHC class II gene-expression signatures. In each of the 4 panels, DLBCL cases are ordered according to their average expression of the respective signature genes (the case with the lowest expression appears on the left end of the spectrum). Cases with the chromosomal abnormalities shown on the right are marked with a yellow bar. Correlations with a P value less than .05 are shown. If more than one cytoband in one chromosomal arm showed a P value less than .05, the cytoband with the lowest P value is displayed.
Chromosomal imbalances influence the lymph node, proliferation, T-cell, and MHC class II gene-expression signatures. In each of the 4 panels, DLBCL cases are ordered according to their average expression of the respective signature genes (the case with the lowest expression appears on the left end of the spectrum). Cases with the chromosomal abnormalities shown on the right are marked with a yellow bar. Correlations with a P value less than .05 are shown. If more than one cytoband in one chromosomal arm showed a P value less than .05, the cytoband with the lowest P value is displayed.
Gene-expression profiling of DLBCL has led to the notion that this diagnostic category consists of at least 3 diseases that differ with respect to their normal cellular counterparts, clinical outcomes, and oncogenic mechanisms.2-5,22 GCB-DLBCL appears to be derived from germinal center B cells, ABC-DLBCL may be derived from a post-germinal center B-cell undergoing plasmacytic differentiation, and PMBCL may be derived from a thymic B cell.2-5,22 The cure rates of each DLBCL subgroup are significantly different, with ABC-DLBCL, GCB-DLBCL, and PMBCL having 5-year survival rates of 30%, 59%, and 64%, respectively.2-4 The DLBCL subgroups use distinct oncogenic mechanisms: GCB-DLBCL is characterized by frequent REL amplifications and BCL2 translocations, events that never occur in ABC-DLBCL.3,6 ABC-DLBCL and PMBCL have constitutive activation of the NF-κB pathway, which they require for survival.5,8,9
The present analysis of CGH profiles in the DLBCL subgroups strongly supports the concept that they represent distinct disease entities. Several chromosomal abnormalities were found to be frequent in one subgroup of DLBCL but uncommon or absent in other subgroups. For example, in ABC-DLBCL gains of chromosome arm 3q were observed in more than one quarter of the cases, but this abnormality was never observed in GCB-DLBCL and in only 1 PMBCL case. Similarly, gains of 18q21-q22 occurred in one third of ABC-DLBCL cases, but less frequently in GCB-DLBCL (10%) and PMBCL (16%) cases. Genomic gains of 3q and 18q were previously found to be correlated with shorter survival in patients with DLBCL.18 Our present findings provide a clear explanation for this observation, namely that both abnormalities are statistically associated with ABC-DLBCL, which has a worse prognosis than the other DLBCL subgroups. Alternatively, it could be speculated that these genetic alterations themselves contribute, at least in part, to the ABC-DLBCL gene-expression phenotype and its inferior prognosis. Another example of a DLBCL subgroup-specific genomic alteration is gain of 9p21, which was observed in more than one third of PMBCL cases, but was observed in only 6% of ABC-DLBCL cases and never in GCB-DLBCL cases. DLBCL subgroup-specific chromosomal aberrations such as these are likely to contribute to oncogenic pathways that are important for one subgroup of DLBCL but may be irrelevant for other subgroups.
Other chromosomal abnormalities occurred more frequently in one DLBCL subgroup than in others but were not restricted to a single DLBCL subgroup. For example, deletions of 6q21-q22 occurred in 40% of ABC-DLBCL cases and 22% of GCB-DLBCL cases, but never in PMBCL. Gains and amplifications of chromosome 12cen-q15 were observed most frequently in GCB-DLBCL but this abnormality was also observed at a low frequency in ABC-DLBCL and PMBCL. These observations suggest that some oncogenic pathways are shared by the various DLBCL subgroups but nonetheless may be more frequently used in different subgroups.
A primary focus of the current study was to understand the relationship between genomic abnormalities in DLBCL and gene expression in the tumors by obtaining CGH profiles and mRNA profiles from the identical tissue. Recent studies have demonstrated a correlation between gene copy number changes and expression of genes encoded in the involved genomic regions.29-33 We have examined the relationship between chromosome gains/amplification and the expression profile of genes located in 4 chromosomal regions commonly overrepresented in GCB and ABC-DLBCL tumors (2p14-p16, 12q12-q15, 3q27-qter, and 18q21-q22). Overall, a strong impact of genomic gains and amplifications on the expression of genes mapping to the involved chromosomal regions was observed. Of the genes located within these chromosomal segments, 25% to 75% were overexpressed in those tumors with increased DNA copy number. For many genes, the levels of expression increased significantly from cases with a normal DNA profile to cases with gains or amplifications, suggesting a direct effect of the gene copy number on mRNA expression levels.
Impact of genomic gains of 3p11-p12 on survival of patients with DLBCL. Kaplan-Meier survival estimates of patients with DLBCL with genomic gains of 3p11-p12 in comparison to their stratification into survival quartiles based on the gene-expression-based outcome predictor model alone3 (P = .029). Q indicates quartile.
Impact of genomic gains of 3p11-p12 on survival of patients with DLBCL. Kaplan-Meier survival estimates of patients with DLBCL with genomic gains of 3p11-p12 in comparison to their stratification into survival quartiles based on the gene-expression-based outcome predictor model alone3 (P = .029). Q indicates quartile.
However, in line with similar studies in solid tumors,29-33 not all genes in the overrepresented chromosomal regions were more highly expressed, suggesting that either individual genes were not amplified or, alternatively, the functional background of the cell was not appropriate for the expression of the gene. For example, REL was significantly overexpressed in GCB-DLBCL with overrepresentation of 2p14-p16 detected by CGH. Although a slight increase in REL expression was observed in ABC-DLBCL with overrepresentation of the chromosomal region 2p14-p16, the level of expression was not significantly higher than in tumors with a normal genetic profile in this region. Quantitative PCR analysis confirmed that the REL locus was amplified in virtually all GCB-DLBCL in which CGH showed amplification of this region. However, REL was not amplified in any of the ABC-DLBCL cases, indicating that genes other than REL may be targeted by 2p14-p16 gains in this DLBCL subgroup. Interestingly, the mRNA expression of BCL11A, located very close to REL, was not influenced by 2p14-p16 gains in the GCB-DLBCL or ABC-DLBCL subgroups, although, similarly to REL, genomic quantitative PCR analysis showed amplification and gains of the BCL11A locus in both DLBCL subgroups. High-resolution techniques such as array-based CGH or detailed FISH analysis will likely refine the minimally targeted genomic regions in these cases in future studies.
One of the most intriguing findings of the present study was that chromosomal aberrations can have strong influences on the expression of genes that are not encoded in the involved region but that instead reflect differences in tumor biology. Previously, we defined gene-expression signatures in DLBCL that reflect variable features of the malignant cells or the nonmalignant tumor-infiltrating cells.2,3,27 Two gene-expression signatures that reflect variation within the malignant cells are the proliferation signature, which is more highly expressed in proliferating than in quiescent cells, and the MHC class II signature, which reflects the coordinate regulation of all MHC class II genes in the malignant DLBCL cells.3,28 The proliferation signature was increased in DLBCL cases with genomic loss in 6q21 and gains in several bands of chromosome 3, whereas gains of 3p11-p12 were associated with decreased MHC class II expression.
Unexpectedly, genomic abnormalities influenced the expression of 2 other signatures that reflect the nature of the nonmalignant cells in DLBCL tumors, the T-cell signature and the lymph node signature.2,3 The T-cell signature is formed by the coordinate expression of pan-T-cell genes (eg, CD2, CD3γ, LAT) and reflects the infiltration of the tumors by T cells. The lymph node signature reflects a host response that is characterized by abundant expression of extracellular matrix components and infiltration of the tumors with immune cells other than T cells. A significantly lower expression of the T-cell signature was observed in DLBCL with gains of cytobands in chromosomes 7, 11, 12, and X as well as losses in 6q and 17p. Xp21 gains were associated with an increased expression of the lymph node signature whereas tumors with gains in several cytobands of chromosome 3 had lower expression of this signature.
Two general models can be envisaged to explain the association of particular genomic abnormalities with changes in gene signature expression. One possibility is that the involved genomic region encodes a key regulator that significantly alters tumor-cell biology and therefore the expression of a gene signature. For example, the proliferation signature might be affected by a chromosomal abnormality that leads to overexpression or underexpression of a key cell-cycle regulator encoded in the involved region. Chromosome aberrations could cause changes in lymph node or T-cell signature expression if the genomic region encodes a cytokine, chemokine, or other immune regulator that can dramatically alter the profile of infiltrating immune cells. We undertook a search for key regulator genes in the regions delineated in this study. However, due to the resolution limit of conventional CGH, these regions still encompass dozens to hundreds of genes, and no obvious key regulators were observed. A second general model would suggest that DLBCL can arise by distinct pathogenetic pathways, and that each pathway may involve the accumulation of different chromosomal abnormalities and other oncogenic events. From this point of view, a particular chromosome abnormality may be a surrogate marker for all of the oncogenic events that contribute to a given pathogenetic pathway. In this model, therefore, the association between a chromosomal abnormality and the expression of a gene signature may reflect the fact that DLBCLs that arise by different pathogenetic pathways could differ broadly in tumor biology. What is important is that these associations between genomic abnormalities and gene-expression signatures are frequent and statistically robust features of our data that point to hitherto unrecognized variation in DLBCL pathogenesis.
Previous genetic studies in DLBCL suggested that several genetic alterations have prognostic importance.10,18 In our study, however, only gains in several regions of chromosome 3 were significantly associated with inferior survival of the patients after adjusting for multiple variable comparisons. Importantly, the prognostic value of genomic gains involving 3p11-p12 was found to be independent of the survival prediction based on an optimal gene-expression-based model.3 Therefore, the integration of this genetic alteration into the gene-expression-based survival model may improve the ability to predict survival in patients with DLBCL.
Appendix
The Lymphoma/Leukemia Molecular Profiling Project is an international consortium of 9 institutions: the National Cancer Institute; University of Nebraska Medical Center; British Columbia Cancer Agency; Southwest Oncology Group; University of Wuerzburg in Germany; Hospital Clinic, University of Barcelona, Spain; Norwegian Radium Hospital in Oslo; and St. Bartholomew's Hospital in London.
Prepublished online as Blood First Edition Paper, July 26, 2005; DOI 10.1182/blood-2005-04-1399.
Supported by the Spanish Comisión Interministerial de Ciencia y Tecnologiáa (CICYT) SAF02-3261, Instituto de Salud Carlos III, Red Temática de Cáncer (G03/10), Red temática limfomas (G03/179; E.C.), by the Interdisciplinary Center for Clinical Research (IZKF) of the University of Würzburg, Germany (A.R.), and by a Director's Challenge grant (UO1-CA84 967) from the National Cancer Institute, Bethesda, MD.
L.M.S., E.C., and A.R. contributed equally to this work. S.B. and A.Z. contributed equally to this work.
A complete list of the member institutions of the Lymphoma/Leukemia Molecular Profiling Project appears in the “Appendix.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal