Abstract
CD40 is expressed on various immune cells, including macrophages and microglia. Aberrant expression of CD40 is associated with autoimmune inflammatory diseases such as multiple sclerosis and rheumatoid arthritis. Interaction of Toll-like receptor-4 (TLR4) with the Gram-negative bacteria endotoxin lipopolysaccharide (LPS) results in the induction of an array of immune response genes. In this study, we describe that LPS is a strong inducer of CD40 expression in macrophages and microglia, which occurs at the transcriptional level and involves the activation of the transcription factors nuclear factor-κB (NF-κB) and signal transducer and activator of transcription 1α (STAT-1α). LPS-induced CD40 expression involves the endogenous production of the cytokine interferon-beta (IFN-β), which contributes to CD40 expression by the activation of STAT-1α. Blocking IFN-β–induced activation of STAT-1α by IFN-β–neutralizing antibody reduces LPS-induced CD40 gene expression. Furthermore, LPS induces acetylation and phosphorylation of histones H3 and H4 and the recruitment of NF-κB, STAT-1α, and RNA polymerase II on the CD40 promoter in vivo in a time-dependent manner, all events important for CD40 gene transcription. These results indicate that both LPS-induced NF-κB activation and endogenous production of IFN-β that subsequently induces STAT-1α activation play critical roles in the transcriptional activation of the CD40 gene by LPS.
Introduction
CD40 is a member of the tumor necrosis factor receptor (TNFR) superfamily that is expressed by cells, including B cells, macrophages, microglia, dendritic cells, endothelial cells, and tumor cells. The interaction between CD40 and its cognate ligand, CD40L (CD154), is critical for a productive immune response.1,2 Upregulation of various cell surface molecules, such as class II major histocompatibility complex, CD40, CD80, and CD86, occurs on CD40-CD154 contact, as well as the production of numerous cytokines and chemokines (interleukin 1 [IL-1], IL-6, IL-10, tumor necrosis factor α [TNF-α] and macrophage inflammatory protein 1 [MIP-1]) and cytotoxic radicals.2 CD40 has been implicated in participating in many human diseases, particularly autoimmune diseases.3,4 Blocking the interaction between CD40-CD154 with anti-CD154 or anti-CD40 antibody is beneficial in animal models of autoimmune diseases.5 These findings illustrate the importance of CD40-CD154 interactions for homeostasis of immune responses. We have previously shown that macrophages and microglia, the endogenous macrophage of the brain, constitutively express CD40 at a low level, which is greatly enhanced by the cytokine interferon γ (IFN-γ). IFN-γ–induced CD40 expression involves IFN-γ–activated signal transducer and activator of transcription 1α (STAT-1α) and nuclear factor-κB (NF-κB) activation through an autocrine response to IFN-γ–induced TNF-α production.6-9
The immune response to microbial pathogens relies on both innate and acquired immunity. Innate immunity is determined by the interaction between potential pathogens and their cognate binding partners (receptors) on phagocytes, which is performed by a group of proteins, the Toll-like receptor (TLR) family.10,11 At least 10 TLRs (TLR1-TLR10) recognize specific molecular patterns that are present in microbial components. Stimulation of different TLRs induces distinct patterns of gene expression, which not only leads to the activation of innate immunity but also instructs the development of antigen-specific acquired immunity.12,13 The TLR family signals via shared downstream signaling molecules, including the adapter molecule myeloid differentiation primary-response protein 88 (MyD88), interleukin-1 receptor-associated protein kinases (IRAK1 and IRAK4), transforming growth factor β (TGF-β)–activated kinase (TAK1), TAK1-binding protein 1 (TAB1), TAB2, and tumor necrosis factor receptor-associated factor 6.10,11 TAK1 is a member of the mitogen-activated protein kinase kinase kinase (MAPKKK) family, which is essential for IL-1, TNF-α, and lipopolysaccharide (LPS)–induced activation of NF-κB and MAPKs.10 The NF-κB family of transcription factors is composed of 5 members, p65 (Rel-A), Rel-B, c-Rel, p50, and p52, which function as homodimers and heterodimers. NF-κB transcription factors are present in the cytoplasm in an inactive state, complexed with inhibitory IκB proteins. The activation process is mediated by the IκB kinase (IKK) complex; activation of IKK by a stimulus such as LPS leads to the phosphorylation and degradation of IκB proteins and subsequent activation of NF-κB.14
LPS, a Gram-negative cell wall component recognized by the specific receptor TLR4, is an adjuvant for the adaptive immune response, which up-regulates costimulatory molecules on antigen-presenting cells.12,15 Studies of MyD88-deficient mice demonstrated that MyD88 is essential for LPS induction of a broad range of genes, including TNF-α and IL-6. However, there are also MyD88-independent pathways that occur in response to LPS.16 The MyD88-independent pathway, which is unique for TLR3 and TLR4 signaling pathways, leads to IFN-β production.15,17,18 Two pathways have been reported for LPS-TLR4–induced signaling: one is through the adaptor proteins MyD88 and MyD88 adaptor-like (MAL) that contribute to LPS-induced activation of the transcription factor NF-κB,10,19 and the other is through adaptors named TIR (Toll/interleukin-1 receptor) domain-containing adaptor inducing interferon-β (TRIF)/TIR-containing adaptor molecule-1 (TICAM-1) and TRAM (TRIF-related adaptor molecule) to induce expression of the transcription factor IFN regulatory factor-3 (IRF-3), which induces IFN-β expression.15,18,20 This endogenous production of IFN-β subsequently induces activation of the Janus kinase (JAK)–STAT pathway and a number of IFN-β–dependent genes.12,21-23
In this study, we examined the molecular mechanism of LPS-induced CD40 gene expression. We demonstrate that LPS induces CD40 mRNA and protein expression in both murine and human macrophages and microglia. Analysis of the CD40 promoter demonstrates that both NF-κB and IFN-γ activation site (GAS) elements are important for LPS-induced CD40 gene transcription. LPS activates the NF-κB signaling pathway, including IKK-α/β, IκBα, and NF-κB p65, through the process of phosphorylation. In addition, LPS induces endogenous production of IFN-β, which subsequently leads to activation of STAT-1α. Blocking IFN-β–induced STAT-1α activation by use of neutralizing antibody partially reduces LPS-induced CD40 gene expression. Chromatin immunoprecipitation (ChIP) assays demonstrate that LPS induces modification of histones H3 and H4 in the CD40 promoter and recruitment of the transcription factors NF-κB p65, p50, and STAT-1α to the CD40 promoter in a time-dependent manner in vivo. RNA polymerase II (Pol II) is also involved in LPS-induced CD40 gene transcription. These data indicate that direct LPS induction of NF-κB activation and LPS-induced production of IFN-β, which subsequently activates STAT-1α, are important events for CD40 gene expression in macrophages and microglia.
Materials and methods
Recombinant proteins and reagents
Escherichia coli LPS was purchased from Sigma (St Louis, MO), murine IFN-β was purchased from Biosource International (Camarillo, CA), and murine IFN-γ was from Genzyme (Boston, MA). Neutralizing antibodies to IFN-α, IFN-β, and IFN-γ were from R&D Systems (Minneapolis, MN). Rat anti–mouse phycoerythrin (PE)–conjugated CD40 antibody (clone 3/23), mouse anti–human fluorescein isothiocyanate (FITC)–conjugated CD40 antibody (clone 5C3) and rat PE-conjugated immunoglobulin G (IgG) isotype control were purchased from Pharmingen (San Diego, CA). Antibodies against phospho–NF-κB p65Ser536, phospho–IKK-α/βSer180/Ser181, phospho–IκBαSer32, and phospho–STAT-1αTyr701 and –STAT-1αSer727 were purchased from Cell Signaling Technology (Beverly, MA). Antibodies against CD40, NF-κB p65, NF-κB p50, STAT-1α, and actin were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against acetylated H3 (Ac-H3), Ac-H4, AcK9-H3, MeK9-H3, and pS10AcK14-H3 were from Upstate Biotechnology (Lake Placid, NY). Antibody against Pol II was from Covance (Princeton, NJ). Expression vectors encoding wild-type and dominant-negative forms of IKK-α and IKK-β were kindly provided by Dr Rudolph Noelle (Dartmouth Medical School, Lebanon, NH).24
Cells
Primary microglia from C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME), and wild-type and STAT-1α–deficient mice on the 129S6/SvEv background (Taconic, Germantown, NY) were prepared as described previously.8 Primary human macrophages were generated by plating 10 × 106 peripheral blood mononuclear cells/well and culturing the cells in Dulbecco modified Eagle medium (DMEM) as described previously.25 The purity of the macrophage cultures is 95% or higher, as determined by staining for CD14. The murine microglia cell line EOC13 and macrophage cell line RAW264.7 were maintained in DMEM supplemented with 10% fetal bovine serum as described previously.6
Immunofluorescence flow cytometry
Cells were plated at 2 × 105 cells/well into 12-well plates and treated with medium, LPS, or IFN-γ for 36 to 48 hours and then incubated with 100 μL 2.4G2 hybridoma supernatant (which contains rat anti–mouse Fcγ receptor [FcγR] antibody [Ab]) for blocking FcγR. Murine cells were then incubated with 10 μg/mL PE-conjugated anti–mouse CD40 Ab and analyzed on the FACStar (Becton Dickinson, Mountain View, CA) as previously described.7 Human macrophages were incubated with 10 μg/mL FITC-conjugated anti–human CD40 Ab. Negative controls were incubated with IgG isotype-matched antibody. Fold change of CD40 expression was calculated by dividing the value of mean fluorescence intensity (MFI) of treated samples by the value of untreated samples.
RNA isolation, riboprobes, and ribonuclease protection assay
Total cellular RNA was isolated from unstimulated or LPS-stimulated cells. The riboprobes for murine CD40, TNF-α, IFN-β, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and human CD40 and GAPDH were prepared as described previously.7 Total RNA (20 μg) was hybridized with CD40, TNF-α, IFN-β, and GAPDH riboprobes at 42°C overnight. The hybridized mixture was then treated with RNase A/T1 (1:200) and then analyzed by 5% denaturing (8 M urea) polyacrylamide gel electrophoresis. Values for CD40, TNF-α, and IFN-β mRNA expression were normalized to GAPDH mRNA levels for each experimental condition.
CD40 promoter constructs
Transient transfection and luciferase assays
CD40 promoter constructs (0.2 μg) were transiently transfected into 5 × 105 RAW264.7 cells in 6-well plates using the LipofectAMINE Plus method as described previously.7 Transfected cells were treated with LPS for 12 hours. The luciferase activity of each sample was normalized to the total protein concentration of each well. Luciferase activity from the untreated sample was set at 1 for calculation of induction. For transfections with the IKK-α and IKK-β expression constructs (0.1 μg), the difference in amount of total DNA transfected was normalized by using empty pcDNA3 vector.8
Immunoblotting
Cell lysate (50 μg) was separated on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and probed with the specific antibodies. Cytoplasmic and nuclear proteins were extracted as described.26 Nuclear and cytoplasmic extracts (50 μg) were analyzed by immunoblotting and probed with the respective phosphorylation-specific antibodies. Membranes were stripped at 50°C in buffer containing 100 mM 2-ME (2-mercaptoethanol), 2% SDS, and 62.5 mM Tris (tris(hydroxymethyl)aminomethane)–HCl (pH 6.7) with occasional shaking and reprobed for total STAT-1α protein, total NF-κB p65, and actin.
ELISA
Supernatants were collected from unstimulated or LPS-stimulated RAW264.7 cells and assayed by enzyme-linked immunosorbent assay (ELISA) for secretion of IFN-β (PBL Biomedical Laboratories, Piscataway, NJ). IFN-β levels were normalized to total protein levels.
Chromatin immunoprecipitation (ChIP) assays
ChIP analysis was done as described.26,27 RAW264.7 or EOC13 cells were stimulated with LPS for up to 4 hours and fixed with 1% formaldehyde for 15 minutes at room temperature, and nuclei were isolated. Chromatin was sheared by sonication, and samples were precleared for 2 hours at 4°C with salmon sperm DNA-saturated protein A/G Sepharose. Chromatin solutions were precipitated overnight at 4°C with 5 μg antibodies or isotype-matched control IgG. Input and immunoprecipitated chromatin were incubated at 65°C overnight to reverse crosslinks. After proteinase K digestion, DNA was extracted with the Qiagen Miniprep Kit (Valencia, CA). Purified DNA was analyzed by polymerase chain reaction (PCR) with Taq polymerase. The primer pair 5′-CTACAGCCTCTGGATGGAGC-3′ and 5′-TGCAGAACCGAAAGCGTCTC-3′ was used to amplify a 250-bp region in the mouse CD40 promoter containing functional NF-κB and GAS elements. Densitometry was used to quantify the PCR results, and all results were normalized by the respective input values.
The study protocol was approved by an institutional review board at the University of Alabama at Birmingham.
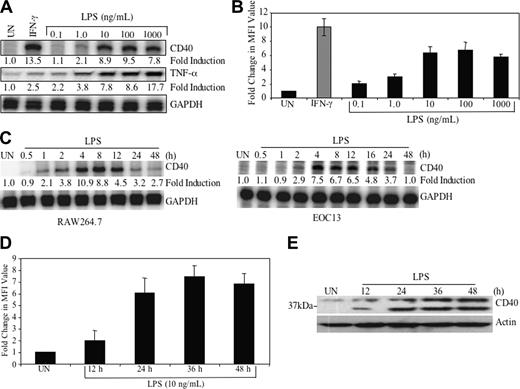
LPS induces CD40 expression in a dose- and time-dependent manner. (A) RAW264.7 cells were treated with medium or varying concentrations of LPS (0.1-1000 ng/mL) or IFN-γ (10 ng/mL) for 8 hours, then total RNA was isolated and analyzed by ribonuclease protection assay (RPA) for CD40, TNF-α, and GAPDH mRNA. The basal level of the untreated sample was set as 1.0, and fold induction on LPS or IFN-γ treatment was compared with that. (B) RAW264.7 cells were treated in the absence or presence of LPS or IFN-γ 36 hours, then stained with either anti-CD40 or isotype-matched control antibody. Cells were subjected to fluorescence-activated cell sorting (FACS) analysis. Samples were analyzed by measuring MFI. Fold change in CD40 MFI value was calculated and shown as the mean ± SD of 3 experiments. (C) RAW264.7 and EOC13 cells were treated with 10 ng/mL LPS for up to 48 hours, then RNA was isolated and subjected to RPA analysis for CD40 and GAPDH mRNA. Fold induction on LPS treatment was calculated as in panel A. (D) RAW264.7 cells were treated in the absence or presence of LPS (10 ng/mL) for up to 48 hours. CD40 protein expression was analyzed by FACS analysis. Fold change of LPS-induced CD40 MFI value was calculated and shown as the mean ± SD of 3 experiments. (E) RAW264.7 cells were treated in the absence or presence of LPS (10 ng/mL) for up to 48 hours. Protein lysates were prepared and subjected to immunoblotting with anti-CD40 antibody, then stripped, and reprobed with antiactin antibody as a loading control. UN indicates untreated. Representative of 3 experiments.
LPS induces CD40 expression in a dose- and time-dependent manner. (A) RAW264.7 cells were treated with medium or varying concentrations of LPS (0.1-1000 ng/mL) or IFN-γ (10 ng/mL) for 8 hours, then total RNA was isolated and analyzed by ribonuclease protection assay (RPA) for CD40, TNF-α, and GAPDH mRNA. The basal level of the untreated sample was set as 1.0, and fold induction on LPS or IFN-γ treatment was compared with that. (B) RAW264.7 cells were treated in the absence or presence of LPS or IFN-γ 36 hours, then stained with either anti-CD40 or isotype-matched control antibody. Cells were subjected to fluorescence-activated cell sorting (FACS) analysis. Samples were analyzed by measuring MFI. Fold change in CD40 MFI value was calculated and shown as the mean ± SD of 3 experiments. (C) RAW264.7 and EOC13 cells were treated with 10 ng/mL LPS for up to 48 hours, then RNA was isolated and subjected to RPA analysis for CD40 and GAPDH mRNA. Fold induction on LPS treatment was calculated as in panel A. (D) RAW264.7 cells were treated in the absence or presence of LPS (10 ng/mL) for up to 48 hours. CD40 protein expression was analyzed by FACS analysis. Fold change of LPS-induced CD40 MFI value was calculated and shown as the mean ± SD of 3 experiments. (E) RAW264.7 cells were treated in the absence or presence of LPS (10 ng/mL) for up to 48 hours. Protein lysates were prepared and subjected to immunoblotting with anti-CD40 antibody, then stripped, and reprobed with antiactin antibody as a loading control. UN indicates untreated. Representative of 3 experiments.
Results
LPS induces CD40 gene expression in macrophage and microglia
Our previous studies demonstrated that IFN-γ is a potent inducer of CD40 expression in macrophages and microglia.6-8 We have recently made the observation that LPS is also a strong inducer of CD40 expression in these cells. Murine macrophage RAW264.7 cells were treated with LPS (0.1-1000 ng/mL) or IFN-γ (5 ng/mL) as a positive control for 8 hours, and then CD40 mRNA expression was analyzed. The results demonstrate that LPS induces CD40 mRNA expression in a dose-dependent manner, with optimal induction using 10 to 100 ng/mL (8.9- to 9.5-fold) (Figure 1A). TNF-α gene expression is shown as a positive control for LPS responsiveness (Figure 1A). LPS-induced CD40 protein expression was also observed in a dose-dependent manner (Figure 1B).
Kinetics analysis of CD40 mRNA expression was analyzed in both RAW264.7 cells and EOC13 cells, a murine microglial cell line. CD40 mRNA expression was first detected 1 hour after addition of LPS, peaked at 4 hours (10.9-fold induction), and persisted out to 48 hours in the RAW264.7 cells (Figure 1C, left). In the EOC13 cells, LPS-induced CD40 mRNA expression was detected at 2 hours and peaked at 4 hours (7.5-fold induction), and expression persisted up to 16 to 24 hours following LPS treatment (Figure 1C, right). CD40 protein expression was detectable 12 hours after LPS stimulation and peaked at 36 hours (∼ 7.5-fold) in the RAW264.7 cells as assessed by flow cytometry (Figure 1D). Immunoblotting was performed as another measurement of CD40 protein expression. LPS-induced CD40 protein expression occurred in a time-dependent manner (Figure 1E), in agreement with the flow cytometry data (Figure 1D). Two isoforms of the CD40 protein are detected (45 and 27 kDa), which are the result of alternative splicing.28 These results collectively demonstrate that LPS induces CD40 mRNA and protein expression in a dose- and time-dependent manner in both murine macrophage and microglial cell lines.
We examined whether CD40 expression was regulated by LPS in primary cultures of macrophages and microglia. In human primary macrophage cultures, low constitutive expression of CD40 mRNA and protein was observed, which was enhanced at 4 to 8 hours at the mRNA level, and at 36 hours at the protein level by LPS (Figure 2A). In primary murine microglia, CD40 mRNA was strongly inducible by LPS at 4 to 8 hours, followed by expression of the CD40 protein (Figure 2B). These data demonstrate that LPS induction of CD40 expression occurs in primary macrophages and microglia.
NF-κB and GAS elements are required for LPS-induced CD40 promoter activity
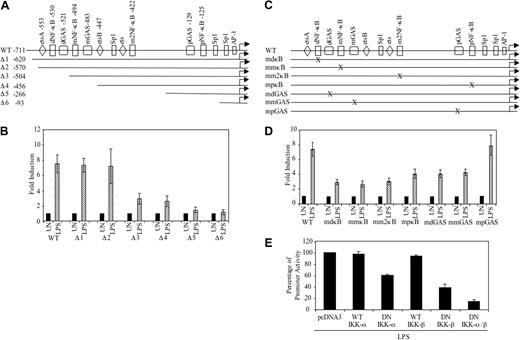
We have previously described that the GAS and NF-κB regulatory elements are conserved between the human and mouse CD40 promoters, as is the spacing between these sites.7,8 The conservation of these identified elements suggests that they are likely to function in a similar manner in both mouse and human cells for CD40 expression. To define the cis-acting elements necessary for LPS-induced CD40 gene transcription, serial deletion and site-directed mutation constructs of the human CD40 promoter were tested in RAW264.7 cells, because these cells are amenable to transfection. The wild-type (WT) CD40 promoter (–711) and serial deletion constructs (Figure 3A) were transiently transfected into RAW264.7 cells, treated with medium or LPS for 12 hours, and then assayed for luciferase activity. LPS stimulation led to an approximate 7.5-fold induction of CD40 promoter activity. Deletion of the –570 to –504 region (Δ3) reduced promoter activity by approximately 80% (Figure 3B). Further deletion to –93 bp (Δ6) abolished LPS-induced promoter activity (Figure 3B). These results demonstrate that the CD40 promoter region of –570 to –504 is critical for LPS-induced CD40 promoter activity.
LPS induces CD40 expression in primary macrophages and microglia. Human primary macrophages (A) or murine primary microglia (B) were treated with LPS (10 ng/mL) for 36 hours, and then cells were subjected to FACS analysis for CD40 protein expression. ISO indicates isotype antibody. In addition, cells were treated with LPS for 4 and 8 hours, then total RNA was isolated and analyzed by RPA for CD40 and GAPDH mRNA expression. The cell numbers are mapped on the y-axis. Representative of 2 experiments.
LPS induces CD40 expression in primary macrophages and microglia. Human primary macrophages (A) or murine primary microglia (B) were treated with LPS (10 ng/mL) for 36 hours, and then cells were subjected to FACS analysis for CD40 protein expression. ISO indicates isotype antibody. In addition, cells were treated with LPS for 4 and 8 hours, then total RNA was isolated and analyzed by RPA for CD40 and GAPDH mRNA expression. The cell numbers are mapped on the y-axis. Representative of 2 experiments.
Within the CD40 promoter, there are 4 NF-κB sites that are designated as dκB, mκB, m2κB, and pκB, and 3 GAS elements designated as dGAS, mGAS, and pGAS.7,8 The CD40 promoter site-directed mutant constructs (Figure 3C) were created by site-directed mutagenesis as previously described.7,8 We wanted to analyze the potential role of each of the NF-κB and GAS elements in LPS-induced CD40 promoter activation. Transient transfection analysis in RAW264.7 cells showed that all the mutations of the dκB, mκB, m2κB, and pκB elements lead to an inhibition of LPS-induced CD40 promoter activity (∼ 45% to ∼ 60%) (Figure 3D). Regarding the GAS elements, mutation of the dGAS and mGAS elements led to an approximate 45% inhibition of CD40 promoter activity, whereas mutation of the pGAS element did not affect LPS activation of the CD40 promoter (Figure 3D). These results suggest that all 4 NF-κB sites and 2 of the 3 GAS elements play important roles in LPS-induced CD40 promoter activity.
Because of the importance of NF-κB activation in LPS-induced signaling,10,11,29-31 the involvement of NF-κB in LPS-induced CD40 gene transcription was further investigated. DN expression constructs of IKK-α or IKK-β that contain substitutions of alanine for an essential lysine in the adenosine triphosphate (ATP)–binding site, rendering the proteins catalytically inactive,24 were used. The DN constructs, as well as WT IKK-α and IKK-β constructs, were cotransfected with the WT CD40 promoter construct into RAW264.7 cells and then treated with medium or LPS. Cotransfection with WT IKK-α or IKK-β constructs did not affect LPS-induced CD40 promoter activity. The IKK-α DN partially suppressed LPS-induced CD40 promoter activity (∼ 40%), whereas the IKK-β DN construct suppressed LPS-induced CD40 promoter activity to a greater extent (∼ 60%). Inclusion of both IKK-α and IKK-β DN constructs inhibited LPS-induced CD40 promoter activity by approximately 85% (Figure 3E). These data indicate that activation of NF-κBis critical for the induction of CD40 promoter activity by LPS.
LPS induces activation of NF-κB and STAT-1α signaling pathways
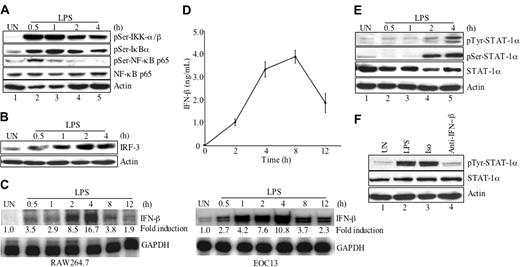
To further investigate activation of the NF-κB signaling pathway, RAW264.7 cells were treated in the absence or presence of LPS for up to 4 hours, then protein lysates were analyzed for the phosphorylation status of IKK-α/β, IκBα, and NF-κB (Figure 4A). Expression levels of total NF-κB p65 and actin were used as loading controls. IKK-α/βSer180/Ser181 was strongly phosphorylated after 0.5 to 1 hour of LPS treatment (lanes 2-3), and the intensity of the band gradually decreased over time, although phosphorylation was still detectable at 2 to 4 hours (lanes 4-5). Phosphorylation of IκBαSer32 was detected at 0.5 hour of LPS treatment (lane 2), peaked at 1 hour (lane 3), then decreased at 2 and 4 hours (lanes 4-5). Phosphorylation of NF-κB p65Ser536 was detected at 0.5 hour after LPS treatment (lane 2), decreased at 1 to 2 hours (lanes 3-4), and was undetectable at 4 hours (lane 5). These results indicate that LPS activates several components of the NF-κB signaling pathway.
Recent studies have demonstrated that TLR4 possesses a MyD88-independent pathway that uses an adaptor protein TRIF to activate IRF-3 expression and subsequent IFN-β production.21-23 The endogenously produced IFN-β activates the JAK-STAT pathway and leads to expression of IFN-β responsive genes.23,30 As the CD40 promoter analysis demonstrated an involvement of GAS elements, which bind STAT proteins, we examined the potential involvement of IRF-3, IFN-β, and STAT-1α in LPS-induced CD40 expression. First, the ability of LPS to induce IRF-3 protein expression was examined. IRF-3 protein expression was increased 1 hour after LPS stimulation, peaked at 2 hours, and was still detectable at 4 hours (Figure 4B). To examine IFN-β mRNA expression, RAW264.7 and EOC13 cells were treated in the absence or presence of LPS for up to 12 hours and then analyzed for IFN-β mRNA levels. LPS-induced IFN-β mRNA expression was detected 0.5 hour after LPS treatment, peaked at 4 hours, and then decreased at 8 hours in both RAW264.7 and EOC13 cells (Figure 4C). LPS-induced IFN-β protein expression in RAW264.7 cells was detected 2 hours after LPS treatment, peaked at 8 hours, and then declined at 12 hours (Figure 4D). These experiments were repeated in EOC13 cells, and similar results were obtained (data not shown).
NF-κB and GAS elements are important for LPS-induced activation of the CD40 promoter. (A) Deletion constructs of the human CD40 promoter. (B) RAW264.7 cells were transiently transfected with 0.2 μg of the indicated constructs, then treated with medium or LPS (10 ng/mL) for 12 hours and analyzed for luciferase activity. Values were normalized to total protein, and fold induction was calculated by dividing the LPS treatment values by UN levels. Data are presented as mean ± SD of 3 experiments. (C) Site-directed mutant constructs of NF-κB and GAS elements in the human CD40 promoter. (D) RAW264.7 cells were transiently transfected with 0.2 μg of the indicated constructs, allowed to recover for 4 hours, then were treated with medium or LPS (10 ng/mL) for 12 hours and analyzed for luciferase activity. Fold induction was calculated and presented as mean ± SD of 3 experiments. (E) RAW264.7 cells were transiently cotransfected with the WT CD40 promoter construct (0.2 μg) and expression vectors containing WT or double negative (DN) of IKK-α or IKK-β cDNA (0.1 μg), then they were treated with LPS (10 ng/mL) for 12 hours and analyzed for luciferase activity. Differences in the amount of DNA were adjusted with the empty vector pcDNA3. Data are presented as mean ± SD of 3 experiments.
NF-κB and GAS elements are important for LPS-induced activation of the CD40 promoter. (A) Deletion constructs of the human CD40 promoter. (B) RAW264.7 cells were transiently transfected with 0.2 μg of the indicated constructs, then treated with medium or LPS (10 ng/mL) for 12 hours and analyzed for luciferase activity. Values were normalized to total protein, and fold induction was calculated by dividing the LPS treatment values by UN levels. Data are presented as mean ± SD of 3 experiments. (C) Site-directed mutant constructs of NF-κB and GAS elements in the human CD40 promoter. (D) RAW264.7 cells were transiently transfected with 0.2 μg of the indicated constructs, allowed to recover for 4 hours, then were treated with medium or LPS (10 ng/mL) for 12 hours and analyzed for luciferase activity. Fold induction was calculated and presented as mean ± SD of 3 experiments. (E) RAW264.7 cells were transiently cotransfected with the WT CD40 promoter construct (0.2 μg) and expression vectors containing WT or double negative (DN) of IKK-α or IKK-β cDNA (0.1 μg), then they were treated with LPS (10 ng/mL) for 12 hours and analyzed for luciferase activity. Differences in the amount of DNA were adjusted with the empty vector pcDNA3. Data are presented as mean ± SD of 3 experiments.
IFN-β activates STAT-1α and STAT-2 through the JAK-STAT pathway.32 To examine the activation of STAT-1α on LPS treatment, RAW264.7 cells were treated in the absence or presence of LPS for up to 4 hours, and STAT-1α tyrosine and serine phosphorylation was examined. Phosphorylation was first detected 2 hours after LPS addition and was enhanced at 4 hours (Figure 4E, lanes 4-5). These experiments were repeated in the EOC13 cells, and similar results were obtained (data not shown). These results indicate that LPS induces IRF-3 and IFN-β expression and subsequent activation of the transcription factor STAT-1α. To determine whether LPS-induced STAT-1α phosphorylation may be due to endogenous IFN-β production, we tested whether IFN-β–neutralizing antibodies could inhibit this response. RAW264.7 cells were incubated with medium, LPS, LPS plus 10 μg/mL IFN-β–neutralizing antibody, or isotype control antibody and then were analyzed for STAT-1α tyrosine phosphorylation. The results shown in Figure 4F demonstrate that inclusion of isotype antibody had no effect on LPS-induced STAT-1α phosphorylation (lane 3), whereas the IFN-β–neutralizing antibody substantially inhibited LPS-induced STAT-1α phosphorylation (lane 4). Identical results were obtained in EOC13 cells (data not shown).
Involvement of IFN-β in LPS-induced CD40 gene expression
The potential involvement of exogenously produced IFN-β in LPS-induced CD40 gene expression was next addressed. RAW264.7 and EOC13 cells were incubated with medium or LPS in the presence of 10 μg/mL neutralizing antibody against IFN-γ, IFN-α, IFN-β, or isotype control antibody for 4 hours, and then RNA was analyzed for CD40 and GAPDH mRNA expression. The results indicate that inclusion of isotype antibody or neutralizing antibodies against IFN-γ or IFN-α does not affect LPS-induced CD40 mRNA expression (Figure 5A). However, IFN-β–neutralizing antibody inhibited LPS-induced CD40 mRNA expression by approximately 60% in RAW264.7 cells and by approximately 50% in EOC13 cells (Figure 5A). To examine the effect of the endogenously produced IFN-β on CD40 protein expression, RAW264.7 cells were incubated with medium or LPS in the presence of 10 μg/mL IFN-β–neutralizing antibody or isotype control antibody for 36 hours, then CD40 surface protein expression was assessed by FACS analysis. Inclusion of the isotype control antibody did not affect LPS-induced CD40 protein expression, whereas the IFN-β–neutralizing antibody inhibited LPS-induced CD40 protein expression by approximately 50% (Figure 5B), comparable to the inhibition seen at the mRNA level (Figure 5A).
NF-κB and STAT-1α signaling pathways are activated by LPS. (A) RAW264.7 cells were incubated in the absence or presence of LPS (10 ng/mL) for up to 4 hours. Protein lysates were prepared and subjected to immunoblotting with anti–phospho-IKK-α/βSer180/Ser181, anti–phospho-IκBαSer32, and anti–phospho-NF-κB p65Ser536, stripped and reprobed with anti–NF-κB p65 and antiactin as loading controls. (B) RAW264.7 cells were incubated in medium or LPS (10 ng/mL) for up to 4 hours, lysed, and assayed for expression of IRF-3 protein. Actin protein expression was used as a loading control. (C) RAW264.7 or EOC13 cells were treated with LPS (10 ng/mL) for up to 12 hours, then RNA was isolated and subjected to RPA analysis for IFN-β and GAPDH mRNA expression. (D) RAW264.7 cells were treated with LPS (10 ng/mL) for up to 12 hours, then supernatants were collected and subjected to ELISA analysis for IFN-β protein expression. Data are presented as the mean ± SD of 3 experiments. (E) RAW264.7 cells were incubated in medium or LPS (10 ng/mL) for up to 4 hours, then cell lysates were prepared and subjected to immunoblotting with anti–phospho-STAT-1αTyr701 and anti–phospho-STAT-1αSer727, stripped, and reprobed with anti–STAT-1α and antiactin as loading controls. (F) RAW264.7 cells were treated with medium or LPS (10 ng/mL) in the absence or presence of 10 μg/mL isotype antibody or IFN-β–neutralizing antibody for 4 hours. Cell lysates were prepared and subjected to immunoblotting with anti–phospho-STAT-1αTyr701. Total STAT-1α and actin protein expression were used as loading controls. Representative of 3 experiments.
NF-κB and STAT-1α signaling pathways are activated by LPS. (A) RAW264.7 cells were incubated in the absence or presence of LPS (10 ng/mL) for up to 4 hours. Protein lysates were prepared and subjected to immunoblotting with anti–phospho-IKK-α/βSer180/Ser181, anti–phospho-IκBαSer32, and anti–phospho-NF-κB p65Ser536, stripped and reprobed with anti–NF-κB p65 and antiactin as loading controls. (B) RAW264.7 cells were incubated in medium or LPS (10 ng/mL) for up to 4 hours, lysed, and assayed for expression of IRF-3 protein. Actin protein expression was used as a loading control. (C) RAW264.7 or EOC13 cells were treated with LPS (10 ng/mL) for up to 12 hours, then RNA was isolated and subjected to RPA analysis for IFN-β and GAPDH mRNA expression. (D) RAW264.7 cells were treated with LPS (10 ng/mL) for up to 12 hours, then supernatants were collected and subjected to ELISA analysis for IFN-β protein expression. Data are presented as the mean ± SD of 3 experiments. (E) RAW264.7 cells were incubated in medium or LPS (10 ng/mL) for up to 4 hours, then cell lysates were prepared and subjected to immunoblotting with anti–phospho-STAT-1αTyr701 and anti–phospho-STAT-1αSer727, stripped, and reprobed with anti–STAT-1α and antiactin as loading controls. (F) RAW264.7 cells were treated with medium or LPS (10 ng/mL) in the absence or presence of 10 μg/mL isotype antibody or IFN-β–neutralizing antibody for 4 hours. Cell lysates were prepared and subjected to immunoblotting with anti–phospho-STAT-1αTyr701. Total STAT-1α and actin protein expression were used as loading controls. Representative of 3 experiments.
To determine the importance of IFN-β–induced STAT-1α activation in LPS-induced CD40 gene expression, primary microglia from STAT-1α–deficient mice were examined. The absence of STAT-1α led to a partial inhibition (∼ 40%) of LPS-induced CD40 mRNA and protein expression (Figure 5C-D). These experiments demonstrate that LPS-induced endogenous IFN-β and subsequent signaling through STAT-1α is responsible, in part, for CD40 gene expression.
Endogenous IFN-β is important for optimal LPS-induced CD40 expression. (A) RAW264.7 or EOC13 cells were treated with medium or LPS (10 ng/mL) in the presence of 10 μg/mL isotype antibody or neutralizing antibodies against IFN-γ, IFN-α, or IFN-β for 4 hours (RAW264.7 cells) or 8 hours (EOC13 cells). RNA was harvested and analyzed by RPA for CD40 and GAPDH mRNA. Representative of 3 experiments. (B) RAW264.7 cells were treated with medium or LPS (10 ng/mL) in the absence or presence of 10 μg/mL isotype antibody or IFN-β–neutralizing antibody for 36 hours. CD40 protein expression was detected by flow cytometry. Samples were analyzed by measuring MFI. Fold change in CD40 MFI value was calculated and shown as the mean ± SD of 3 experiments. (C) Primary microglia from WT and STAT-1α–deficient mice were treated with LPS (10 ng/mL) for 4 hours, and then mRNA was analyzed by RPA for CD40 and GAPDH expression. (D) Primary microglia from WT and STAT-1α–deficient mice were treated with LPS for 36 hours, then cells were subjected to FACS analysis for CD40 protein expression. Fold change in CD40 MFI value was calculated and shown as the mean ± SD of 3 experiments.
Endogenous IFN-β is important for optimal LPS-induced CD40 expression. (A) RAW264.7 or EOC13 cells were treated with medium or LPS (10 ng/mL) in the presence of 10 μg/mL isotype antibody or neutralizing antibodies against IFN-γ, IFN-α, or IFN-β for 4 hours (RAW264.7 cells) or 8 hours (EOC13 cells). RNA was harvested and analyzed by RPA for CD40 and GAPDH mRNA. Representative of 3 experiments. (B) RAW264.7 cells were treated with medium or LPS (10 ng/mL) in the absence or presence of 10 μg/mL isotype antibody or IFN-β–neutralizing antibody for 36 hours. CD40 protein expression was detected by flow cytometry. Samples were analyzed by measuring MFI. Fold change in CD40 MFI value was calculated and shown as the mean ± SD of 3 experiments. (C) Primary microglia from WT and STAT-1α–deficient mice were treated with LPS (10 ng/mL) for 4 hours, and then mRNA was analyzed by RPA for CD40 and GAPDH expression. (D) Primary microglia from WT and STAT-1α–deficient mice were treated with LPS for 36 hours, then cells were subjected to FACS analysis for CD40 protein expression. Fold change in CD40 MFI value was calculated and shown as the mean ± SD of 3 experiments.
Recruitment of NF-κB p65 and p50 and STAT-1α to the CD40 promoter, and modification of H3 and H4 histones in response to LPS
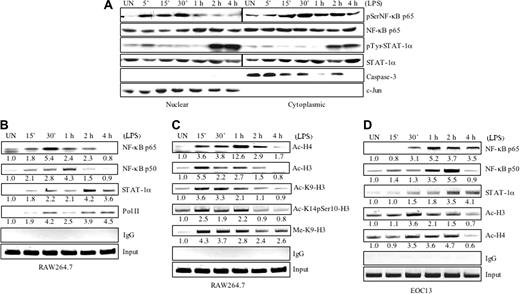
Although we have shown that NF-κB and STAT-1α transcription factors play critical roles in CD40 gene transcription,8 it is still unknown what transcription factors are recruited to the endogenous CD40 promoter in vivo, and how this is coupled to CD40 gene transcription. To study this, nuclear translocation of NF-κB and STAT-1α was investigated. RAW264.7 cells were incubated in the absence or presence of LPS for up to 4 hours, then nuclear and cytoplasmic fractions were isolated for protein analysis. LPS treatment lead to nuclear accumulation of phospho–NF-κB p65 at 5 to 30 minutes, then diminished at 1 to 4 hours (Figure 6A, top). Tyrosine-phosphorylated STAT-1α was detected in the nuclear fraction at 2 and 4 hours after LPS treatment (Figure 6A, middle), in agreement with the phosphorylation status shown previously (Figure 4D). Immunoblotting with anti–c-Jun and anti–caspase-3 antibodies showed a clean separation of nuclear and cytoplasmic fractions, respectively (Figure 6A, bottom).
Gene transcription in eukaryotic cells is controlled by protein complexes, including transcription factors, coregulators, chromatin-remodeling complexes, and the complexes responsible for signal-specific histone modifications on promoters.33 To monitor transcription factor binding in vivo, RAW264.7 and EOC13 cells were incubated in the absence or presence of LPS for up to 4 hours, and ChIP assays were performed using antibodies against NF-κB p65, NF-κB p50, STAT-1α, or normal rabbit IgG (as a negative control). PCR analysis of the positive control (input) indicates that the soluble chromatin samples obtained from each time point had equal amounts of chromatin fragments containing the CD40 promoter (Figure 6B-D). The transcription factors NF-κB and STAT-1α were recruited to the CD40 promoter in a time-dependent manner on LPS treatment. NF-κB p65 was weakly associated with the CD40 promoter in untreated RAW264.7 cells. Increased binding of p65 was observed at 15 minutes after LPS addition, and the peak of NF-κB p65 recruitment was at 30 minutes, although binding was still observed at 1 to 2 hours (Figure 6B). NF-κB p50 was also weakly associated with the CD40 promoter in untreated RAW264.7 cells. Increased binding of p50 was observed at 15 minutes and 30 minutes after LPS treatment, with the peak of NF-κB p50 recruitment at 1 hour, then diminishing at the 2-hour time point (Figure 6B). In the EOC13 cells, peak NF-κB p65 and p50 recruitment were observed at 1 to 2 hours (Figure 6D). STAT-1α was not associated with the CD40 promoter in untreated cells, and a small increase in binding of STAT-1α was observed at 15 minutes to 1 hour after LPS treatment in both RAW264.7 and EOC13 cells (Figure 6B-D). Optimal recruitment of STAT-1α to the CD40 promoter was observed between 2 and 4 hours of LPS treatment (Figure 6B,D). ChIP assays were performed for the binding of RNA Pol II to the CD40 promoter. Pol II was not associated with the CD40 promoter in untreated RAW264.7 cells. Recruitment of Pol II was observed 15 to 30 minutes after LPS addition, diminished slightly at 1 hour, and then reassociated with the CD40 promoter at 2 to 4 hours (Figure 6B).
LPS stimulates nuclear translocation and recruitment of transcriptional activators to the CD40 promoter and histone modifications. (A) RAW264.7 cells were incubated in the absence or presence of LPS (10 ng/mL) for up to 4 hours. Cytoplasmic and nuclear fractions were prepared and assayed with anti–phospho-NF-κB p65Ser536 and anti–phospho-STAT-1αTyr701. The cytoplasmic and nuclear fractions were also probed with anti–caspase-3, anti–c-Jun, anti–NF-κB p65, or anti–STAT-1α antibodies. (B) RAW264.7 cells were treated with LPS (10 ng/mL) for up to 4 hours, then the cells were crosslinked with formaldehyde. Soluble chromatin was subjected to immunoprecipitation with anti–NF-κB p65, anti–NF-κB p50, anti–STAT-1α, and anti–Pol II antibody, or normal rabbit IgG. The basal level of the untreated sample was set as 1.0, and fold induction on LPS treatment was compared with that. Representative of 3 to 4 experiments. (C) RAW264.7 cells were treated with LPS (10 ng/mL) for up to 4 hours, then the cells were crosslinked with formaldehyde. Soluble chromatin was subjected to immunoprecipitation with antibodies against histone acetylation, methylation, and phosphorylation. Fold induction on LPS treatment was calculated as in panel B. Representative of 3 experiments. (D) EOC13 cells were treated with LPS (10 ng/mL) for up to 4 hours, then the cells were crosslinked with formaldehyde. Soluble chromatin was subjected to immunoprecipitation with antibodies against NF-κB p65, anti–NF-κB p50, STAT-1α, Ac-H3, and Ac-H4. Fold induction on LPS treatment was calculated as in panel B. Representative of 3 experiments.
LPS stimulates nuclear translocation and recruitment of transcriptional activators to the CD40 promoter and histone modifications. (A) RAW264.7 cells were incubated in the absence or presence of LPS (10 ng/mL) for up to 4 hours. Cytoplasmic and nuclear fractions were prepared and assayed with anti–phospho-NF-κB p65Ser536 and anti–phospho-STAT-1αTyr701. The cytoplasmic and nuclear fractions were also probed with anti–caspase-3, anti–c-Jun, anti–NF-κB p65, or anti–STAT-1α antibodies. (B) RAW264.7 cells were treated with LPS (10 ng/mL) for up to 4 hours, then the cells were crosslinked with formaldehyde. Soluble chromatin was subjected to immunoprecipitation with anti–NF-κB p65, anti–NF-κB p50, anti–STAT-1α, and anti–Pol II antibody, or normal rabbit IgG. The basal level of the untreated sample was set as 1.0, and fold induction on LPS treatment was compared with that. Representative of 3 to 4 experiments. (C) RAW264.7 cells were treated with LPS (10 ng/mL) for up to 4 hours, then the cells were crosslinked with formaldehyde. Soluble chromatin was subjected to immunoprecipitation with antibodies against histone acetylation, methylation, and phosphorylation. Fold induction on LPS treatment was calculated as in panel B. Representative of 3 experiments. (D) EOC13 cells were treated with LPS (10 ng/mL) for up to 4 hours, then the cells were crosslinked with formaldehyde. Soluble chromatin was subjected to immunoprecipitation with antibodies against NF-κB p65, anti–NF-κB p50, STAT-1α, Ac-H3, and Ac-H4. Fold induction on LPS treatment was calculated as in panel B. Representative of 3 experiments.
Covalent histone modifications are indicators of the recruitment of histone-modifying complexes, such as histone acetyltransferases, histone deacetylases, and histone methyltransferases, and also have functional roles in gene transcriptional regulation.34-36 To study histone modifications during LPS-induced CD40 gene transcription, ChIP assays were performed. Enhanced acetylation of histone H4 was increased at 15 to 30 minutes following LPS treatment, peaked at 1 hour, then diminished over time in RAW264.7 cells (Figure 6C). The acetylation of histone H3 peaked at 15 minutes following LPS treatment and then diminished over the 4-hour time course (Figure 6C). In EOC13 cells, enhanced acetylation of H3 and H4 histones was observed at 30 minutes, persisted for up to 2 hours, then diminished at the 4-hour time point (Figure 6D). More specific changes in acetylation, phosphorylation, and methylation of H3 were examined in RAW264.7 cells by ChIP assays. Enhanced acetylation of K9-H3 was observed 15 minutes to 1 hour after LPS stimulation and then decreased (Figure 6C). Phosphorylation of serine 10 and acetylation of lysine 14 of H3 was increased at 15 minutes to 1 hour of LPS stimulation and then diminished (Figure 6C). Methylation of K9-H3 was observed at 15 minutes after LPS treatment and persisted over the 4-hour time course (Figure 6C). These results suggest that LPS induces histone modifications on the CD40 promoter concurrent with activation of the CD40 gene.
Discussion
In this study, we have presented data that LPS is a potent inducer of CD40 gene expression in macrophages and microglia. LPS induction of CD40 expression occurs at the transcriptional level and involves activation of the transcription factors NF-κB and STAT-1α. LPS directly activates NF-κB, and also induces endogenous production of IFN-β, which then leads to STAT-1α activation, and ultimately to CD40 gene expression. We also investigated the dynamic formation of transcriptional complexes during LPS-induced activation of the CD40 gene. Activators and RNA Pol II were recruited to the CD40 promoter in vivo in a stepwise and coordinated order, which was dependent on activation of the NF-κB and STAT-1α signaling pathways. LPS also induced histone H3 and H4 permissive modifications that were concurrent with activation of the CD40 gene. These findings illustrate that the coordination of cell signaling, histone modifications, and stepwise recruitment of transcription regulators is critical to LPS-induced CD40 gene expression.
CD40 has been implicated in participating in many human diseases, particularly autoimmune and inflammatory diseases.37-39 CD40 expression by microglia is critical for the infiltration and retention of inflammatory leukocytes into the central nervous system (CNS).40 Given the importance of CD40 and CD154 in neurologic diseases, and the contribution of macrophages/microglia to various CNS diseases such as multiple sclerosis and Alzheimer disease, we have been particularly interested in the regulation of CD40 expression in these cells. We initially made the observation that IFN-γ is a potent stimulator of CD40 expression and demonstrated that this induction was mediated by IFN-γ activation of STAT-1α and constitutively expressed PU.1, Spi-B, or both.6,7 STAT-1α bound to GAS elements within the CD40 promoter, whereas PU.1, Spi-B, or both bound to Ets elements.6,7 In addition, we and others demonstrated that IFN-γ induced production of TNF-α and subsequent activation of NF-κB is a critical aspect of IFN-γ-induced CD40 expression.8,41
The LPS induction of CD40 expression involves both GAS and NF-κB elements within the CD40 promoter, similar to IFN-γ; however, the intracellular signaling pathways leading to LPS-induced CD40 expression differ from those of IFN-γ. LPS directly activates NF-κB (p65 and p50), leading to binding at the CD40 promoter. In addition, LPS induces the expression of the transcription factor IRF-3, leading to IFN-β production and subsequent activation of STAT-1α. The partial inhibition of LPS-induced CD40 expression on inclusion of IFN-β–neutralizing antibody demonstrates the importance of this signaling pathway in CD40 gene expression. In addition, the importance of IFN-β–induced STAT-1α activation was verified using STAT-1α–deficient microglia; LPS-induced CD40 expression was partially inhibited in these cells. These findings reveal for the first time that LPS induces the recruitment of NF-κB and STAT-1α to the CD40 promoter in a time-dependent manner, which correlates with the kinetics of LPS-induced CD40 gene transcription in these cells. In a subsequent study, we have analyzed the influence of exogenous IFN-β on CD40 expression. IFN-β activation is associated with rapid recruitment of STAT-1α to the CD40 promoter (within 30 minutes), followed by acetylation of H3 and H4 histones. This response is independent of NF-κB because IFN-β does not activate the NF-κB pathway in RAW264.7 and EOC13 cells, and DN-IKK constructs do not inhibit IFN-β–induced CD40 promoter activity (H.Q., C.A.W., S.J.L., and E.N.B., manuscript submitted).
It has been shown that phorbol 12-myristate 13-acetate–activated matrix metalloproteinase-9 and IFN-γ–activated class II transactivator gene transcription is dependent on recruitment of Pol II to promoters.27,42 The LPS-induced recruitment kinetics of RNA Pol II correlates with transcriptional activation of the CD40 gene, suggesting that Pol II is important for LPS-initiated transcription of the CD40 gene. Concurrent with transcription complex formation, permissive histone modifications generally correlate with gene activation.27 We demonstrated that LPS-induced modification of histones H3 and H4 also correlated with activation of the CD40 gene. However, we did note that acetylation of H3 and H4, and acetylation of lysine 9 of H3, declined at 2 to 4 hours after LPS treatment, just as STAT-1α was being recruited to the CD40 promoter. Longer kinetic analysis of H3 and H4 acetylation will be performed to determine whether these permissive histone modifications occur again after binding of STAT-1α and the continuation of CD40 gene transcription. It has been suggested that chromatin-remodeling complexes may serve as “translators” to remodel nucleosomes corresponding to the instruction of the histone code.43 The interaction of transcription factors with cotransactivators such as CBP (CREB [cAMP (cyclic adenosine monophosphate) response element-binding protein]–binding protein) and p300, as well as coactivator-associated arginine methyltransferase (CARM1), and chromatin remodeling, may also be important for LPS-induced CD40 gene transcription. Studies on the transcriptional complexes involved in LPS-induced CD40 gene activation are ongoing.
Proposed model of LPS-induced CD40 gene expression. LPS activates NF-κB, which leads to subsequent nuclear translocation and binding of p65 and p50 to NF-κB elements in the CD40 promoter. Concurrently, LPS induces IRF-3 expression, which activates IFN-β expression. This leads to the subsequent activation of STAT-1α, which then dimerizes, translocates into the nucleus, and binds to GAS elements in the CD40 promoter with delayed kinetics compared with NF-κB p65 and p50. Concurrent with NF-κB and STAT-1α recruitment, LPS leads to modifications in H3 and H4 and to recruitment of RNA Pol II. The sequential recruitment of transcription factors and Pol II to the CD40 promoter, in conjunction with permissive histone modifications, results in transcriptional activation of the CD40 gene. See “Discussion” for details. MD2 indicates the cofactor for TLR4; TIRAP, Toll-IL-1R domain-containing adapter protein; TYK2, tyrosine kinase-2.
Proposed model of LPS-induced CD40 gene expression. LPS activates NF-κB, which leads to subsequent nuclear translocation and binding of p65 and p50 to NF-κB elements in the CD40 promoter. Concurrently, LPS induces IRF-3 expression, which activates IFN-β expression. This leads to the subsequent activation of STAT-1α, which then dimerizes, translocates into the nucleus, and binds to GAS elements in the CD40 promoter with delayed kinetics compared with NF-κB p65 and p50. Concurrent with NF-κB and STAT-1α recruitment, LPS leads to modifications in H3 and H4 and to recruitment of RNA Pol II. The sequential recruitment of transcription factors and Pol II to the CD40 promoter, in conjunction with permissive histone modifications, results in transcriptional activation of the CD40 gene. See “Discussion” for details. MD2 indicates the cofactor for TLR4; TIRAP, Toll-IL-1R domain-containing adapter protein; TYK2, tyrosine kinase-2.
Previous studies indicate that the time course of gene expression and ordered recruitment is accomplished by gene-specific transcription programs.27,44,45 In the case of LPS-induced CD40 gene transcription, LPS activates NF-κB p65 and p50, likely through the MyD88-dependent pathway, leading to their rapid association with the CD40 promoter. Concurrently, through the adaptor proteins TRIF and TRAM, LPS induces IRF-3 expression, which activates IFN-β gene expression, and then subsequently activates the STAT-1α transcription factor, leading to association with the CD40 promoter with delayed kinetics. Concurrent with NF-κB and STAT-1α recruitment, histone acetylation, methylation, and phosphorylation modifications occur, as well as RNA Pol II recruitment, which leads to CD40 gene expression (Figure 7). Therefore, cell-specific factors and signaling pathways regulate transcriptional activation of the CD40 gene by modulating its gene-specific transcriptional program.
Prepublished online as Blood First Edition Paper, July 14, 2005; DOI 10.1182/blood-2005-02-0759.
Supported by the National Institutes of Health (grants NS45290 and NS36765) (E.N.B.) and grant AR48311 (H.Q.).
H.Q. designed and performed many of the experiments in this study and wrote the first draft of the manuscript; C.A.W. assisted with many of the experiments in the study and contributed to “Materials and methods”; S.J.L. and X.Z. contributed to the data shown in the manuscript; and E.N.B. provided support for this project, helped with the experimental design, and finalized the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Rudolph Noelle (Dartmouth Medical School, Lebanon, NH) for providing the expression vectors of wild-type and dominant-negatives of IKK-α and IKK-β, Dr Vincent Nguyen (University of Texas, Galveston) for generation of the CD40 promoter constructs, and Dr Olaf Kutsch (University of Alabama at Birmingham) for providing primary human macrophage cultures. We thank Drs Max Cooper and Duane Wesemann (University of Alabama at Birmingham) for providing the STAT-1α–deficient mice.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal