Abstract
Using laser tweezers, we measured for the first time the forces of individual knob-into-hole interactions underlying fibrin polymerization. Exposure of A-knobs in desA-fibrin or its fragment from the central part of the molecule (N-terminal disulphide knot, NDSK) resulted in strong interactions with fibrinogen or fragment D (containing only a- and b-holes), producing a binding strength of approximately 125 to 130 pN. The interactions were not present in the absence of either knobs or holes and were abrogated by a specific inhibitor of fibrin polymerization, a peptide mimic of the A-knob (GPRPam). Exposure of both the A- and B-knobs in desAB-fibrin or desAB-NDSK did not change the rupture force spectra compared with the desA molecules, and their interactions with fibrinogen remained highly sensitive to GPRPam but not to GHRPam (B-knob), suggesting that neither A:b nor B:b nor B:a contacts contributed significantly to binding strength in addition to A:a contacts. The A:a interactions had a relatively small zero-force off-rate of approximately 10–4 s–1 and tight knob-to-hole contacts characterized by a transition state distance of approximately 0.3 nm. The results demonstrate that the knob-hole binding during thrombin-induced fibrin polymerization is driven by strong, stable, and highly specific A:a bonding, whereas A:b, B:b, or B:a interactions were not detected.
Introduction
Formation of insoluble fibrin polymer from its soluble precursor protein, fibrinogen, is the final stage of blood clotting. In vertebrates, fibrinogen conversion to fibrin occurs in 3 ordered steps: limited proteolysis by thrombin, self-assembly of newly formed fibrin monomers, and crosslinking of assembled fibrin to strengthen the clot. Thrombin splits off 2 pairs of fibrinopeptides A and B (FpA and FpB) from the fibrinogen polypeptide chains Aα and Bβ, respectively, whereas the γ-chains remain unaltered. Cleavage of the fibrinopeptides occurs in the central N-terminal part of the fibrinogen molecule called the E-region and results in exposure of pairs of “A” and “B” binding sites (Figure 1). These sites, also called A- and B-knobs, interact with complementary and constitutively accessible “a” and “b” sites (or a- and b-holes) located in the γ-modules and β-modules, respectively, of the D-regions at the ends of fibrinogen molecules1,2 (knob/hole terminology in this paper is from Russell F. Doolittle,3 who had insight on the nature of these binding interactions long before crystal structures were available). Although there are several other potential sites of mutual recognition, the interplay between D- and E-regions belonging to different fibrin monomer molecules is critical for fibrin self-assembly.4
Thrombin cleaves FpA more rapidly than FpB and first uncovers the new N-terminal α-chain motif Gly-Pro-Arg (GPR), the main functional amino acid sequence in the A-knob. Release of FpA and exposure of the GPR motif is sufficient to initiate fibrin polymerization, which is inhibited in the presence of free GPRPamide peptide by competing with the A-knobs for the a-holes. Snake venom enzymes such as batroxobin cleave FpA without cleaving FpB to produce desA-fibrin monomer. The polymerized desA-fibrin (or α-fibrin) in many respects is similar to normal desAB-fibrin except for some differences in ultrastructure.5 It is unknown whether A-knobs can form A:b contacts in addition to well-defined A:a binding, although structurally this does not seem impossible because the GPRPamide peptide binds to the b-holes (Laudano et al6 and O.V.G., unpublished data, April 15, 2005).
Because of the substantial delay, most FpBs are cleaved after the process of fibrin assembly has started and fibrin polymers have been formed.7,8 FpB release leads to exposure of the new N-terminal β-chain motif Gly-His-Arg-Pro (GHRP), which constitutes a major part of the B-knobs presumed to bind the b-holes. When co-crystallized with fragment DD, the GHRPamide peptide was found bound to the b-hole.9 However, there is no evidence that GHRP peptide would competitively inhibit normal fibrin formation.10 The role of B:b interactions was studied using either variant fibrinogens with impaired release of FpA or specific enzymes so that it was possible to remove FpB to obtain desB-fibrin monomer with exclusively or predominantly exposed B-knobs. These monomers were able to form a clot (β-fibrin) only at the temperature of 15°C or lower.11-13 Moreover, studies with recombinant fibrinogen suggest that polymerization might occur because of the B:a rather than the B:b interactions.14 FpB release from growing protofibrils enhances their lateral aggregation into thicker fibers.12,15 However, the mechanism by which this occurs is not clear. At least part of the effect of FpB cleavage arises from the consequent release of αC-domains and their intermolecular interactions that bring protofibrils together.16 Other aspects of lateral aggregation have also been proposed from the intermolecular packing observed in the crystal structures of fibrinogen fragments.17
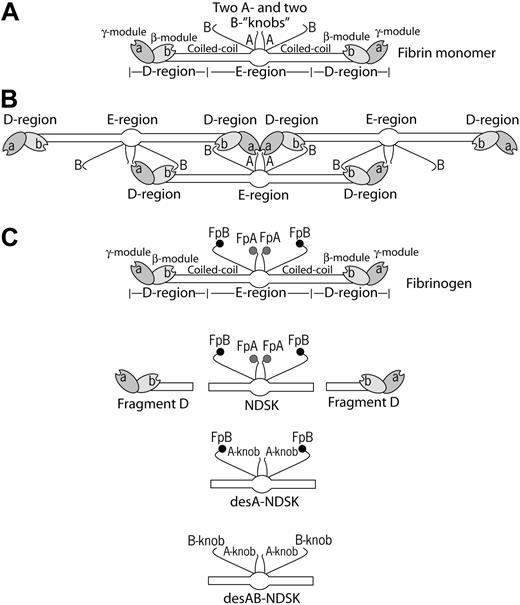
Schematic representation of fibrin molecules bearing knobs and holes that bind each other in the course of fibrin polymerization. (A) A fibrin monomer is 45 nm long and consists of 3 parts, namely 2 D-regions and 1 E-region. The D-regions contain the distal portions of the coiled-coil and the C-terminal β- and γ-modules. The E-region contains the central N-terminal part of the molecule and the proximal portions of both sets of coiled-coils. The E-region has 2 pairs of binding sites named A- and B-knobs that are exposed after cleavage of FpA and FpB by thrombin. The D-regions have constitutively open a- and b-holes located in the γ- and β-modules, respectively. (B) The driving force of fibrin polymerization is the complementary binding of the A-knobs and a-holes and perhaps of the B-knobs and b-holes resulting in formation of a half-staggered 2-strand protofibril. The cartoon is based on the crystallographic data and represents approximately the relative positions and dimensions of the molecular parts. The A- and B-knobs are highly flexible and hence have not been visualized in the crystal structure to date. (C) Cartoons of the molecules used in this study (fibrinogen, fragment D, and 3 types of the fragment called N-terminal disulphide knot, NDSK). The gray and black circles represent fibrinopeptides A (FpA) and B (FpB), respectively.
Schematic representation of fibrin molecules bearing knobs and holes that bind each other in the course of fibrin polymerization. (A) A fibrin monomer is 45 nm long and consists of 3 parts, namely 2 D-regions and 1 E-region. The D-regions contain the distal portions of the coiled-coil and the C-terminal β- and γ-modules. The E-region contains the central N-terminal part of the molecule and the proximal portions of both sets of coiled-coils. The E-region has 2 pairs of binding sites named A- and B-knobs that are exposed after cleavage of FpA and FpB by thrombin. The D-regions have constitutively open a- and b-holes located in the γ- and β-modules, respectively. (B) The driving force of fibrin polymerization is the complementary binding of the A-knobs and a-holes and perhaps of the B-knobs and b-holes resulting in formation of a half-staggered 2-strand protofibril. The cartoon is based on the crystallographic data and represents approximately the relative positions and dimensions of the molecular parts. The A- and B-knobs are highly flexible and hence have not been visualized in the crystal structure to date. (C) Cartoons of the molecules used in this study (fibrinogen, fragment D, and 3 types of the fragment called N-terminal disulphide knot, NDSK). The gray and black circles represent fibrinopeptides A (FpA) and B (FpB), respectively.
To study the molecular interactions during fibrin formation at the single molecule level, we have developed a model system to measure the binding strength between individual knobs and holes. Using laser tweezers-based methodology, we measured the rupture forces between fibrinogen, fibrin, and their fragments (Figure 1C). Fibrinogen and fibrinogen fragment D were used as the source of a- and b-holes, and fibrin monomer and fibrin monomer fragment NDSK (N-terminal disulfide knot) were the source of A- and B-knobs. The results based on rupture force profiles obtained under different experimental conditions indicate that the knob-hole binding is determined exclusively by strong, stable, tight, and highly specific A:a interactions without detectable A:b, B:b, or B:a binding.
Materials and methods
Purification of recombinant human fibrinogen
Normal recombinant fibrinogen was purified from the Chinese hamster ovary (CHO) cell culture medium (Cell Culture Center, Minneapolis, MN) by a 2-step procedure outlined previously.18 Briefly, fibrinogen was concentrated by ammonium sulfate precipitation followed by immunoaffinity chromatography using fibrinogen-specific antibody IF-1 (Iatron, Tokyo, Japan). Purified fibrinogen was dialyzed against 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.4, 150 mM NaCl (HEPES-buffered saline [HBS]) with 1 mM CaCl2 followed by extensive dialysis against HBS. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) confirmed high purity and proper polypeptide chain composition of the protein. Aliquots of purified protein were stored at –80°C.
Preparation of fibrinogen fragment D
Normal recombinant fibrinogen (8 mg) in HBS with 20 mM CaCl2 (0.61 mg/mL final concentration) was consumed to produce fragment D as described previously.19 Digestion of fibrinogen was initiated by adding 100 μL immobilized TPCK Trypsin (Pierce, Rockford, IL) which then proceeded at room temperature over a 4-day period while the course of reaction was monitored by SDS-PAGE. When only fragments D and E bands were visible by SDS-PAGE, the digestion was stopped by removing the trypsin-coated beads by filtering through a 0.22-μm filter (Costar, Corning, NY). The fragment D was purified as described19 using the GPRPAA affinity column. Fractions containing fragment D were pooled, dialyzed against HBS, and stored frozen at –80°C.
Preparation of NDSK fragments of fibrin(ogen)
The NDSK fragment, obtained by digestion with CNBr (cyanogen bromide), is composed of 2 of each chain Aα1-51, Bβ1-118, and γ1-78 linked together by 11 disulfide bonds.20,21 The active forms of NDSK without FpA and FpB contain the A- and B-knobs (Figure 1C) enabling the NDSK to bind specifically to fibrinogen, fibrin, and their fragments possessing a- and b-holes.22-24
Three types of NDSK fragment were purified based on the procedure described elsewhere.20 (1) To obtain the NDSK fragment with uncleaved FpA and FpB, intact plasma fibrinogen (10 mg/mL) was dialyzed against 70% formic acid and then CNBr (16 mg/mL) was added. The reaction was allowed to proceed under a layer of nitrogen for 36 hours at room temperature. The CNBr was then removed by dialysis against 5% acetic acid/0.1 M NaCl, and the digest was concentrated in a centrifugal filter device (50 kDa molecular weight [MW] cutoff; Millipore, Bedford, MA) to a final absorbance of A280 = 60. The NDSK fragment was separated from the rest of the CNBr digestion products with a fast protein liquid chromatography (FPLC) system (Amersham Biosciences, Piscataway, NJ) equipped with 2 sequentially connected Superdex 200 HR16/50 columns, equilibrated with 5% acetic acid/0.1 M NaCl at a flow rate of 1 mL/min. The CNBr digest, 4 mL A280 = 60, was injected into the column, and 1.8-mL fractions were collected. The peak with NDSK fragment was identified by the protein's ability to release FpA and FpB in the reaction with thrombin. Pooled fractions of NDSK were dialyzed against HBS, concentrated in a centrifugal filter device, and frozen at –80°C. The NDSK concentration was measured at 280 nm assuming an extinction coefficient 0.8 (calculated from the amino acid composition of NDSK, including the molecular weight of carbohydrates). The purified NDSK fragment ran as a single band on SDS-PAGE with an apparent molecular weight of 65 kDa and purity of greater than 95%. (2) The desA-NDSK (only FpA cleaved) and (3) desAB-NDSK fragments (FpA and FpB cleaved) were obtained from CNBr digests of desA- and desAB-fibrin, respectively, in the same way as with the fibrinogen CNBr digest. A fibrin clot was obtained by adding to 10 mg/mL fibrinogen either batroxobin (0.1 BU/mL, Batroxobin moojeni; CenterChem, Stamford, CT) or α-thrombin (0.1 U/mL; Enzyme Research Laboratories, South Bend, IN) and allowing clotting for 2 hours in HBS. The resulting clot was wrapped around a glass rod, washed in 0.1 M NaCl, dissolved in 70% formic acid, digested with CNBr, and purified with FPLC. The purity of desA and desAB-NDSK fragments as determined by SDS-PAGE was greater than 95%.
Coating surfaces with the proteins
Surfaces coated with the interacting proteins were prepared as described previously25 with some modifications. Each of the 3 NDSK fragments was bound covalently to spheric silica pedestals 5 μm in diameter anchored to the bottom of a chamber. Pedestals coated with a thin layer of polyacrylamide were activated with 10% glutaraldehyde, after which the proteins were immobilized overnight at 4°C from 1 mg/mL solution in 20 mM HEPES pH 7.4 containing 150 mM NaCl. After washing to remove noncovalently adsorbed protein, 2 mg/mL bovine serum albumin (BSA) in 0.055 M borate buffer pH 8.5 containing 1 mM CaCl2 was added as a blocker. To form desA- or desAB-fibrin–coated pedestals, fibrinogen was first immobilized as described above and then treated (1 BU/mL, 37°C, 1 hour) with batroxobin (B moojeni) or human thrombin (1 U/mL, 37°C, 1 hour), respectively, followed by washing of the chambers with 20 volumes of 100 mM HEPES pH 7.4 containing 150 mM NaCl, 3 mM CaCl2, 2 mg/mL BSA, and 0.1% (vol/vol) Triton X-100 before the measurements. Note that 1 unit of batroxobin activity (BU) was calibrated to be equal to 1 unit of thrombin activity (U) in terms of the rate of FpA release.
Fibrinogen or fragment D was bound covalently to carboxylate-modified 1.87 μm latex beads using N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (Sigma, Saint Louis, MO) as a crosslinking agent. BSA was used as a blocker. The immobilization step lasted 15 minutes at 4°C in 0.055 M borate buffer pH 8.5 containing 150 mM NaCl and 3 mM CaCl2. When immobilized from 20 μg/mL solution containing 100% fibrinogen labeled with I125, the surface density of I125-fibrinogen was determined to be about 11 ± 2 × 10–9 μg/μm2, which approaches the point of surface-binding saturation. Fibrinogen and fragment D were both used at the same concentration (20 μg/mL) in the binding solution.
The binding experiments were performed at room temperature in 100 mM HEPES pH 7.4 containing 150 mM NaCl, 3 mM CaCl2 with 2 mg/mL BSA and 0.1% (vol/vol) Triton X-100 added to reduce nonspecific interactions.
Brief description of the model system to quantify bimolecular interactions
We used a laser tweezers-based model system to study individual protein-protein interactions. A laser tweezers or gradient optical trap is formed by focusing a laser beam with a microscope objective to a spot in the specimen plane. The electric field of the light beam induces a dipole moment in dielectric particles and this attracts them to the center of the focused beam where the field is most intense. This system permits the measurement of discrete rupture forces produced by surface-bound molecular pairs during repeated intermittent contact.25 For these studies, a protein containing the E-region of fibrin (fibrin monomer or the NDSK fragment) was covalently bound to stationary pedestals anchored to the inner surface of a flow chamber. Latex beads coated covalently with fibrinogen or fragment D were then flowed into the chamber. One of the latex beads was trapped by a focused laser beam and moved in an oscillatory manner so that the bead was intermittently in contact with a stationary pedestal. The tension produced when fibrinogen (or fragment D) on the latex bead interacted with a complementary molecule on the anchored pedestal was sensed and displayed as a force signal that was proportional to the strength of protein-protein binding (Figure 2A). Rupture forces from many interactions were collected and displayed as normalized force spectra histograms for each experimental condition.
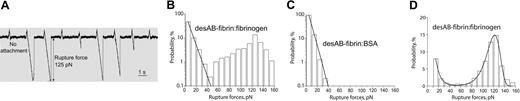
Rupture forces of individual protein molecules registered as multiple calibrated signals and arranged into force distribution histograms. (A) Data trace of 9 successive signals produced during repeated contacts of a desAB-fibrin–coated pedestal and fibrinogen-coated latex bead. At the moment of contact the laser trap exerts a small positive, compressive force on the pedestal and the bead. When the pedestal and the bead bind, either specifically or nonspecifically, the force on the bead increases in the negative direction until the pedestal-bead bond is ruptured, and the force rapidly returns to zero. If no attachment occurs, there is no negative rupture force. (B) DesAB-fibrin–fibrinogen rupture forces displayed as a normalized force distribution with 2 force regimes bordering at about 50 pN. The total number of contacts (n = 10 865) is taken to be 100%. (C) A control histogram of exponentially decreasing rupture forces 0 to 40 pN produced by nonspecifically interacting desAB-fibrin–and BSA-coated surfaces (n = 3689). (D) A sample of rupture force distribution of desAB-fibrin–fibrinogen interactions fitted to the Bell function combined with exponential decay for nonspecific forces less than 40 pN (n = 1378). Signals that appeared as forces below 10 pN were considered nonbinding events or zero.
Rupture forces of individual protein molecules registered as multiple calibrated signals and arranged into force distribution histograms. (A) Data trace of 9 successive signals produced during repeated contacts of a desAB-fibrin–coated pedestal and fibrinogen-coated latex bead. At the moment of contact the laser trap exerts a small positive, compressive force on the pedestal and the bead. When the pedestal and the bead bind, either specifically or nonspecifically, the force on the bead increases in the negative direction until the pedestal-bead bond is ruptured, and the force rapidly returns to zero. If no attachment occurs, there is no negative rupture force. (B) DesAB-fibrin–fibrinogen rupture forces displayed as a normalized force distribution with 2 force regimes bordering at about 50 pN. The total number of contacts (n = 10 865) is taken to be 100%. (C) A control histogram of exponentially decreasing rupture forces 0 to 40 pN produced by nonspecifically interacting desAB-fibrin–and BSA-coated surfaces (n = 3689). (D) A sample of rupture force distribution of desAB-fibrin–fibrinogen interactions fitted to the Bell function combined with exponential decay for nonspecific forces less than 40 pN (n = 1378). Signals that appeared as forces below 10 pN were considered nonbinding events or zero.
Measurement of rupture forces, data processing, and analysis
The position of the optical trap and hence a fibrinogen- or fragment D–coated latex bead was oscillated in a triangular waveform at 0.5 Hz with the pulling velocity of 1.8 μm/s which corresponded to the loading rate of 400 pN/s. All experiments were conducted at a trap stiffness of 0.22 ± 0.01 pN/nm, as computed from the bandwidth of Brownian motion. Contact duration between interacting surfaces varied from 10 to 200 ms. Rupture forces were collected at 2000 scans per second (0.5 ms time resolution). The results of many experiments under similar conditions were averaged so that each rupture force histogram represented from 103 to 1.5 × 104 repeated contacts of more than 10 different bead-pedestal pairs. Individual forces measured during each contact-detachment cycle were collected into 10 pN-wide bins. The number of events in each bin was plotted against the average force for that bin after normalizing for the total number of interaction cycles (Figure 2B-C). The percentage of events in a particular force range (bin) represents the probability of rupture events at that tension. Optical artifacts observed with or without trapped latex beads produce signals that appeared as forces below 10 pN. Accordingly, rupture forces in this range were not considered when the data were analyzed.
Data analysis was based on the Bell model for the rupture under load of single-molecule interactions.26 In the model, the probability P(f) that a complex will come unbound at a force f during linearly increased load (loading rate, rF) is given by:
where k0 is the dissociation rate in the absence of a pulling force, γ is the transition state position, and kB and T are Boltzmann constant and temperature.27,28
To account separately for nonspecific interactions observed at low forces, we modified this equation to the form,
in which A1 and A2 are scaling constants, and τ is an additional constant to fit the force-dependence of the nonspecific interactions.
The model given by the second equation was fit to the data using a least squares fitting analysis in Origin 7.5 (OriginLab, Northampton, MA) with τ, γ, k0, A1, and A2 as free parameters and rF = 400 pN/s, kB = 1.38 × 10–23 J/K, and T = 290 K as fixed parameters. Error bars represent the Origin-estimated standard error for each parameter.
Results
Rupture force spectra of the specific fibrin-fibrinogen binding versus nonspecific protein-protein interactions
Surfaces coated with thrombin-treated preimmobilized fibrinogen (desAB-fibrin monomer) were highly reactive with surfaces coated with untreated fibrinogen. During repetitive binding/unbinding they generated a wide range of individual rupture force signals varying from 10 to 160 pN (Figure 2A). The force values were arranged into histograms with 10 pN–wide bins, which were normalized by the total number of contacts between a pedestal and a bead (Figure 2B). The spectrum could be visually segregated into 2 force regimes. One of them covered the 10- to 40-pN force range with an exponentially decreasing binding probability, and the other formed a pronounced asymmetric peak with a maximum at 120 to 130 pN and a gently sloped shoulder over the lower forces of 40 to 100 pN (Figure 2B).
To discriminate specific binding from nonspecific protein-protein interactions, the rupture force measurement was repeated with BSA-coated beads contacting desAB-fibrin–coated pedestals (Figure 2C). The results suggest that forces ranging from 0 to approximately 40 pN are required to rupture nonspecific protein-protein binding, consistent with nonspecific rupture forces reported previously for a number of biologically inert protein pairs.25,29 Although shown in the subsequent experimental histograms, nonspecific forces less than 40 pN were excluded from the analysis and interpretation of the forces reflecting specific knob-hole binding.
The rupture forces greater than 40 pN that represent specific fibrin-fibrinogen interactions were well fitted with a Bell model function describing the dependence of rupture force spectra on the thermodynamics and kinetics of single receptor-ligand pairs26 (Figure 2D). The fitting analysis enabled the extraction of 2 molecular parameters of the interaction, namely, the zero-force off-rate (k0) and the distance in the direction of the applied force between the minimum free energy of the bound state and the free energy of the transition state between attached and detached states (transition state distance, γ). For desAB-fibrin–fibrinogen interactions, k0 = 0.000 98 ± 0.000 60 s–1 and γ= 0.29 ± 0.04 nm (Table 1). Figure 2D is an example of a typical force distribution for the specific interactions and illustrates the fitting function that was applied to analyze experimental data presented here.
Off-rate (k0) and transition-state distance (γ) of the A:a interactions measured using fibrin(ogen) molecules and their fragments
Interacting molecular pairs . | k0 (s-1) . | γ (nm) . |
|---|---|---|
| desA-fibrin/fibrinogen | 0.00067 ± 0.00063 | 0.34 ± 0.03 |
| desA-NDSK/fibrinogen | 0.00086 ± 0.00059 | 0.33 ± 0.06 |
| desA-NDSK/fragment D | 0.00089 ± 0.00037 | 0.33 ± 0.05 |
| desAB-fibrin/fibrinogen | 0.00098 ± 0.00060 | 0.29 ± 0.04 |
| desAB-NDSK/fibrinogen | 0.00135 ± 0.00072 | 0.33 ± 0.02 |
| desAB-NDSK/fragment D | 0.00034 ± 0.00052 | 0.37 ± 0.05 |
Interacting molecular pairs . | k0 (s-1) . | γ (nm) . |
|---|---|---|
| desA-fibrin/fibrinogen | 0.00067 ± 0.00063 | 0.34 ± 0.03 |
| desA-NDSK/fibrinogen | 0.00086 ± 0.00059 | 0.33 ± 0.06 |
| desA-NDSK/fragment D | 0.00089 ± 0.00037 | 0.33 ± 0.05 |
| desAB-fibrin/fibrinogen | 0.00098 ± 0.00060 | 0.29 ± 0.04 |
| desAB-NDSK/fibrinogen | 0.00135 ± 0.00072 | 0.33 ± 0.02 |
| desAB-NDSK/fragment D | 0.00034 ± 0.00052 | 0.37 ± 0.05 |
Data are expressed as mean ± standard error of the mean.
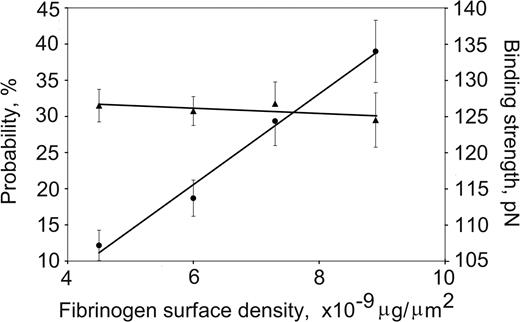
DesA-NDSK/fibrinogen binding probability and strength plotted against the fibrinogen surface density. The fibrinogen surface densities were nonsaturating in this range, and the binding probabilities (•) increased linearly, whereas the binding strengths (▴) remained unchanged. Error bars indicate standard deviation.
DesA-NDSK/fibrinogen binding probability and strength plotted against the fibrinogen surface density. The fibrinogen surface densities were nonsaturating in this range, and the binding probabilities (•) increased linearly, whereas the binding strengths (▴) remained unchanged. Error bars indicate standard deviation.
Discriminating single versus multiple desA-NDSK/fibrinogen bonds
The effect of the fibrinogen surface density on the strength of fibrinogen binding to desA-NDSK was tested by measuring rupture force distributions of beads coated with fibrinogen at protein densities of 4.5 × 10–9 μg/μm2, 6 × 10–9 μg/μm2, 7.3 × 10–9 μg/μm2, and 8.6 × 10–9 μg/μm2 successively flowed into a chamber containing desA-NDSK–coated pedestals. The bead-pedestal binding strength was unchanged, whereas the cumulative probability for specific binding increased linearly with fibrinogen surface density (Figure 3) consistent with the interactions of desA-NDSK binding to single fibrinogen molecules.30
Interactions of desA-fibrin and desA-NDSK with fibrinogen and fragment D
To study particular pairs of knob-hole interactions, combinations of fibrin(ogen) molecules and their fragments bearing D- and E-regions were bound to pedestals and beads. DesA-fibrin and desA-NDSK, which were obtained by batroxobin-induced cleavage of FpA, are suitable to study A:a and other potential A-mediated interactions because they have only A-knobs exposed.
The molecules of desA-fibrin and fibrinogen had an average rupture force distribution with the peak at 127 ± 1 pN and a cumulative binding probability of 49% ± 18% (Figure 4A; Table 2). In the presence of GPRP the binding probability was reduced 12-fold (Figure 4B; Table 2) and in the absence of cleavage with batroxobin fibrinogen-coated pedestals bound fibrinogen-coated beads with probability 8-fold lower than with batroxobin treatment (Figure 4C), suggesting that the desA-fibrin and fibrinogen interactions were specific. The off-rate (k0 = 0.000 67 ± 0.000 63 s–1) and transition state distance (γ= 0.340 ± 0.032 nm) for desA-fibrin and fibrinogen were similar to the desAB-fibrin–fibrinogen interactions (Table 1).
Effects of GPRPam and GHRPam on the binding strength and probability of knob-hole interactions
Interacting molecular pairs and conditions . | Binding strength, pN . | Cumulative probability, % . |
|---|---|---|
| desA-fibrin/fibrinogen | 127 ± 1 | 49 ± 18 |
| desA-fibrin/fibrinogen + GPRPam | Indeterminable | 3.9 ± 1.3 |
| desA-NDSK/fibrinogen | 127 ± 2 | 44 ± 13 |
| desA-NDSK/fibrinogen + GPRPam | Indeterminable | 1.3 ± 0.6 |
| desA-NDSK/fragment D | 127 ± 2 | 28 ± 13 |
| desA-NDSK/fragment D + GPRPam | Indeterminable | 5.4 ± 2.6 |
| desAB-fibrin/fibrinogen | 122 ± 2 | 52 ± 16 |
| desAB-fibrin/fibrinogen + GPRPam | 129 ± 1 | 4.9 ± 1.8 |
| desAB-fibrin/fibrinogen + GHRPam | 124 ± 2 | 37 ± 13 |
| desAB-NDSK/fibrinogen | 126 ± 1 | 35 ± 10 |
| desAB-NDSK/fibrinogen + GPRPam | Indeterminable | 0.6 ± 0.2 |
| desAB-NDSK/fibrinogen + GHRPam | 124 ± 1 | 37 ± 7 |
| desAB-NDSK/fragment D | 124 ± 2 | 26 ± 10 |
| desAB-NDSK/fragment D + GPRPam | Indeterminable | 1.2 ± 0.3 |
| desAB-NDSK/fragment D + GHRPam | 122 ± 1 | 34 ± 13 |
Interacting molecular pairs and conditions . | Binding strength, pN . | Cumulative probability, % . |
|---|---|---|
| desA-fibrin/fibrinogen | 127 ± 1 | 49 ± 18 |
| desA-fibrin/fibrinogen + GPRPam | Indeterminable | 3.9 ± 1.3 |
| desA-NDSK/fibrinogen | 127 ± 2 | 44 ± 13 |
| desA-NDSK/fibrinogen + GPRPam | Indeterminable | 1.3 ± 0.6 |
| desA-NDSK/fragment D | 127 ± 2 | 28 ± 13 |
| desA-NDSK/fragment D + GPRPam | Indeterminable | 5.4 ± 2.6 |
| desAB-fibrin/fibrinogen | 122 ± 2 | 52 ± 16 |
| desAB-fibrin/fibrinogen + GPRPam | 129 ± 1 | 4.9 ± 1.8 |
| desAB-fibrin/fibrinogen + GHRPam | 124 ± 2 | 37 ± 13 |
| desAB-NDSK/fibrinogen | 126 ± 1 | 35 ± 10 |
| desAB-NDSK/fibrinogen + GPRPam | Indeterminable | 0.6 ± 0.2 |
| desAB-NDSK/fibrinogen + GHRPam | 124 ± 1 | 37 ± 7 |
| desAB-NDSK/fragment D | 124 ± 2 | 26 ± 10 |
| desAB-NDSK/fragment D + GPRPam | Indeterminable | 1.2 ± 0.3 |
| desAB-NDSK/fragment D + GHRPam | 122 ± 1 | 34 ± 13 |
Values are mean ± SEM.
To avoid potential D/D interactions, the desA-fibrin monomer experiments were repeated with desA-NDSK. The rupture force spectrum for desA-NDSK and fibrinogen peaked at 127 ± 2 pN with a cumulative probability of 44% ± 13% (Figure 4D; Table 2). In the presence of GPRP (Figure 4E) the desA-NDSK/fibrinogen interactions were almost totally abrogated with the initial force spectrum being completely recovered after the peptide was washed out (not shown), suggesting that the desA-NDSK and fibrinogen interactions were specific. The off-rate and transition state distance for the desA-NDSK/fibrinogen binding were almost the same as for the desA-fibrin and fibrinogen (Table 1). The spectrum of rupture forces for NDSK/fibrinogen interactions did not show a specific peak and was very different from desA-NDSK/fibrinogen (Figure 4F). This force spectrum was unaffected by either GPRP or GHRP (not shown), suggesting that the interactions were not caused by binding at A- or B-sites.
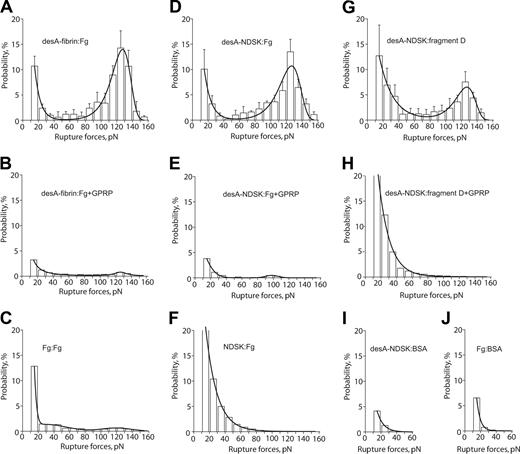
The panel of rupture force spectra demonstrating the interactions of desA-fibrin and desA-NDSK with fibrinogen and fragment D along with control data for nonspecific protein-protein binding. (A,D,G) Pure interactions between desA-fibrin and fibrinogen, desA-NDSK and fibrinogen, desA-NDSK and fragment D, respectively; (B,E,H) the same interactions in the presence of 1 mM GPRPam; (C,F,I,J) negative controls with one or both interacting surfaces coated with the proteins lacking complementary D/E binding sites: fibrinogen versus fibrinogen (C), NDSK versus fibrinogen (F), desA-NDSK versus BSA (I), fibrinogen versus BSA (J). Error bars indicate standard deviation.
The panel of rupture force spectra demonstrating the interactions of desA-fibrin and desA-NDSK with fibrinogen and fragment D along with control data for nonspecific protein-protein binding. (A,D,G) Pure interactions between desA-fibrin and fibrinogen, desA-NDSK and fibrinogen, desA-NDSK and fragment D, respectively; (B,E,H) the same interactions in the presence of 1 mM GPRPam; (C,F,I,J) negative controls with one or both interacting surfaces coated with the proteins lacking complementary D/E binding sites: fibrinogen versus fibrinogen (C), NDSK versus fibrinogen (F), desA-NDSK versus BSA (I), fibrinogen versus BSA (J). Error bars indicate standard deviation.
To test whether the D-region of surface-bound fibrinogen was responsible for the interaction with desA-NDSK, we used fragment D for comparison. The rupture force distribution of the desA-NDSK/fragment D binding was quite similar to that of desA-NDSK/fibrinogen, with the same binding strength of 127 ± 2 pN (Figure 4G; Table 2). The kinetic parameters of the interactions with fragment D (the off-rate and the transition state position) were indistinguishable from desA-NDSK and fibrinogen (Table 1). Susceptibility of the desA-NDSK/fragment D interactions to the inhibitory effect of the GPRP peptide was almost as high as for the desA-NDSK/fibrinogen interactions (Figure 4H; Table 2) but the fraction of GPRP-insensitive binding events with rupture forces less than 40 pN was greater (17% versus 5% on average). This may be due to the random orientation of immobilized fragment D molecules, leading to increased nonspecific protein-protein interactions observed in controls at the desA-NDSK/BSA (Figure 4I) and fibrinogen/BSA (Figure 4J) interfaces. GHRP did not affect rupture force distributions of desA-fibrin and desA-NDSK interacting with fibrinogen and fragment D (data not shown).
Interactions of desAB-fibrin and desAB-NDSK with fibrinogen and fragment D
DesAB-fibrin and desAB-NDSK with FpA and FpB cleaved have both A- and B-knobs exposed and can potentially participate in several types of interactions with a- and b-holes.
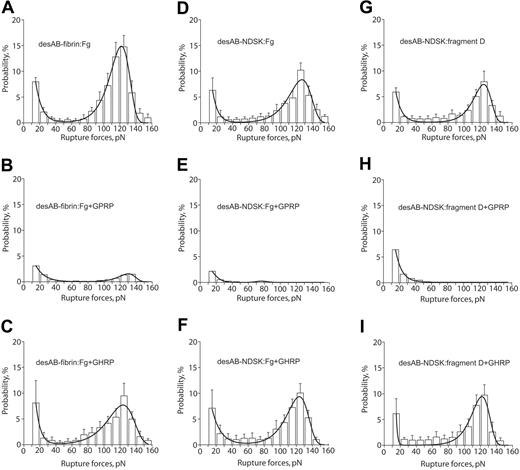
The rupture force spectra for interactions of desAB-fibrin or desAB-NDSK with fibrinogen or fragment D were similar to the desA-fibrin–fibrinogen spectra (Figure 5A,D,G; Table 2). The GPRP peptide reduced both components of the spectra (Figure 5B,E,H), whereas GHRP did not significantly affect the force spectra (Figure 5C,F,I), the kinetic parameters (Table 2), or the binding probabilities (Table 1).
The panel of rupture force spectra showing effects of the GPRPam and GHRPam peptides on the interactions of desAB-fibrin and desAB-NDSK with fibrinogen and fragment D. (A,D,G) Pure interactions between desAB-fibrin and fibrinogen, desAB-NDSK and fibrinogen, desAB-NDSK and fragment D, respectively; (B,E,H) the same interactions in the presence of 1 mM GPRPam; (C,F,I) the same interactions in the presence of 1 mM GHRPam. Error bars indicate standard deviation.
The panel of rupture force spectra showing effects of the GPRPam and GHRPam peptides on the interactions of desAB-fibrin and desAB-NDSK with fibrinogen and fragment D. (A,D,G) Pure interactions between desAB-fibrin and fibrinogen, desAB-NDSK and fibrinogen, desAB-NDSK and fragment D, respectively; (B,E,H) the same interactions in the presence of 1 mM GPRPam; (C,F,I) the same interactions in the presence of 1 mM GHRPam. Error bars indicate standard deviation.
Discussion
Repeated touching and separation of surfaces coated with fibrin(o-gen) molecules and their fragments require a wide range of forces up to 160 pN. To distinguish between specific and nonspecific binding, 2 types of control experiments were performed in which the specific interactions were suppressed by either replacing one of the interacting molecules with an inert protein (Figures 2C and 4C,F,I,G) or adding the peptide GPRPam, an inhibitor of fibrin polymerization (Figures 4B,E,H and 5B,E,H; Table 2). The data suggest that nearly all of the nonspecific interactions are limited to ruptures occurring at less than 40 pN, whereas more than 92% of the ruptures occurring at greater than 40 pN are specific. Accordingly, the analysis was focused on ruptures at forces greater than 40 pN.
On the basis of several criteria that have been proposed to test whether the observed ruptures were due to the attachments of single or multiple pairs of molecules,25,29 our data indicate that under the experimental conditions studied the rupture events were due to single bimolecular attachments. First, the rupture of multiple attachments should occur in a sequence of multiple steps, whereas the rupture of a single attachment should always occur in one step. Typically 90% to 99% of the ruptures occurred in one step (as in Figure 2A) and less than 10% occurred in 2 or more steps manifesting themselves as jagged signals (not shown). Only the single-step interactions were included in the analysis. Second, the distribution of rupture forces of multiple identical attachments should appear as a series of peaks that are multiples of a single value of force and have probabilities inversely proportional to the number of bonds. For interactions with rupture forces greater than 40 pN we observed only a single well-defined peak in the force histograms (Figures 2D, 4, and 5). Third, if the interactions between beads and pedestals are because of the multiple attachments, then the binding strength will increase at increased surface densities of the interacting proteins. This did not occur for any of the fibrin(ogen) and fibrin(ogen) fragment interactions (Figure 3). Fourth, if the interactions are due to multiple molecular pairs then the probability of an interaction occurring will be strongly dependent in a nonlinear manner on the surface density of the interacting proteins. The probability of interactions increased linearly with protein density for the binding strength of desA-NDSK/fibrinogen interactions (Figure 3), which is consistent with the attachment of single molecular pairs.
Fibrin and fibrinogen molecules readily form complexes in solution,4 so it was not surprising that surfaces coated with desA- or desAB-fibrin monomers were highly reactive with surface-bound fibrinogen (Figures 4A and 5A). Pairs of fibrin(ogen) molecules have several potential specific interactions such as the knob-hole, end-to-end, side-to-side,17,31 αC:central region, and αC:αC.32,33 In addition, contacts between large, long, and flexible molecules may result in numerous nonspecific interactions because of their large surface areas. However because the interactions of the NDSK fragments with fibrinogen have the same binding characteristics as the interactions of parent molecules (Figures 4D and 5D; Tables 1 and 2), it is likely that the knob-hole interactions dominate over all other specific interactions. When fibrinogen was replaced with fragment D and allowed to interact with desA-NDSK or desAB-NDSK, the binding parameters remained essentially unchanged (Figures 4G and 5G; Tables 1 and 2), confirming that specific binding between D- and E-regions of surface-bound molecules are responsible for the interactions between desA- or desAB-fibrin and fibrinogen.
To investigate the binding-site specificity of the D:E interactions and to identify particular knob-hole pairs involved in these interactions we used 2 approaches. In the first, the effects on the force distribution of batroxobin-induced cleavage of FpA, exposing only A-knobs, was compared with thrombin-induced cleavage of both FpA and FpB, exposing both A- and B-knobs. In the second approach either a- or b-holes were selectively blocked using GPRPam or GHRPam peptide, respectively. In the experiments using desA-fibrin and desA-NDSK exposed to fibrinogen or fragment D there are 2 possible combinations of the knob-hole pairs, either A:a or A:b. Addition of the GPRP almost completely but reversibly abrogated the rupture forces greater than 40 pN, indicating that they reflected specific A:a interactions (Figure 4B,E,H; Table 2). Addition of the GHRPam peptide, in contrast to the GPRPam, did not affect force spectra produced by the A-knob–containing proteins, suggesting that there were no detectable A:b interactions.
For desAB-fibrin or desAB-NDSK interacting with fibrinogen or fragment D, the possible combinations of the knob-hole pairs are A:a, A:b, B:b, and B:a. In our model system the B:b interactions may be blocked because the b-hole in the bound fibrinogen molecule is oriented in the opposite direction from the a-hole,17,32 which might cause steric hindrance. To correct for this possibility we used D fragment because it can bind to the surface such that either the a- or b-hole is properly oriented toward the interacting counterparts: A- and B-knobs. Because the GHRPam peptide, again, did not quantitatively or qualitatively change the force spectrum (Figure 5C,F,I; Table 2), it is likely that the only type of knob-hole interaction is the A:a binding.
There are at least 2 possible explanations for the absence of B:b interactions in our model system. First, the A:a bonds may form more quickly than B:b bonds, and the brief bead-pedestal contacts are too short for B:b bond formation. An a-hole is capable of fast interactions with GPRPam (the A-knob analog) and does not undergo discernible conformational changes on binding, whereas the b-hole undergoes slow conformational changes involving movement of the residues βE397 and βD398 when binding GHRPam (the B-knob analog).9,19,34 Second, the B:b contacts may require specific orientation of the interacting sites achieved only at the D/E/D interface when a fibrin polymer is preformed via A:a binding.35 The latter assumption is consistent with the model in which initial interactions of the D- and E-regions via A:a binding result in dissociation of the β-module from the coiled coil and subsequent completion of the b-polymerization pocket enabling the B:b interactions.36
Because the rupture force spectra produced by different molecular pairs containing E- and D-regions of fibrin(ogen) represent exclusively the interactions between the A-knobs and the a-holes, they can be analyzed in terms of single-molecule mechanics and kinetics in relation to the known composition and structure of these sites.
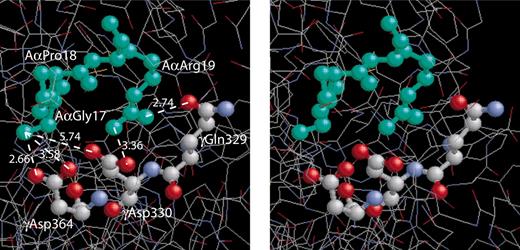
A stereo figure showing the residues GPR, corresponding to the fibrin α-chain N-terminus, bound to the polymerization pocket in the globular γ-chain portion of fragment D19and depicting the actual bond lengths (Å). The GPR sequence of the GPRPam peptide mimics the working part of A-knob composed of the amino acid residues (Aα17-19) exposed after cleavage of FpA. The a-hole is represented by 3 selected amino acid residues (γ329, γ330, and γ364) that are directly involved in the interaction.1,9 Considering GPRP the “surrogate A-knob”17 it is likely that the driving force of the A:a binding are the multiple electrostatic and hydrogen bonds between the α-amino group of AαGly17 and the guanidinium group of AαArg19 of an A-knob, on the one hand, and γAsp330 and γAsp364 along with carboxyamide of γGln329 of an a-hole, on the other hand. This aggregate of bonds stabilizing the A:a coupling is abruptly broken as soon as the knob and the hole are pulled apart to a distance of about 3 Å.
A stereo figure showing the residues GPR, corresponding to the fibrin α-chain N-terminus, bound to the polymerization pocket in the globular γ-chain portion of fragment D19and depicting the actual bond lengths (Å). The GPR sequence of the GPRPam peptide mimics the working part of A-knob composed of the amino acid residues (Aα17-19) exposed after cleavage of FpA. The a-hole is represented by 3 selected amino acid residues (γ329, γ330, and γ364) that are directly involved in the interaction.1,9 Considering GPRP the “surrogate A-knob”17 it is likely that the driving force of the A:a binding are the multiple electrostatic and hydrogen bonds between the α-amino group of AαGly17 and the guanidinium group of AαArg19 of an A-knob, on the one hand, and γAsp330 and γAsp364 along with carboxyamide of γGln329 of an a-hole, on the other hand. This aggregate of bonds stabilizing the A:a coupling is abruptly broken as soon as the knob and the hole are pulled apart to a distance of about 3 Å.
The binding between an A-knob and an a-hole is relatively strong and is characterized by a yield force of about 125 to 130 pN when the applied load is increased at the rate of 400 pN/s. It is stronger than the bimolecular integrin αIIbβ3-fibrinogen complex37 and most of the other specific protein-protein pairs measured at similar loading rates.38 It is comparable with the binding strength of streptavidin to biotin, thought to be the strongest noncovalent interaction in biology.39 Because a fibrin molecule is bivalent and has an A-knob and an a-hole reacting in each of 2 interacting partners (Figure 1), the measured binding strength may be doubled to 250 to 260 pN for bimolecular fibrin-fibrin interactions. To our knowledge, the only work attempting to quantify the single-molecule fibrin monomer-monomer binding strength reported a value on the order of 400 pN,40 which was calculated indirectly from the parameters of shear-induced detachment of fibrin-coated beads from the fibrin-coated surfaces. Although this value has the same order of magnitude, it may not be comparable because rupture forces are dependent on the loading rate, which was not reported in this earlier study.
From the Bell theory of forced molecular unbinding26 that was further modified by Evans and Ritchie,27 we have analyzed the rupture force spectra to extract 2 binding parameters. One of them is the off-rate (k0) extrapolated to spontaneous force-free dissociation. For the A:a interactions the value of k0 was found to be on the order of 10–3 to 10–4 s–1 (Table 1). For comparison, the reported k0 values are 5.4 × 10–6 s–1 for streptavidin-biotin,41 approximately 7 × 10–3 s–1 for avidin-biotin,42 whereas for P-selectin/PSGL-1 the k0 determined using laser tweezers-based force spectroscopy was equal to 1.39 to 4.3 s–1.43 Thus, despite inherent difficulties in the measurement of this constant and the high degree of variability, the k0 of approximately 10–3 to 10–4 s–1 indicates that single A:a interactions are relatively stable and very slowly reversible because of low unbinding rate.
Another important parameter of a binding interaction is the transition state distance (γ), which can be interpreted as the distance of molecular separation at which the bond fails. As calculated from the fitting of the rupture force distribution with the equation based on the Bell theory, the transition state distance for the A:a unbinding is approximately 0.3 nm (Table 1). In other words, the system of electrostatic and hydrogen bonds stabilizing the A:a binding pocket (Figure 6)1,9,19 starts to break when the Aα-chain knob and the γ-chain hole are separated to the distance of about 3 Å.
In conclusion, the data quantitatively characterize at the single-molecule level the highly specific A:a knob-hole interactions between the N-terminal part of the α-chain (A-knob) and the C-terminal part of the γ-chain (a-hole) belonging to interacting fibrin monomers and provide new evidence for the crucial role of these interactions in fibrin formation. We see only A:a binding even when B-knobs and b-holes are available as well, but the reason for this needs to be further explored. A protein complex resulting from the A:a binding is mechanically strong and must be capable of withstanding rupture forces of several hundred piconewtons which may be greater than the platelet contractile forces,44 forces of the physiologic blood shear inducing clot deformation,45 wound tension,46 and perhaps other mechanical forces applied to a forming clot in vivo.47 Taken together, these data provide the molecular mechanisms for mechanical strength and thermodynamic stability of fibrin fibers enabling the accomplishment of physiologic functions of a fibrin clot.
Prepublished online as Blood First Edition Paper, July 5, 2005; DOI 10.1182/blood-2005-05-2039.
Supported by the National Institutes of Health (grant HL30954) (J.W.W.) and the American Heart Association (grant 0365340U) (O.V.G).
All laser tweezer experiments were done by R.I.L.; all proteins used were prepared by O.V.G.; all authors participated in the design of the experiments and in the analysis and interpretation of results.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal