Abstract
Fluorescence in situ hybridization (FISH) is more sensitive than conventional cytogenetics for recognizing chromosomal changes. Several FISH-detected abnormalities have been associated with inferior prognosis, including deletion of chromosomes 17 and 13 (Δ13) and t(4;14)(p16.3;q32). We analyzed the prognostic value of FISH testing in 238 patients who received high-dose therapy between January 1990 and September 2001. All patients had pretransplantation cytoplasmic immunoglobulin FISH done on cytospin slides from bone marrow aspirates for t(11;14), t(4;14), and -17(p13.1) (TP53). Time to progression and overall survival were significantly shorter for patients with t(4;14) and those with -17(p13.1) but were not affected by t(11;14). Overall survival was significantly shorter for patients with both t(4;14) and Δ13 abnormalities than for those with Δ13 alone (26.8 vs 18.8 months). In a multivariable analysis of the effect of Δ13 and t(4;14), the risk ratio for t(4;14) was greater than for Δ13 (2.6 vs 1.5). For high-dose therapy patients, -17(p13) and t(4;14) have clinical importance for estimating time to progression and overall survival. The presence of t(4;14) identifies a subset of patients whose time to progression is only 8.2 months. These patients receive minimal benefit from autologous stem cell transplantation and are candidates for novel therapeutic approaches. (Blood. 2005;106:2837-2840)
Introduction
Chromosome translocations involving the immunoglobulin heavy chain (IgH) gene locus define distinct entities of multiple myeloma (MM) with clinical, morphologic, immunophenotypic, and therapeutic implications. The 2 most common IgH translocations are t(4;14)(p16.3;q32) and t(11;14)(q13;q32). These translocations result from illegitimate IgH rearrangements.1 The presence of these rearrangements has been correlated with morphology (plasmablastic with t(4;14)(p16.3;q32) and lymphoplasmacytic or small mature with t(11;14)(q13;q32))2 and tumor mass.3 The presence of t(11;14) has also been associated with the presence of CD20 expression on the surface of the cells.4 The t(11;14)(q13;q32) results in up-regulation of cyclin D1 and is the most common translocation detected in myeloma.5 It has been suggested that patients with t(11;14) have better survival6 and response to treatment, particularly high-dose therapy and stem cell support.
In contrast, patients with t(4;14)(p16.3;q32) have an inferior outcome, regardless of the mode of treatment (conventional or high dose).7 The purpose of our study was to evaluate these translocations with fluorescence in situ hybridization (FISH) using patient samples from a single institution where all the patients were treated with high-dose chemotherapy and stem cell transplantation. The study addresses the clinical implications of these chromosome-14 translocations and the -17(p13.1), TP53, deletion on time to progression and overall survival. Multivariable analysis was used to determine whether these abnormalities provide information about patient prognosis not currently available from detection of Δ13 and the biologic variables β2-microglobulin (B2M) and plasma cell labeling index (PCLI).8
Patients and methods
All patients receiving high-dose chemotherapy with stem cell replacement had a pretransplantation bone marrow biopsy performed. All patients gave written informed consent for the collection of additional cells at the time of the bone marrow examination, including separate authorization for genetic studies and separate consent for review of their charts in accord with both U.S. federal regulations and the Health Insurance Portability and Account-ability Act guidelines and the Declaration of Helsinki. The consent form was approved by the institutional review board of Mayo Foundation. In most cases, slides were made with the collected cells, and in some cases, new slides were made from cells frozen in liquid nitrogen. All patients from whom available samples were obtained were included in the analysis. The minimal follow-up for survivors was 36 months. Interphase FISH9 was combined with immunofluorescence for cytoplasmic immunoglobulin FISH to identify exclusively the plasma cell population10 and to ensure that all chromosomal abnormalities detected were part of the plasma cell clone. For the detection of these chromosome abnormalities, we used the commercially available probes LSI IgH/FGFR3 (t(4;14)(p16.3;q32)), LSI IgH/CCND1 XT (t(11;14)(q13;q32)), and LSI p53 (17p13.1 deletion) in combination with CEP17 (Vysis, Downers Grove, IL) under the same conditions previously published by us.11 Trisomies and abnormalities such as c-MAF (16q23), MAFB (20q11), or cyclin D3 (6p21) were not analyzed as part of this study.
Descriptive statistics were used to characterize the patients. The Fisher exact test was used to test for association between translocations and clinical characteristics for continuous variables. The Wilcoxon rank sum test was used to test for differences between patient groups on the basis of their translocation status. The Fisher exact test was used to test differences among levels of categoric variables among patients with FISH abnormalities. Distributions for survival and progression-free survival were estimated with the Kaplan-Meier method. Log-rank testing was used to assess differences in survival for significance. Multivariable analysis was performed by the Cox method.
Results
A total of 238 patients were studied between January 1990 and September 2001. Their characteristics are given in Table 1. The outcomes of patients with the various translocations are summarized in Table 2.
Characteristics of the 238 study patients
Characteristic . | Data . |
|---|---|
| Men, no. | 142 |
| Median age, y (range) | 56 (30-71) |
| Median creatinine, mg/dL (range) | 1.1 (0.7-3.5)* |
| Median β2-microglobulin, mg/dL (range) | 2.53 (0.9-11.2)† |
| Median bone marrow labeling index, % (range) | 0.4 (0-8.0) |
| Median bone marrow plasma cells, % (range) | 20 (0-95) |
| Median serum M protein, g/dL (range) | 1.6 (0-10.4)‡ |
| Median urine protein, g/d (range) | 0.09 (0.007-10.4) |
| Median time from diagnosis to bone marrow transplantation, mo (range) | 15.3 (3.7-87.5) |
| Status at transplantation, no. of patients | |
| Plateau | 70 |
| Primary induction failure | 33 |
| Relapse off therapy | 86 |
| Relapse on therapy (resistant relapse) | 49 |
Characteristic . | Data . |
|---|---|
| Men, no. | 142 |
| Median age, y (range) | 56 (30-71) |
| Median creatinine, mg/dL (range) | 1.1 (0.7-3.5)* |
| Median β2-microglobulin, mg/dL (range) | 2.53 (0.9-11.2)† |
| Median bone marrow labeling index, % (range) | 0.4 (0-8.0) |
| Median bone marrow plasma cells, % (range) | 20 (0-95) |
| Median serum M protein, g/dL (range) | 1.6 (0-10.4)‡ |
| Median urine protein, g/d (range) | 0.09 (0.007-10.4) |
| Median time from diagnosis to bone marrow transplantation, mo (range) | 15.3 (3.7-87.5) |
| Status at transplantation, no. of patients | |
| Plateau | 70 |
| Primary induction failure | 33 |
| Relapse off therapy | 86 |
| Relapse on therapy (resistant relapse) | 49 |
To convert to μM, multiply values by 88.4.
To convert to nM, multiply values by 85.
To convert to g/L, multiply values by 10.
Summary of FISH findings for 11;14, 4;14, and p53
. | Successful determination, no. of patients . | Patients with translocation or deletion, no. (%) . | Overall survival with/without abnormality, median no. . | Progression-free survival with/without abnormality, median mo. . |
|---|---|---|---|---|
| t(11;14)(q13;q32) | 197 | 34 (17) | 36.6/34.8 | 20.1/15.3 |
| −17p13.1 (TP53) | 168 | 18 (11) | 15.1/38.8* | 8.7/16.1* |
| t(4;14)(p16.3;q32) | 153 | 26 (17) | 18.8/43.9† | 8.2/17.8† |
| t(4;14)+(p16;q32)Δ13 | 84 | 22 (26) | 18.8/26.8* | 8.2/12.9† |
. | Successful determination, no. of patients . | Patients with translocation or deletion, no. (%) . | Overall survival with/without abnormality, median no. . | Progression-free survival with/without abnormality, median mo. . |
|---|---|---|---|---|
| t(11;14)(q13;q32) | 197 | 34 (17) | 36.6/34.8 | 20.1/15.3 |
| −17p13.1 (TP53) | 168 | 18 (11) | 15.1/38.8* | 8.7/16.1* |
| t(4;14)(p16.3;q32) | 153 | 26 (17) | 18.8/43.9† | 8.2/17.8† |
| t(4;14)+(p16;q32)Δ13 | 84 | 22 (26) | 18.8/26.8* | 8.2/12.9† |
FISH indicates fluorescence in situ hybridization.
p ≤.01.
p < .001.
t(11;14)(q13;q32)
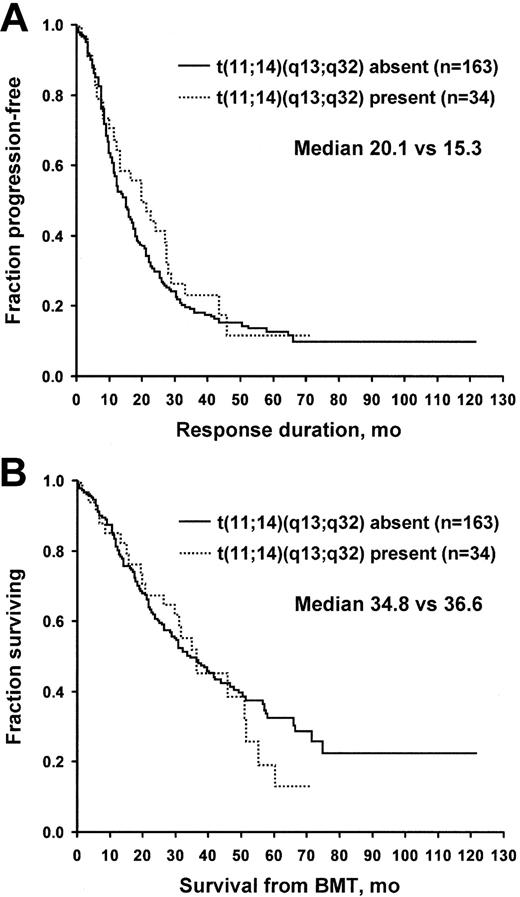
For the t(11;14)(q13;q32), samples were available to study 197 specimens. This translocation was detected in 34 patients (17%). No differences were found between the patients with and those without the t(11;14)(q13;q32) for age, C-reactive protein level, bone marrow PCLI, serum creatinine, lactate dehydrogenase (LDH), B2M, status at stem cell transplantation, and percentage of bone marrow plasma cells. Overall survival and freedom from progression were not different for patients with t(11;14)(q13;q32). Freedom from progression was 20.1 versus 15.3 months, and overall survival was 36.6 versus 34.8 months (Figure 1). A separate analysis was performed for t(11;14) patients stratified for the presence or absence of Δ13. No survival advantage was found for t(11;14) in Δ13-negative patients.
t(4;14)(p16.3;q32)
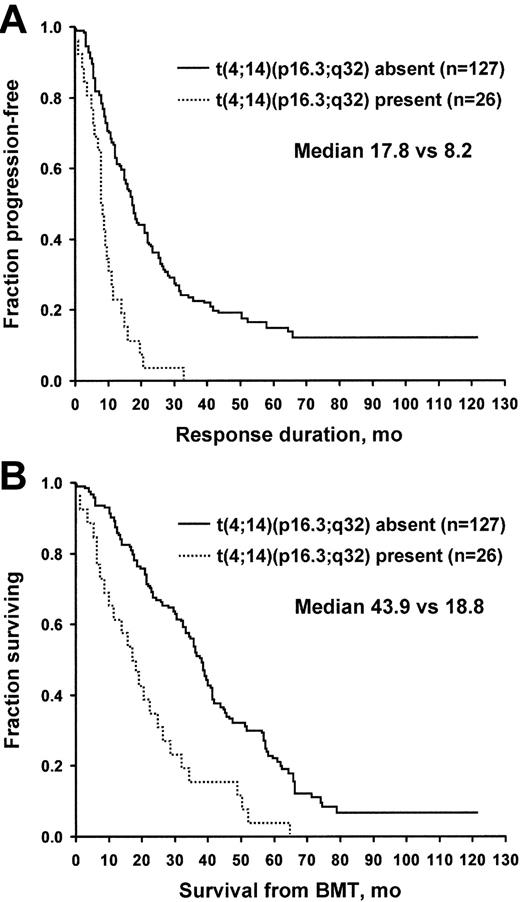
A successful determination was made in 153 patients. Twenty-six patients (17%) had t(4;14)(p16.3;q32). This chromosome translocation had a profound effect on both time to progression and overall survival. Time to progression for patients with and for those without t(4;14)(p16.3;q32) was 8.2 versus 17.8 months (P = .001), and overall survival was 18.8 versus 43.9 months (P = .001) (Figure 2). Patients with t(4;14)(p16.3;q32) had a higher C-reactive protein, PCLI, and percentage of bone marrow plasma cells (all P = .04). Age, creatinine, LDH, and B2M were not significantly different between the 2 groups. There were no differences in the frequency of t(4;14)(q13;q32) among the patients who underwent transplantation at different phases of their disease. No association was found between t(4;14) and heavy chain type (IgA vs not), light chain type (κ vs λ), or the presence of MM bone disease, although only 10% of the patient population had no myeloma bone disease. When the analysis was restricted to the 70 patients who had transplantation upfront in first response, t(4;14)(q13;q32) retained its significance. The median survival rates were 75 months for patients with t(4;14)(p16.3;q32) and 29 months for those without this translocation (P = .01).
Patients with and without t(11;14)(q13;q32) translocation. (A) Time to progression. (B) Overall survival. BMT indicates bone marrow transplantation.
Patients with and without t(11;14)(q13;q32) translocation. (A) Time to progression. (B) Overall survival. BMT indicates bone marrow transplantation.
TP53
Of 168 patients for whom analysis was possible, -17p13.1 (TP53) was detected in 18 (11%). No differences in TP53 status—positive or negative—were detected for age, creatinine, PCLI, LDH, B2M, bone marrow plasma cells, or status at the time of transplantation. The presence of the TP53 deletions was significant for both the time to progression and overall survival, 8.7 versus 16.1 months and 15.1 versus 38.8 months (P < .01), respectively (Figure 3).
Interaction between abnormalities
To separate the specific effect of t(4;14)(p16.3;q32) from that of Δ13, an analysis was performed by stratifying Δ13 among patients with t(4;14)(p16.3;q32). In the larger cohort of 212 patients, FISH detected Δ13 in 111 (52%). Of these 111 patients positive for Δ13, 84 had successful studies for t(4;14)(p16.3;q32), of whom 22 (26%) had the abnormality. Survival analysis performed only for patients with Δ13 showed that the presence of t(4;14)(p16.3;q32) had a significant effect on outcome. When both t(4;14)(p16.3;q32) and Δ13 were present, overall survival was 18.8 months, significantly worse than for patients who had Δ13 but not t(4;14)(p16.3;q32) (26.8 months, P = .001). The median progression-free survival times were 12.9 versus 8.2 months, respectively (P = .001), for Δ13-positive patients without and those with t(4;14). Conversely, in the t(4;14)-positive cohort, the presence or absence of Δ13 had no effect on survival (19.4 vs 18.8 months).
Patients with and without t(4;14)(p16.3;q32) translocation. (A) Freedom from progression. (B) Overall survival. BMT indicates bone marrow transplantation.
Patients with and without t(4;14)(p16.3;q32) translocation. (A) Freedom from progression. (B) Overall survival. BMT indicates bone marrow transplantation.
-17p13.1, TP53. (A) Freedom from progression. (B) Overall survival. BMT indicates bone marrow transplantation.
-17p13.1, TP53. (A) Freedom from progression. (B) Overall survival. BMT indicates bone marrow transplantation.
We constructed a hybrid variable comprising patients who had TP53 deletions, t(4;14)(p16.3;q32), or Δ13 (n = 120) with those lacking all of the 3 FISH abnormalities (n = 69). Median survival was 26.3 months for those with any of the abnormalities and 51.5 months for those without any abnormality (P = .005).
Assessment of univariate effect of various characteristics
Univariate effect on freedom from progression and overall survival was examined for 203 patients in whom studies were performed for t(4;14)(p16.3;q32), t(11;14)(q13;q32), TP53 deletion, and Δ13 (Table 3). For freedom from progression, the percentage of plasma cells in bone marrow, PCLI, Δ13, TP53 deletions, t(4;14)(p16.3; q32), and the status at stem cell transplantation were all significant (P < .05), and for “overall” survival, B2M, percentage of plasma cells in bone marrow, PCLI, Δ13, TP53 deletions, t(4;14)(p16.3; q32), and status at stem cell transplantation were significant. A Cox proportional hazards model was constructed with the characteristics found to be significant in the univariate analysis (Table 4). In this model, the most important characteristic predicting freedom from progression was whether the patient received the stem cell transplant at the time of relapse (P = .001). In this model, the presence or absence of t(4;14)(p16.3;q32) was significant (P = .001), as was Δ13 (P = .05). The risk ratio for patients who received a transplant at the time of relapse was 2.3 (compared with transplant in plateau) and 2.6 for the presence of t(4;14)(p16.3; q32). In the survival model, the most important predictors of survival were PCLI and patient status at the time of the stem cell transplantation, with risk ratios of 1.3 and 2.1, respectively. In this model, t(4;14)(p16.3;q32) was significant to .006, with a risk ratio of 2.2. Δ13 was significant at 0.02, with a risk ratio of 1.7.
Univariate predictors of outcome for 203 patients
Characteristic . | Overall survival log-rank P value . | Freedom from progression log-rank P value . |
|---|---|---|
| Sex | .21 | .56 |
| Serum M spike | .9 | .82 |
| Urine total protein | .1 | .25 |
| β2-microglobulin | .016 | .14 |
| Lactate dehydrogenase | .48 | .6 |
| C-reactive protein | .07 | .64 |
| Bone disease | .11 | .49 |
| % plasma cells in bone marrow | .001 | .001 |
| Plasma cell labeling index | .001 | .001 |
| Creatinine | .42 | .88 |
| Age | .77 | .77 |
| Δ13q | .004 | .017 |
| −17p13.1 (TP53) | .04 | .003 |
| t(11;14)(q13;q32) | .7 | .35 |
| t(4;14)(p16.3;q32) | .001 | .001 |
| Status at stem cell transplantation | .001 | .001 |
Characteristic . | Overall survival log-rank P value . | Freedom from progression log-rank P value . |
|---|---|---|
| Sex | .21 | .56 |
| Serum M spike | .9 | .82 |
| Urine total protein | .1 | .25 |
| β2-microglobulin | .016 | .14 |
| Lactate dehydrogenase | .48 | .6 |
| C-reactive protein | .07 | .64 |
| Bone disease | .11 | .49 |
| % plasma cells in bone marrow | .001 | .001 |
| Plasma cell labeling index | .001 | .001 |
| Creatinine | .42 | .88 |
| Age | .77 | .77 |
| Δ13q | .004 | .017 |
| −17p13.1 (TP53) | .04 | .003 |
| t(11;14)(q13;q32) | .7 | .35 |
| t(4;14)(p16.3;q32) | .001 | .001 |
| Status at stem cell transplantation | .001 | .001 |
Cox (multivariable) analysis of characteristics affecting time to progression and overall survival
. | Time to progression . | . | Overall survival . | . | ||
|---|---|---|---|---|---|---|
| Characteristic . | P . | Risk ratio . | P . | Risk ratio . | ||
| Status at stem cell transplantation | .001 | 2.3 | .003 | 2.1 | ||
| t(4;14)(p16.3;q32) | .001 | 2.6 | .006 | 2.2 | ||
| −17p13.1 (TP53) | .08 | 1.9 | .5 | 1.3 | ||
| Δ13 | .05 | 1.5 | .02 | 1.7 | ||
| Plasma cell labeling index | .01 | 1.2 | .001 | 1.3 | ||
| β2-microglobulin | .53 | 1.0 | .19 | 1.1 | ||
. | Time to progression . | . | Overall survival . | . | ||
|---|---|---|---|---|---|---|
| Characteristic . | P . | Risk ratio . | P . | Risk ratio . | ||
| Status at stem cell transplantation | .001 | 2.3 | .003 | 2.1 | ||
| t(4;14)(p16.3;q32) | .001 | 2.6 | .006 | 2.2 | ||
| −17p13.1 (TP53) | .08 | 1.9 | .5 | 1.3 | ||
| Δ13 | .05 | 1.5 | .02 | 1.7 | ||
| Plasma cell labeling index | .01 | 1.2 | .001 | 1.3 | ||
| β2-microglobulin | .53 | 1.0 | .19 | 1.1 | ||
When the multivariable analysis was restricted to the 132 patients who had successful analysis for deletion 13, TP53, and t(4;14)(q13;q32) (ie, no missing data), status at transplantation was most significant (P = .001) but t(4;14)(q13;q32) retained its predictive value as well (P = .03), whereas deletion 13 was marginal (P = .08). When the impact of TP53 and t(4;14)(q13;q32) was restricted only to patients who had a transplant more than 12 months after diagnosis (late transplant), both features retained statistical significance in the univariate model, 29.8 versus 16.4 and 29.8 versus 14.2 months for presence or absence of t(4;14) (q13;q32) and TP53, respectively; neither retained significance in the multivariable model.
Discussion
The t(11;14)(q13;q32) results in up-regulation of cyclin D1 in MM, as in the case of mantle cell lymphoma.3 In an Eastern Cooperative Oncology Study Group (ECOG) protocol with 336 evaluable patients, 16% had t(11;14)(q13;q32)11 compared with 17% of our high-dose therapy patients, suggesting that the current study had no selection bias. However, in the ECOG study, patients with t(11;14)(q13;q32) appeared to have borderline improvement in survival and response to therapy, whereas our study did not demonstrate any effect on survival or time to progression. Moreau et al6 have suggested that high-dose therapy for patients with t(11;14)(q13;q32) markedly improves survival. Our results do not corroborate this observation.
In our study, deletions of 17p13.1 at the TP53 locus were clinically important (although not independent) for estimating of overall survival and time to progression. Similar observations have been made for patients who received conventional therapy and those who received high-dose therapy. Chang et al12 found that patients with TP53 deletions had significantly shorter progression-free (median 7.9 vs 25.7 months, P = .032) and overall (median 14.7 vs 48.1 months, P = .001) survival than patients without a TP53 deletion. In their study, the abnormality retained its independent value in the multivariable model.
Although FISH detection of Δ13 was associated with a poor prognosis in our cohort, the presence of t(4;14)(p16.3;q32) was more powerful and provided additional information not provided by the presence of Δ13 alone. The t(4;14)(p16.3;q32) results in the overexpression of fibroblast growth factor receptor 3 and multiple myeloma SET domain.13-15 The negative prognostic effect of t(4;14)(p16.3;q32) appears to be independent of the actual level of fibroblast growth factor receptor 3 expression.7
In our study, the 2 most important variables were the PCLI of bone marrow (a measure of the fraction of plasma cells in the S phase) and the status of the patient at stem cell transplantation (ie, whether the transplantation was performed at the time of relapse). Because of the low prevalence of these abnormalities (15%), a much larger study is needed to elucidate the actual prognostic implications of these chromosome abnormalities.
Our patients with t(4;14)(p16.3;q32) had a median time to progression of only 8.2 months after stem cell therapy and an overall survival of 18.8 months. This suggests that high-dose therapy, as currently practiced, has minimal benefit for these patients. Whether the addition of nonmyeloablative strategies will overcome these therapeutic limitations is not known; thus, the strategies cannot be deemed the best therapy for this high-risk group of patients. Also, the dismal outcome for t(4;14)(p16.3;q32) after a stem cell transplantation raises the question of the role of allogeneic transplantation in suitable younger candidates. Currently, preclinical and early clinical investigations are being developed for the treatment of patients with IgH chain translocations. Until these therapies are available, patients should be considered candidates for novel therapeutic approaches.
At Mayo Clinic, a FISH panel is routinely performed for all patients with newly diagnosed MM and includes FISH assessment for 4p16.3 (FGFR3), 11q13 (CCND1 XT), 13q14 (Rb1), 13q34 (LAMP1), 14q32 (IGH5′, IGH3′), 14q32 (IGH XT), 16q23 (c-MAF), 17p12 (p53), and 17cen (D17Z1). The presence of TP53 deletion, t(4;14), or Δ13 halves survival from 51.5 to 26.3 months.
Prepublished online as Blood First Edition Paper, June 23, 2005; DOI 10.1182/blood-2005-04-1411.
Supported in part by Hematologic Malignancies Fund of Mayo Clinic. R.F. is a Clinical Investigator of the Damon Runyon Cancer Research Fund. Supported by grants R01 CA83724-01, SPORE P50 CA100707-01, and P01 CA62242 from the National Cancer Institute (NCI) and the Fund to Cure Myeloma, and by NCI grant CA21115-25C.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


Comments
Nomenclature error