Abstract
Transcriptional control has been identified as a key mechanism regulating the formation and subsequent behavior of hematopoietic stem cells. We have used a comparative genomics approach to identify transcriptional regulatory elements of the LMO2 gene, a transcriptional cofactor originally identified through its involvement in T-cell leukemia and subsequently shown to be critical for normal hematopoietic and endothelial development. Of the 2 previously characterized LMO2 promoters, the second (proximal) promoter was highly conserved in vertebrates ranging from mammals to fish. Real-time reverse transcriptase–polymerase chain reaction (RT-PCR) expression analysis identified this promoter as the predominant source of transcription in hematopoietic tissue. Transient and stable transfections indicated that the proximal promoter was active in hematopoietic progenitor and endothelial cell lines and this activity was shown to depend on 3 conserved Ets sites that were bound in vivo by E74-like factor 1 (Elf1), Friend leukemia integration 1 (Fli1), and erythroblastosis virus oncogene homolog E twenty-six–1 (Ets1). Finally, transgenic analysis demonstrated that the LMO2 proximal promoter is sufficient for expression in endothelial cells in vivo. No hematopoietic expression was observed, indicating that additional enhancers are required to mediate transcription from the proximal promoter in hematopoietic cells. Together, these results suggest that the conserved proximal promoter is central to LMO2 transcription in hematopoietic and endothelial cells, where it is regulated by Ets factors.
Introduction
A fundamental question in developmental biology concerns the molecular mechanisms by which pluripotent stem cells become committed to single lineages. The decision to follow a specific developmental pathway is believed to be reflected in different transcriptional programs of various cell types.1 Hematopoietic stem cells (HSCs) have long provided a model system for the study of stem-cell specification and subsequent differentiation. Research in the last 2 decades has shown that transcriptional regulation is central for the development of blood and endothelial cells and resulted in the identification of a number of transcription factors that are indispensable for hematopoiesis (for review see Kluger et al2 and Cantor and Orkin3 ).
One protein with a crucial role in blood and endothelial development is Lin-1, Isl-1, Mec-3 (LIM) domain only 2 (LMO2), a member of the LIM-only zinc finger protein family.4 Mice lacking Lmo2 die around embryonic day 10.5 because of a complete absence of erythropoiesis.5 Furthermore, studies of chimeric mice produced from Lmo2–/– embryonic stem (ES) cells have shown that Lmo2 is also required for the formation of adult hematopoietic cells6 as well as for vascular endothelial remodeling.7 These data suggest that Lmo2 may function at the early stages of hematopoietic and endothelial development, possibly at the level of the hemangioblast, a bipotent precursor of both cell types. Consistent with the knock-out results, LMO2 has been shown to be expressed in early extraembryonic mesoderm and blood islands,8 fetal liver,9 hematopoietic progenitors,5 long-term repopulating HSCs,10 and in endothelial cells.7 Following differentiation of HSCs, Lmo2 expression is maintained in erythroid cells but is repressed in terminally differentiated granulocytes, macrophages, T cells, and mature B cells,5 with the exception of germinal center B cells.11 This transcriptional down-regulation appears to be particularly important in T lymphocytes where aberrant expression of LMO2 results in T-cell leukemia.9,12-14
The LMO2 gene is located on chromosome 11 in band p13,4,9 a recurrent site of T-cell acute lymphoblastic leukemia (T-ALL)–specific translocations.15,16 Overexpression of LMO2 has been reported in several patients with T-ALL with LMO2-associated chromosomal translocations t(11;14)(p13;q11) involving the T-cell receptor (TCR)δ locus.4,9,12 Ectopic expression of Lmo2 in thymocytes of transgenic mice has confirmed a role for this protein in the etiology of T-ALL.13,14 Transcriptional activation, as a consequence of retroviral vector integration into the LMO2 locus, has also been implicated in the development of clonal T-cell proliferation in 2 patients undergoing gene therapy for X-linked severe combined immunodeficiency (X-SCID).17 In addition, LMO2 has recently been linked to diffuse large B-cell lymphoma, where high levels of expression were found to be the strongest predictor of survival.18 Taken together, these studies indicate that appropriate transcriptional control of LMO2 is crucial for the formation and subsequent behavior of blood cells.
Unlike many transcription factors that interact directly with DNA, LMO2 is believed to act as a bridging molecule involved in the assembly of multiprotein complexes. In erythroid cells, Lmo2 is present in a multimeric complex containing Scl, E2A, Gata-1, and Ldb1.19 In T-cell tumors from Lmo2 transgenic mice, Lmo2 was shown to form a complex with E2A, Scl, and Ldb1.20 Lastly, in multipotent hematopoietic progenitors, a complex containing Lmo2, stem cell leukemia (Scl), E2A, LIM domain binding protein 1 (Ldb1), and Gata-2 has been proposed.21 As part of these complexes, Lmo2 has been found to regulate the promoters of KIT,22 Epb4.2,23 GYPA,24 and Hba-a121 in different hematopoietic lineages. While the identity of several LMO2 target genes has recently been determined,21-24 the transcription factors involved in the regulation of LMO2 itself remain largely unknown.
LMO2 is transcribed from 2 alternate promoters leading to 2 mRNA isoforms that differ in their 5′ untranslated region (5′UTR) but encode the same protein.25 A proximal promoter is present in the vicinity of exon 3 while a distal promoter is located nearly 25 kb upstream.25,26 Activity of the distal promoter appears to be mostly restricted to hematopoietic cells.25,26 Moreover, the distal promoter was shown to be regulated in reporter assays by a proline- and acidic amino acid–rich (PAR) domain transcription factor binding site situated in intron 1,26 as well as GATA sites located near the transcriptional start site, which can be activated by overexpression of GATA-1.27 In addition, an upstream negative regulatory element has been shown to repress the activity of the distal promoter in luciferase reporter assays.28 In contrast, the regulation of the proximal promoter has not been investigated.
Comparative genomics has emerged in recent years as a powerful method to locate transcriptional regulatory sequences. Critical functional elements are subject to strong selective pressure during evolution and exhibit a slower rate of substitution. These elements frequently display a higher degree of nucleotide identity when compared with neighboring sequences in multispecies alignments of distantly related genomes, and can thus be identified.29-31 This approach, termed phylogenetic footprinting,32 has been used to effectively isolate regulatory elements for several genes,30,31 including enhancers that control expression of the LMO2 interacting partner, SCL.33-35
In the present study, we used a comparative genomics approach to identify functionally important sequences for the transcriptional regulation of LMO2. Our results demonstrate that the proximal promoter region of LMO2 is conserved throughout vertebrate evolution, is sufficient for transcriptional activity in hematopoietic and endothelial cell lines, and directs reporter gene expression to endothelium in transgenic mouse embryos. Promoter activity was dependent on 3 conserved Ets factor binding sites, which were bound in vivo by the Ets family of transcription factors, E74-like factor 1 (Elf1), Friend leukemia integration 1 (Fli1), and erythroblastosis virus oncogene homolog E twenty-six–1 (Ets1) in both blood progenitor and endothelial cells.
Materials and methods
Sequence analysis
Genomic LMO2 sequences from various species were downloaded from the Ensembl (www.ensembl.org) and UCSC (http://genome.ucsc.edu/) genome browsers. Multiple sequence alignments of the LMO2 loci were performed by multi-Lagan36 and displayed using mVista37 and Genedoc. Sequences and feature files used to generate the alignments shown in Figure 1 are provided as supplemental materials, which are available on the Blood website; click on the Supplemental Materials link at the top of the online article. Putative transcription factor binding sites in the conserved motif were predicted using the Transcription Element Search System (TESS; http://www.cbil.upenn.edu/tess/index.html) and Transcription Factor Binding Site (TFBS) search.38
Real-time PCR
Expression analysis of LMO2 was performed by real-time polymerase chain reaction (PCR) using Sybr Green (Applied BioSystems, Warrington, United Kingdom). Transcripts originating from the distal LMO2 promoter were amplified using oligonucleotides located in exon 1, 5′-CAAAGCAGGCAATTAGCCC-3, and exon 2, 5′-CCTCTCCACTAGCTACTGC-3′ for human, and situated in exon 2, 5′-GTAGTGACGATTGGAGAGG-3′, and exon 4, 5′-GCGTGGCTGGCTAGCTG-3′ for mouse. Total LMO2 expression was quantified with forward 5′-TGAGCTGCGACCTCTGTGG-3′ and reverse primers 5′-CACCCGCATTGTCATCTCAT-3′, situated in exons 5 and 6 of the human LMO2 transcript and forward 5′-TCAGCTGTGACCTCTGTGG-3′ and reverse 5′-CACCCGCATCGTCATCTC-3′ oligos for mouse. Amplifications were also performed with actin primers 5′-GCTATCCCTGTACGCCTCTG-3′ and 5′-AGGGCATACCCC TCGTAGAT-3′ for human, and 5′-TCCTGGCCTCACTGTCCAC-3′ and 5′-GTCCGCCTAGAAGCACTTGC-3′ for mouse. Dissociation curves were run to detect nonspecific amplification and it was determined that single products were amplified in each reaction.
To verify the amplification efficiency of both the distal and total LMO2 primer pairs, a common template DNA was generated by reverse transcriptase (RT)–PCR. This was achieved by amplifying human LMO2 cDNAs from bone marrow and K562 cells using oligonucleotides situated in exons 1 and 6 and amplifying mouse Lmo2 transcripts from 416B cells using exon 2 and 6 primers. Serial dilutions of the amplified LMO2 cDNAs were used to determine the primer efficiencies by comparing their respective cycle threshold (CT) values (Supplemental Figure S1).
Comparative sequence analysis reveals a conserved noncoding segment in the proximal promoter region of LMO2. MVista37 graphical representation of a Multi-Lagan36 multiple sequence alignment of vertebrate LMO2 loci, where Hs is Homo sapiens, Mm is Mus musculus, Rn is Rattus norvegicus, Cf is Canis familiaris, Xt is Xenopus tropicalis, Tr is Takifugu rubripes, and Dr is Danio rerio. Conserved regions are displayed relative to their positions in the human genome (horizontal axis). Segments that show more than 70% sequence identity (indicated on the vertical axis) at the nucleotide level over a 100-bp window, are highlighted in pink (noncoding regions), cyan (untranslated regions), or purple (coding exons). Exons are displayed above the comparison plots, and repetitive elements are shown in orange.
Comparative sequence analysis reveals a conserved noncoding segment in the proximal promoter region of LMO2. MVista37 graphical representation of a Multi-Lagan36 multiple sequence alignment of vertebrate LMO2 loci, where Hs is Homo sapiens, Mm is Mus musculus, Rn is Rattus norvegicus, Cf is Canis familiaris, Xt is Xenopus tropicalis, Tr is Takifugu rubripes, and Dr is Danio rerio. Conserved regions are displayed relative to their positions in the human genome (horizontal axis). Segments that show more than 70% sequence identity (indicated on the vertical axis) at the nucleotide level over a 100-bp window, are highlighted in pink (noncoding regions), cyan (untranslated regions), or purple (coding exons). Exons are displayed above the comparison plots, and repetitive elements are shown in orange.
To account for the small differences in primer efficiencies observed, the equations of the generated relative standard curves were used to adjust threshold cycle values for total and distal LMO2 amplicons in each tissue or cell line. These corrected levels of amplified LMO2 transcripts were then normalized by the adjusted levels of actin obtained for each tissue or cell line. Results were expressed relative to the total level of LMO2 transcripts in bone marrow, which was given an arbitrary value of 1. The relative level of transcripts arising from the proximal promoter was inferred by subtracting the percentage of amplified transcripts detected using the distal primer pairs from the total level of LMO2.
Real-time PCR expression analysis of murine Ets factors was performed as described for LMO2 in this section, using primers 5′-GGAGTATGACCACATGAATGG-3′ and 5′-GACTCTCCGTTCGTTGGTG-3′ for Fli1,5′-CAAGTAACGGCATGGAGGAC-3′ and 5′-CTGTAAGGGTGATGTCGTC-3′ for Elf1, 5-GATATCCTGTGGGAGCATCTAG-3′ and 5′-GAAGTAAACCGAGGTGTAACAG-3′ for Ets1, and 5′-GGGAACATCTAGAGCAGATG-3′ and 5′-GTCCAGGAGATTGTCTTTGG-3′ for Ets2.
Plasmid constructs
All promoter constructs were designed by cloning the distal promoter or segments of the proximal promoter of LMO2 into the XhoI/HindIII sites of the pGL2 basic luciferase vector (Promega, Southampton, United Kingdom). Promoter fragments were generated by PCR using primers listed in Table 1 where the positions are relative to the translation start site of LMO2. Mutant promoter constructs were created by PCR using oligonucleotides with mismatches as underlined in Table 1. Plasmids were purified using the Plasmid Maxi Kit (Qiagen, Crawley, United Kingdom) and verified by sequencing.
Primers for LMO2 distal and proximal promoter constructs
Position (mutation) . | Sequence (5′-3′) . |
|---|---|
| -23025 | ccgctcgagGGAGAAGTAAATACAGGCTG |
| -22076 | cccaagcttGAGTGGTCTCCCTTTGTGG |
| -661 | ccgctcgagGGACCGGGCAGCTGTCTCT |
| -661 (E box) | ccgctcgagGGACCGGGAAGCTATCTCTTTAAATGT |
| -661 (i) | ccgctcgagGGACCGGGCAGCTGTCTCTACAAATGTGATTTC |
| -661 (Ets1) | ccgctcgagGACCGGGCAGCTGTCTCTTTAAATGTGATTTCTTCTATTGTATTTG |
| -661 (ii) | ccgctcgagGGACCGGGCAGCTGTCTCTTTAAATGTGATTTCCTTCTAATATATTTG |
| -582 | ccgctcgagGAGACAGAGGGAAGCTGAGCG |
| -572 | cccaagcttCCCTCTGTCTCTGGTTTCATTT |
| -572 (iii) | cccaagcttCCCTCTGTCTCTGGTTTCATTTCCTTTTTCCTGATAATGATTCAAA |
| -572 (Ets2) | cccaagcttCCCTCTGTCTCTGGTTTCATTTCCTTTTTCTTGATCACGATTC |
| -572 (Ets3) | cccaagcttCCCTCTGTCTCTGGTTTCATTTCTTTTTTCCTGATCACGATTC |
| -572 (Ets2,3) | cccaagcttCCCTCTGTCTCTGGTTTCATTTCTTTTTTCTTGATCACGATTC |
| -572 (iv) | cccaagcttCCCTCTGTCTTTTGTTTCATTTCCTTTTTCCTGATCACGATTC |
| -572 (v) | cccaagcttCACTATGTCTCTGGTTTCATTTCCTTTTTCCTGATCACGATTC |
| -313 | cccaagcttCAGGACTTAACCTTCCATCCC |
Position (mutation) . | Sequence (5′-3′) . |
|---|---|
| -23025 | ccgctcgagGGAGAAGTAAATACAGGCTG |
| -22076 | cccaagcttGAGTGGTCTCCCTTTGTGG |
| -661 | ccgctcgagGGACCGGGCAGCTGTCTCT |
| -661 (E box) | ccgctcgagGGACCGGGAAGCTATCTCTTTAAATGT |
| -661 (i) | ccgctcgagGGACCGGGCAGCTGTCTCTACAAATGTGATTTC |
| -661 (Ets1) | ccgctcgagGACCGGGCAGCTGTCTCTTTAAATGTGATTTCTTCTATTGTATTTG |
| -661 (ii) | ccgctcgagGGACCGGGCAGCTGTCTCTTTAAATGTGATTTCCTTCTAATATATTTG |
| -582 | ccgctcgagGAGACAGAGGGAAGCTGAGCG |
| -572 | cccaagcttCCCTCTGTCTCTGGTTTCATTT |
| -572 (iii) | cccaagcttCCCTCTGTCTCTGGTTTCATTTCCTTTTTCCTGATAATGATTCAAA |
| -572 (Ets2) | cccaagcttCCCTCTGTCTCTGGTTTCATTTCCTTTTTCTTGATCACGATTC |
| -572 (Ets3) | cccaagcttCCCTCTGTCTCTGGTTTCATTTCTTTTTTCCTGATCACGATTC |
| -572 (Ets2,3) | cccaagcttCCCTCTGTCTCTGGTTTCATTTCTTTTTTCTTGATCACGATTC |
| -572 (iv) | cccaagcttCCCTCTGTCTTTTGTTTCATTTCCTTTTTCCTGATCACGATTC |
| -572 (v) | cccaagcttCACTATGTCTCTGGTTTCATTTCCTTTTTCCTGATCACGATTC |
| -313 | cccaagcttCAGGACTTAACCTTCCATCCC |
The linker sequence to facilitate cloning is shown in lowercase, and the mutated sequence is underlined.
Cell culture and transfection conditions
The endothelial MS1 cells were maintained in Dulbecco minimal essential media (DMEM) supplemented with 10% fetal calf serum and antibiotics. The hematopoietic progenitor 416B cell line was maintained in RPMI supplemented with 10% fetal calf serum and antibiotics.
Transient and stable transfections of 416B and MS1 cells were performed using electroporation. Typically, for transient analysis, cells were cotransfected with 10 μg plasmid DNA and 3 μg of the lacZ vector pEF Bos LacZ. Transfected cells were washed 24 hours later in phosphatebuffered saline (PBS) and harvested and assayed as previously described.39 The data were normalized to the internal control and expressed with respect to pGL2b (basic). For stable transfections, 10 μg linearized plasmid DNA and 1 μg pGK neo (416B) or 2 μg of pGK puro (MS1) were coelectroporated. G418 was added to the cells 24 hours after transfection at a concentration of 0.75 mg/mL of media while puromycin was added at a concentration of 1 μg/mL. Transfected cells were assayed 7 to 10 (416B) days or 14 to 20 (MS1) days later for luciferase activity.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as previously described by Forsberg et al.40 Briefly, 416B and MS1 cells were treated with formaldehyde and the cross-linked chromatin was sonicated in order to obtain fragments of 500–base pair (bp)–averaged size. Immunoprecipations were performed with Santa Cruz Biotechnology (Santa Cruz, CA) antibodies against Fli1(sc-356x), Elf1 (sc-631x), Ets1 (sc-350x), and Ets2 (sc-351x) to recover the DNA-bound transcription factors. Enrichment was measured by real-time PCR as described earlier with the LMO2 proximal promoter forward primer 5′-CGCTATTTGGATTGCTAATCC-3′ and reverse primer 5′-CTTCCCTCTGTCTCTGGTTTC-3′ and the control LMO2 exon 6 forward primer 5′-CCATCTTTCTGTTGTCCCC-3′ and reverse primer 5′-CACCCGCATCGTCATCTC-3′.
Transgenic analysis
The LMO2 proximal promoter transgenic reporter construct was generated by cloning the –661 to –313 fragment into the XhoI/HindIII site of the pGLac lacZ plasmid (Genbank accession number U19930). This plasmid was linearized with XhoI and BamH1 and founder transgenic embryos produced by pronuclear injection as described.41 Embryos were harvested at 12.5 days after coitus (E12.5) and analyzed as described.41
Results
The LMO2 proximal promoter is conserved in all vertebrates
To identify candidate LMO2 regulatory elements, we compared the genomic loci of 4 mammalian species: human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus), and dog (Canis familiaris) with the teleost species pufferfish (Fugu rubripes) and zebrafish (Danio rerio) and the Western clawed frog (Xenopus tropicalis). Multispecies sequence comparisons were performed with Lagan36 and an overview of the alignment is displayed in Figure 1 as a Vista plot,37 using the human sequence as reference. High levels of sequence conservation among the mammalian sequences can be seen across the entire LMO2 genomic region. As expected, the percentage of nucleotide identity is significantly lower following inclusion of the fish or frog lmo2 loci in the alignment. High levels of homology between the human, fish, and frog sequences are restricted to the coding exons 4, 5, and 6 and a stretch of approximately 90 bp upstream of exon 3 (Figure 1).
LMO2 expression is predominantly driven by the proximal promoter in hematopoietic cells. (A) Pictorial representation of the human and mouse LMO2 loci, where the coding exons are represented by black boxes and the untranslated regions by white boxes. The 2 alternative promoter regions, transcribing the long (exon 1-6) and short (exons 4 to 6) mRNA variants are indicated by arrows above the exons. The positions of the oligonucleotides used to quantify the transcript isoforms are indicated underneath the loci by short arrows. (B) Expression analysis of the 2 transcript forms by real-time PCR. Total cDNAs were subjected to real-time PCR using primers that amplified either all LMO2 transcripts or only those with the distal noncoding exons. For both human and mouse, the total level of LMO2 in bone marrow, normalized by the level of actin, was assigned a value of 1. The relative abundance of the transcript variants, normalized by the level of actin in those tissues and cell lines, is depicted by ▪ (distal) and ▦ (proximal) plus or minus standard deviation (SD). BM indicates bone marrow; PB, peripheral blood; FL, fetal liver.
LMO2 expression is predominantly driven by the proximal promoter in hematopoietic cells. (A) Pictorial representation of the human and mouse LMO2 loci, where the coding exons are represented by black boxes and the untranslated regions by white boxes. The 2 alternative promoter regions, transcribing the long (exon 1-6) and short (exons 4 to 6) mRNA variants are indicated by arrows above the exons. The positions of the oligonucleotides used to quantify the transcript isoforms are indicated underneath the loci by short arrows. (B) Expression analysis of the 2 transcript forms by real-time PCR. Total cDNAs were subjected to real-time PCR using primers that amplified either all LMO2 transcripts or only those with the distal noncoding exons. For both human and mouse, the total level of LMO2 in bone marrow, normalized by the level of actin, was assigned a value of 1. The relative abundance of the transcript variants, normalized by the level of actin in those tissues and cell lines, is depicted by ▪ (distal) and ▦ (proximal) plus or minus standard deviation (SD). BM indicates bone marrow; PB, peripheral blood; FL, fetal liver.
The expression of LMO2 has been shown to be under the control of 2 promoters,25 located upstream of the first exon (distal) or overlapping with exon 3 (proximal), which direct the transcription of identical open reading frames (Figure 2A). Interestingly, the conserved noncoding sequence upstream of exon 3 overlaps with the region in which the transcriptional start site for the LMO2 proximal promoter has been reported.9 Therefore, the only noncoding peak of identity detectable in a 7-species comparison corresponds to the LMO2 proximal promoter region.
Most LMO2 transcripts in hematopoietic tissues start at the proximal promoter
It has previously been determined that the distal promoter of LMO2 directs hematopoietic-specific expression, whereas the proximal promoter is active in a wide range of tissues.25,26 This conclusion was based on RT-PCR and Northern blot analyses where it was shown that exons unique to transcripts originating from the distal promoter could only be detected in fetal liver and normal bone marrow, whereas coding exons, shared between both isoforms, could be identified in many tissues.25,26 However, the contribution of the proximal promoter to overall expression in hematopoietic tissues was not assessed in either of these studies. Since our comparative genomic data demonstrated that the proximal promoter was highly conserved, we decided to evaluate the activity of the proximal promoter in blood and endothelium to assess whether the high level of sequence conservation correlates with a major role for the proximal promoter in controlling LMO2 expression.
To investigate promoter usage in different hematopoietic and endothelial tissues and cell lines, expression of the 2 LMO2 transcript variants was quantitated using real-time PCR. We determined the total expression of LMO2, which represents transcripts originating from both the proximal and distal promoters, by amplifying the shared coding exons, and assessed the contribution of the distal promoter by amplifying exons specific to this isoform (Figure 2A) from various human and mouse cDNA sources. As no unique exons are present in the short mRNA form, specific primers that would selectively amplify transcripts originating from the proximal promoter could not be designed. The contribution from the proximal promoter was therefore calculated by comparing the levels of amplification obtained with the total and distal primer pairs. These experiments, shown in Figure 2B, indicate that the vast majority of transcripts in all tissues and cell lines tested initiate from the proximal promoter. In human bone marrow 12% of LMO2 transcripts contained the distal exons whereas 10% of LMO2 mRNAs contained exons 1 and 2 in a human leukemia cell line (K562; Figure 2B). The contribution from the distal promoter was below detectable levels for human peripheral blood. In mouse bone marrow, 9% of mRNAs possessed the distal exons, whereas 5% and 7% of Lmo2 transcripts in mouse fetal liver and a myeloid progenitor cell line (416B) contained these exons. Finally, distal promoter usage was below detectable levels in mouse endothelial and T-cell lines (Figure 2B; data not shown). Taken together, these data confirm that the distal promoter is active in some hematopoietic tissues but also highlight the fact that the proximal promoter is likely the dominant site of transcription in most LMO2-expressing tissues, including those of hematopoietic origin.
The LMO2 proximal promoter region is active in blood and endothelial cell lines
To assess the transcriptional activity of the 2 LMO2 promoters, sequences containing the distal promoter (corresponding to positions –23 025/–22 076; numbering with respect to the ATG start codon in exon 4) and the proximal promoter (corresponding to positions –661/–313) were PCR amplified and subcloned into luciferase reporter plasmids. In transient transfection assays using the hematopoietic progenitor 416B and endothelial MS1 cell lines, the distal promoter construct directed reporter activity at levels 50-fold and 8-fold higher, respectively, than a promoterless luciferase control construct, whereas the proximal promoter fragment (–661/–313) showed activity in both cell lines at levels 50-fold higher than the control construct (Figure 3). Therefore, our results demonstrate that the proximal promoter has equal or higher transcriptional activity than the distal promoter in endothelial and hematopoietic cells, contrasting with a previous report that showed that the distal element was the dominant promoter25 when assayed in nonphysiologic COS cells.
The LMO2 proximal promoter is active in hematopoietic and endothelial cell lines. Shown on the left are the reporter constructs in which the distal promoter or regions of the proximal promoter were inserted upstream of the promoterless pGL2B vector and transfected into 416B and MS1 cell lines. The conserved noncoding sequence, present in the proximal promoter, is depicted by a series of filled circles. LUC indicates luciferase. In the middle panel are the luciferase activities, obtained with each plasmid in transient transfections, corrected for transfection efficiency with the lacZ pEF-BOS LacZ plasmid. On the right are the stable transfection results normalized by cell counts. The luciferase activities are presented as fold-increase over the activity of the basic (pGL2B) vector, which was assigned a value of 1. Each bar is the mean of the relative luciferase activity from at least 2 experiments performed in triplicate, plus or minus SD.
The LMO2 proximal promoter is active in hematopoietic and endothelial cell lines. Shown on the left are the reporter constructs in which the distal promoter or regions of the proximal promoter were inserted upstream of the promoterless pGL2B vector and transfected into 416B and MS1 cell lines. The conserved noncoding sequence, present in the proximal promoter, is depicted by a series of filled circles. LUC indicates luciferase. In the middle panel are the luciferase activities, obtained with each plasmid in transient transfections, corrected for transfection efficiency with the lacZ pEF-BOS LacZ plasmid. On the right are the stable transfection results normalized by cell counts. The luciferase activities are presented as fold-increase over the activity of the basic (pGL2B) vector, which was assigned a value of 1. Each bar is the mean of the relative luciferase activity from at least 2 experiments performed in triplicate, plus or minus SD.
To account for chromatin integration effects on the transcriptional activity of the promoter sequences tested, we also performed stable transfection assays. In stable transfection experiments, the distal promoter (–23 025/–22 076) directed reporter activity at a level 20-fold (416B) and 10-fold (MS1) higher than the basic construct, whereas the activity of the proximal promoter was even stronger at levels of 50-fold over the control plasmid in both cell lines (Figure 3). The promoter transfection studies contrasted with our real-time PCR analysis of promoter usage, which had suggested that the proximal promoter was nearly 20-fold stronger than the distal promoter in 416B cells, and that the distal promoter was not used at all in MS1 cells. This apparent discrepancy might be due to differences in mRNA stability of LMO2 transcripts initiated at the distal and proximal promoters, which would not become apparent when the promoters are fused to the luciferase sequence. Moreover and in contrast to promoter transfection studies, real-time RT-PCR experiments measure transcript levels generated at the endogenous locus in the context of additional cis-regulatory elements. It may be relevant in this context that a negatively acting cis-regulatory element within the vicinity of the LMO2 distal promoter has recently been described.28 Together with the high degree of sequence conservation of the proximal promoter region, these results served to focus our subsequent attention to a detailed analysis of this promoter.
To begin to elucidate which sections of the proximal promoter were important for transcriptional function, 2 new proximal promoter constructs (–661/–572 and –582/–313) were generated. As LMO2 undergoes heterogeneous transcription initiation (UCSC genome browser), both partial fragments harbored LMO2 transcriptional start sites, whereas the 5′ construct also contained the region of sequence conservation shown in Figure 4A. Removal of the conserved sequence from the proximal promoter region (construct –582/–313) resulted in a 2-fold decrease in promoter activity in transient transfections, whereas in stable transfections it reduced the activity by over 90% (Figure 3). The conserved region by itself (–661/–572) drove expression to about half the levels obtained with the full-length construct, at approximately 25-fold over background in transient transfections and importantly retained approximately 75% of the promoter activity in stable transfections. These results demonstrated that both segments of the proximal promoter fragment could independently function as promoters in transient transfection assays. Importantly, in the stable transfection experiments, where reporter constructs are measured after chromatin integration, the conserved sequence alone (–661/–572) maintained nearly full activity of the proximal promoter fragment (–661/–313), whereas the nonconserved portion was barely active (Figure 3). These results focused attention on the 89-bp region that was conserved between mammals, amphibians, and fish and acted as a strong promoter in both blood and endothelial cell lines.
Ets sites are required for transcriptional activity of the proximal promoter conserved region
To define functionally important motifs within the conserved region of the LMO2 proximal promoter, we searched for the occurrence of evolutionarily conserved transcription factor binding sites within this sequence. As shown in Figure 4A, this analysis revealed the presence of an E box in all but the zebrafish sequence. In addition, 3 Ets factor binding sites were conserved among all 6 species analyzed as well as 5 additional 4- to 5-bp motifs that did not correspond to known transcription factor binding consensus sequences (Figure 4A). To examine the relative importance of all conserved sequence motifs, mutations were introduced both individually and in combination (Figure 4B). Mutation of the E box resulted in a small decrease in promoter activity in 416B and MS1 in transient transfections, whereas in stable transfections, expression was lowered by 4-fold in both cell lines (Figure 4B). Mutation of the first Ets site only moderately reduced promoter function. However, mutations of the second, third, both the second and third, or all 3 Ets sites together markedly reduced luciferase activity in both cell lines (Figure 4B). Mutation of the TA-rich motif (labeled as i), which may represent a nonconsensus TATA box, lowered expression by approximately 5-fold in transient transfections and between 10- and 20-fold in stable transfections. Finally, mutations of the 4 other unknown motifs (labeled as ii, iii, iv, and v) did not markedly reduce promoter activity except when stably transfected in 416B cells. Together, these results suggest that regulatory mechanisms conserved between endothelial and hematopoietic progenitor cells appear to be largely dependent on at least 2 of the conserved Ets binding sites and the TTTAAA motif (which were conserved) and possibly to a lesser extent the E box (which was not conserved in all species).
Ets transcription factor binding sites are necessary for promoter activity. (A) Nucleotide sequence of the conserved proximal promoter region in which the predicted E box, Ets sites, and other conserved sequence blocks are marked. Nucleotides in the alignment are shaded in gray if they are conserved at that position in more than 70% of the sequences, in dark gray if conserved in more than 85% of the sequences, or in black if they are identical in all sequences. Species included in the multiple sequence alignment are as described in Figure 1. (B) Reporter assays of a series of proximal promoter mutation constructs in which the predicted transcription factor binding sites were ablated. Constructs were transfected into MS1 and 416B cells as described in Figure 3. • indicates E box motif;  , Ets motif; and ○, unknown motif.
, Ets motif; and ○, unknown motif.
Ets transcription factor binding sites are necessary for promoter activity. (A) Nucleotide sequence of the conserved proximal promoter region in which the predicted E box, Ets sites, and other conserved sequence blocks are marked. Nucleotides in the alignment are shaded in gray if they are conserved at that position in more than 70% of the sequences, in dark gray if conserved in more than 85% of the sequences, or in black if they are identical in all sequences. Species included in the multiple sequence alignment are as described in Figure 1. (B) Reporter assays of a series of proximal promoter mutation constructs in which the predicted transcription factor binding sites were ablated. Constructs were transfected into MS1 and 416B cells as described in Figure 3. • indicates E box motif;  , Ets motif; and ○, unknown motif.
, Ets motif; and ○, unknown motif.
Fli1, Elf1, and Ets1 occupy the proximal promoter conserved region of LMO2 in both endothelial and hematopoietic progenitor cells in vivo
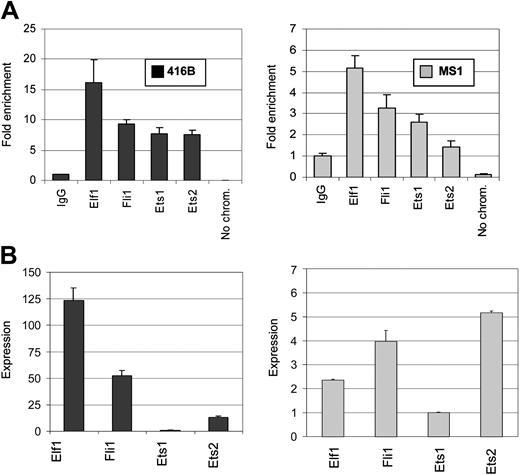
To determine the identity of Ets factors recruited to the LMO2 proximal promoter conserved region, we performed chromatin immunoprecipitation (ChIP) assays with antibodies to Elf1, Fli1, Ets1, and Ets2 in MS1 and 416B cells. These Ets family members were chosen because they were known to be expressed in these cell lines (Göttgens et al42 and B.G., S.K., and J.-R.L., unpublished data, December 2004) and had been shown to regulate other genes in blood progenitors and endothelial cells.42-44 Immunoprecipitated chromatin samples were analyzed by quantitative real-time PCR. As shown in Figure 5A, the conserved region of the proximal promoter was specifically enriched in Elf1, Fli1, Ets1, and Ets2 immunoprecipitates by 16.1-, 9.4-, 7.7-, and 7.5-fold, respectively, relative to the levels obtained with rabbit immunoglobulin G (IgG) antibodies in 416B cells. A similar pattern of enrichment, albeit at lower levels, was also found in MS1 cells, with the exception of Ets2, which was not found to occupy the proximal promoter conserved region in the endothelial cell line (Figure 5B). As a negative control, primers were designed for a region approximately 10 kilobase (kb) downstream, overlapping with exon 6 of LMO2. This region of exon 6 was not significantly enriched in immunoprecipitates obtained with antibodies to Fli1, Elf1, Ets1, or Ets2 (results not shown). These studies demonstrate that the Ets factors Fli1, Elf1, and Ets1 are recruited to the conserved region of the proximal promoter of LMO2 in both endothelial and hematopoietic progenitor cells in vivo.
Elf1, Fli1, and Ets1 bind the conserved noncoding region. (A) Chromatin immunoprecipitation experiments were performed with anti-Elf1, -Fli1, -Ets1, and -Ets2 as well as control IgG antibodies. The DNA content of the immunoprecipitates was analyzed by real-time PCR for the presence of the conserved noncoding sequence in the proximal promoter region. The values plotted represent the level of enrichment with each antibody when compared with the levels obtained with the control IgG, plus or minus SD. (B) Real-time expression analysis of Elf1, Fli1, Ets1, and Ets2 transcripts in 416B and MS1 cell lines. The levels of expression of the different Ets factors are shown relative to Ets1, which was given an arbitrary value of 1 (note that Ets1 was expressed at significant levels in both cell lines) Error bars indicate standard deviation.
Elf1, Fli1, and Ets1 bind the conserved noncoding region. (A) Chromatin immunoprecipitation experiments were performed with anti-Elf1, -Fli1, -Ets1, and -Ets2 as well as control IgG antibodies. The DNA content of the immunoprecipitates was analyzed by real-time PCR for the presence of the conserved noncoding sequence in the proximal promoter region. The values plotted represent the level of enrichment with each antibody when compared with the levels obtained with the control IgG, plus or minus SD. (B) Real-time expression analysis of Elf1, Fli1, Ets1, and Ets2 transcripts in 416B and MS1 cell lines. The levels of expression of the different Ets factors are shown relative to Ets1, which was given an arbitrary value of 1 (note that Ets1 was expressed at significant levels in both cell lines) Error bars indicate standard deviation.
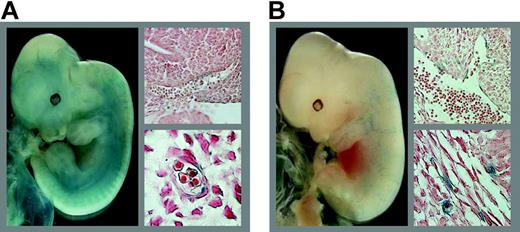
LMO2 proximal promoter directs endothelial expression in transgenic mice. E12.5 transgenic embryos expressing lacZ under the control of the LMO2 proximal promoter. (A) A representative embryo with endothelial staining discernible from whole-mount analysis and confirmed by histologic sections of a blood vessel (top right) and a capillary (bottom right). (B) The founder embryo with scattered β-galactosidase staining subsequently demonstrated to represent endothelial activity by histologic sectioning of the dorsal aorta (top right) and capillaries (bottom right). Whole-mount images were acquired using a Pixera Pro 150ES digital camera (Pixera, Los Gatos, CA) attached to a Nikon SM7800 microscope (Nikon, Kingston upon Thames, United Kingdom). Images of sections were acquired with the same camera attached to an Olympus BX51 microscope (Olympus, Southall, United Kingdom) using Olympus UPlanApo 40 ×/0.85 numeric aperture (NA) and 100 ×/1.35 NA objectives. ImagePro Express version 4.5 (Image Processing Solutions, North Reading, MA) was used for acquisition of both types of images. Digital images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
LMO2 proximal promoter directs endothelial expression in transgenic mice. E12.5 transgenic embryos expressing lacZ under the control of the LMO2 proximal promoter. (A) A representative embryo with endothelial staining discernible from whole-mount analysis and confirmed by histologic sections of a blood vessel (top right) and a capillary (bottom right). (B) The founder embryo with scattered β-galactosidase staining subsequently demonstrated to represent endothelial activity by histologic sectioning of the dorsal aorta (top right) and capillaries (bottom right). Whole-mount images were acquired using a Pixera Pro 150ES digital camera (Pixera, Los Gatos, CA) attached to a Nikon SM7800 microscope (Nikon, Kingston upon Thames, United Kingdom). Images of sections were acquired with the same camera attached to an Olympus BX51 microscope (Olympus, Southall, United Kingdom) using Olympus UPlanApo 40 ×/0.85 numeric aperture (NA) and 100 ×/1.35 NA objectives. ImagePro Express version 4.5 (Image Processing Solutions, North Reading, MA) was used for acquisition of both types of images. Digital images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
To investigate whether the relative occupancy of the 4 Ets factors tested was a result of differences in their abundance, we determined the relative expression of Elf1, Fli1, Ets1, and Ets2 in 416B and MS1 cells by real-time PCR. As shown in Figure 5B, there did not appear to be any direct relationship between the transcript levels of the different Ets factors and the levels of enrichment observed in the ChIP assays. This lack of correlation between transcription factor occupancy and mRNA expression levels would be consistent with the selective recruitment of specific Ets family members to the LMO2 proximal promoter. However, to definitively address this issue one would need to be able to determine the relative levels of Ets family proteins available for binding in 416B and MS1 cell nuclei.
The LMO2 proximal promoter region drives endothelial expression in transgenic mice
To evaluate the in vivo transcriptional activity of the evolutionarily conserved LMO2 proximal promoter, founder transgenic mouse embryos were generated using a construct in which the –661/–313 promoter region was cloned upstream of a promoterless lacZ reporter gene. Reporter expression was analyzed by whole-mount X-gal staining of midgestation (E12.5) embryos. Of 6 transgenic embryos, 3 showed transgene expression. Staining in 2 of these appeared to be localized to blood vessels (Figure 6A; data not shown) whereas expression in the third was weak and scattered and specificity could not be assessed from whole-mount staining (Figure 6B).
To further investigate specificity of transgene expression, all 3 E12.5 transgenic founder embryos with β-galactosidase staining were sectioned for histologic analysis. LacZ expression was observed in a subset of endothelial cells, mostly in capillaries, in all 3 embryos including the embryo with weak and scattered whole-mount staining (Figure 6A-B; data not shown). The endothelial staining observed was reminiscent of the endothelial expression previously observed in transgenic embryos in which a lacZ reporter gene had been knocked into one Lmo2 allele,7 and suggested that the proximal promoter is involved in regulating endothelial-specific expression of LMO2. However, not all endothelial cells stained positive for transgene expression, suggesting that additional sequences, outside the proximal promoter, contribute to LMO2 expression in endothelium. No staining was present in fetal liver and yolk sac hematopoietic cells, indicating that the proximal promoter requires enhancer sequences yet to be identified to function in hematopoietic cells.
Discussion
Understanding the molecular mechanisms controlling expression of LMO2 is likely to provide new insights into the transcriptional control of hematopoietic and endothelial development as well as the pathogenesis of T-lymphoid leukemias. Here, we have used a comparative genomics approach to identify conserved regulatory elements for LMO2. An 89-bp region of the human proximal promoter of LMO2 was found to be maintained in a similar position and orientation in the genome of divergent species such as mouse, dog, frog, and pufferfish. This conserved 89-bp region drove reporter gene expression in both hematopoietic progenitor and endothelial cells and was bound in vivo by the Ets family transcription factors Fli1, Elf1, and Ets1. Finally, the proximal promoter was shown to be the predominant site of transcription in hematopoietic cells and to be active in the endothelium of transgenic mice.
Prior to this report, the proximal promoter of LMO2 was believed to drive transcription ubiquitously, whereas the distal promoter was thought to be primarily active in hematopoietic cells.25,26 The hematopoietic specificity of the distal promoter was confirmed in transgenic studies where a distal promoter fragment directed expression of a cat transgene in bone marrow and blood of adult mice26 ; however, the type of hematopoietic cells expressing the transgene was not investigated. Importantly, none of the previous studies quantitated the relative contribution of the distal and proximal promoters to overall levels of endogenous LMO2 expression. The results presented here, based on real-time amplifications, suggest that the distal promoter's contribution to LMO2 mRNA levels is minor. Instead, the proximal promoter appears to be the major LMO2 transcriptional start site in bone marrow, peripheral blood, fetal liver, hematopoietic and endothelial cell lines. Interestingly, a significant proportion of mapped LMO2 translocations were found to have occurred downstream of exon 1, thus removing the distal promoter from the LMO2 coding sequence.25 Following on from our observation that the proximal promoter is the major site of transcription in hematopoietic cells, it would therefore be interesting to investigate whether most LMO2 transcripts in T-cells of patients with T-ALL with chromosomal translocations 5′ of the distal promoter or in the 2 children with clonal proliferation of T-lymphocytes following gene therapy for X-SCID17 also originate from the conserved proximal promoter.
In this study, we have demonstrated that functionally important Ets motifs in the conserved region of the human LMO2 proximal promoter are occupied in vivo by the Ets family transcription factors Fli1, Elf1, Ets1, and Ets2 in hematopoietic progenitors and Fli1, Elf1, and Ets1 in endothelial cells. All of these proteins were previously shown to be expressed in cells of hematopoietic and endothelial lineages. High levels of Ets1 are present in the blood islands of the yolk sac and the vascular endothelium and the more ubiquitously expressed Ets2 is found in fetal liver hematopoietic cells (reviewed in Oikawa and Yamada45 and Maroulakou and Bowe46 ). Fli1 was shown to be expressed in blood islands, fetal liver, and the developing vasculature, and Elf1 in hematopoietic tissues of adult mice and humans,45,46 as well as developing blood vessels of chicken.47 Moreover, these Ets factors have been found to regulate a number of key players in blood and/or endothelial development. For example, Ets binding sites, critical for activity of the Flk1 promoter in the endothelium of transgenic mice,43 were activated by Ets1 and Ets2 in coexpression experiments.43 Fli1 has been shown to interact and transactivate, together with Ets1, its own promoter.44 Finally, 2 SCL enhancers, +1948 and –3.8,42 have been found to be controlled by Fli1 and Elf1.
The identification of common transcription factors in the regulation of LMO2 and SCL emphasizes the particularly close relationship of these 2 key hematopoietic proteins in the transcription network that governs blood and endothelial development. However, our data suggest that SCL and LMO2 regulation differs at the molecular level since the SCL +19 and –3.8 enhancers can direct expression in blood when assayed in isolation,49,50 whereas the LMO2 proximal promoter cannot. By supplying novel interactions between LMO2 and various Ets proteins, the current study begins to lay the foundation for integrating LMO2, itself a key regulator of blood and endothelial development, into an emerging regulatory network that controls mesodermal differentiation toward hemangioblasts, and subsequently blood and endothelium. Moreover, since LMO2 has recently been proposed as a new target for antiangiogenesis therapy to combat solid tumors,51 understanding the molecular mechanisms responsible for LMO2 expression in endothelium may provide new tumor angiogenesis drug targets.
In summary, we have established that the proximal promoter predominantly transcribes LMO2 in blood and endothelium, where it is regulated by various Ets transcription factors. Although the LMO2 proximal promoter can direct endothelial expression in transgenic mice, it is not sufficient to recapitulate the full spectrum of LMO2 expression in vivo. The identification of additional LMO2 enhancers will prove important in understanding the molecular mechanisms underlying transcriptional regulation in endothelial- and blood-cell lineages.
Prepublished online as Blood First Edition Paper, June 30, 2005; DOI 10.1182/blood-2004-12-4755.
Supported by grants from the Leukaemia Research Fund, Wellcome Trust, the Cambridge-MIT Institute, and a fellowship from the Canadian Institutes of Health Research (J.-R.L.).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Juan Li, George Vasiliou, Peter Campbell, and John Pimanda for human and mouse cDNAs. We acknowledge Paula Braker for coordinating with the Central Biomedical Services.




![Figure 4. Ets transcription factor binding sites are necessary for promoter activity. (A) Nucleotide sequence of the conserved proximal promoter region in which the predicted E box, Ets sites, and other conserved sequence blocks are marked. Nucleotides in the alignment are shaded in gray if they are conserved at that position in more than 70% of the sequences, in dark gray if conserved in more than 85% of the sequences, or in black if they are identical in all sequences. Species included in the multiple sequence alignment are as described in Figure 1. (B) Reporter assays of a series of proximal promoter mutation constructs in which the predicted transcription factor binding sites were ablated. Constructs were transfected into MS1 and 416B cells as described in Figure 3. • indicates E box motif; , Ets motif; and ○, unknown motif. \batchmode \documentclass[fleqn,10pt,legalpaper]{article} \usepackage{amssymb} \usepackage{amsfonts} \usepackage{amsmath} \pagestyle{empty} \begin{document} \(\mathbf{x}\) \end{document} indicates motifs mutated in the various constructs.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/8/10.1182_blood-2004-12-4755/6/m_zh80200585250004.jpeg?Expires=1764977953&Signature=Be1MWTeywX9XuG5gVKlqH965VxfRU6Llu4KvKUpvFYr53KfJxTGUbk8OvmTRAg1j7XTw3gob~51tB1lrTb1MYNdR7-yOoEJycTEx9sE5On3L5nVasVi7yAcDxkLl20YAYd9etSCym3Yj1YcJnjCqFtDY7Lfn-grxN~0A42deI3U7-316OaoCzWmNwgJS72X13Ygsf5rb0QsuIULoaKMe1CL92SIRFMA9saeUbumC-LpjQsbJJ3XNLoEpwFKMheiT6HXajtvnIuHfTpqi2dNyVA2BDEkjUSJgM7iO85aGK79dtVZpeN2bFU7XQob3tgr15Ln24XlnsJO-aAZ~CjB2Gw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal