Abstract
Recent reports linking insertional activation of LMO2 following gene therapy for X-linked severe combined immunodeficiency (X-SCID) have led to a re-evaluation of risks following gene therapy with retroviral vectors. In our analysis of 702 integration sites in rhesus macaques that underwent transplantation up to 7 years earlier with autologous CD34+ cells transduced with amphotropic murine leukemia virus (MLV)-derived retroviral vectors containing marker genes, we detected insertion into one locus, the Mds1/Evi1 region, a total of 14 times in 9 animals. Mds1/Evi1 integrations were observed stably long term, primarily in myeloid cells. We hypothesize that this over-representation likely results from an impact on the self-renewal and engraftment potential of CD34+ progenitor cells via insertional mutagenesis at this specific locus. There is no evidence of ongoing in vivo clonal expansion of the Mds1/Evi1 populations, and all animals are hematologically normal without evidence for leukemia. Characterization of integration sites in this relevant preclinical model provides critical information for gene therapy risk assessment as well as identification of genes controlling hematopoiesis. (Blood. 2005;106:2530-2533)
Introduction
Retroviruses, because they integrate into genomic DNA, may activate nearby proto-oncogenes; however, the risk of insertional mutagenesis using replication-incompetent retroviral vectors for gene therapy has been estimated to be low, assuming random integration and a single hit per cell. The recent report of lymphoproliferation due to insertional activation of the LMO2 gene following gene therapy for X-linked severe combined immunodeficiency (X-SCID) has led to a re-evaluation of insertional mutagenesis.1,2 Nonhuman primates are a relevant model for assessing efficacy and safety.3 We surveyed retroviral insertion sites (RISs) in 22 rhesus macaques engrafted with CD34+ cells transduced with retroviral vectors containing only marker genes, and reported a pattern of murine leukemia virus (MLV) vector integration preferentially near the 5′ end of genes.4 We now report that insertions within the first 2 introns of the Mds1/Evi1 gene locus are found at a high frequency, with 14 Mds1/Evi1 insertions in a total of 9 animals detected in circulating granulocytes long term. These results suggest that perturbation of this specific locus via retroviral insertion uniquely results in enhancement of engraftment and/or in immortalization of progenitor cells.
Study design
Transplantation
All experiments had animal care and use committee approval. Details of mobilization, CD34 enrichment, transduction, and transplantation were as published.5-9
Analysis of integration sites
DNA was isolated from granulocytes and mononuclear cells of 22 rhesus macaques 6 months to 7 years after transplantation. Inverse polymerase chain reaction (PCR) or linear-amplification-mediated (LAM)-PCR were performed as described4,10,11 using primers listed in Table S1 (see the Supplemental Materials link at the top of the online article, at the Blood website). Junctions between genomic regions and 5′ long terminal repeats (LTRs) were purified from agarose gels and cloned with the TOPO TA kit (Invitrogen, Carlsbad, CA). Criteria for genuine retroviral integration sites (RISs) were as described.4 To confirm the presence of insertions, 200 ng DNA underwent a 35-cycle PCR using 5′ insertion-specific primers (Table S1) with the 3′ LTR-R1 primer. A quantity of 0.2% of this product was used as a template for a 35-cycle PCR using a nested 5′ primer with the 3′ LTR-R2 primer. More than 95% pure granulocytes, T cells, and B cells were obtained as described.10
Quantitation of contributions from individual clones over time after engraftment was performed using one genomic primer, one vector LTR primer, and probes spanning the LTR-genomic junction, in comparison to albumin genome number controls. Primers and Taqman probes were designed using Applied Biosystems Primer Express software (Foster City, CA). Plasmid standards containing the probe/primer region generated a curve for determination of absolute copy numbers for specific Mds1 insertions and albumin control sequences. The ABI 7900HT Sequence Detection System (Applied Biosystems) ran 50 cycles of amplification at 95°C for 15 seconds and 60°C for 60 seconds.
Statistical analysis
A Java program simulated insertions using a random number generator, assuming random integration in a genome size of 3 × 109 bp. After generating 702 insertions, the number of integration sites within 30 kb or 50 kb of each other were counted. The process was repeated 10 000 times and the average of expected common integration sites (CISs) was calculated. This frequency was compared to the observed frequency of integration via Poisson statistics.
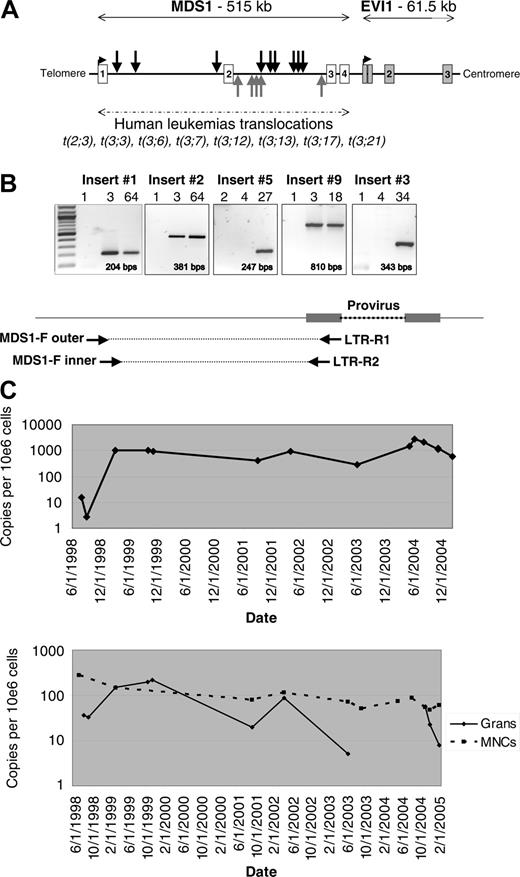
Retroviral integration at the MDS1/EVI1 locus. (A) Location of the 14 independent RISs identified within the Mds1/Evi1 locus. Black arrows indicate RISs with the provirus in the same orientation as transcription; gray arrows indicate RISs with the provirus in the reverse orientation. Locations mapped using July 2003 human genome assembly. (B) RIS-specific PCR using insert-specific 5′ genomic nested forward (F) primers and 3′ LTR-R primers to confirm the presence of the fusion sequence in blood granulocytes from individual animals, shown for insertion nos. 1, 2, 3, 5, and 9 (see Table 1 for list of inserts and Table S1 for primer sequences). MPT indicates months after transplantation. Expected amplification product size is indicated. (C) Taqman PCR measuring the contributions from individual Mds1 clones over time. Animal RC501 underwent transplantation in June 1998. The top panel shows the level of insert no. 1 in granulocytes. The bottom panel shows the level of insert no. 2 in granulocytes and mononuclear cells. A total of 4 additional inserts analyzed by Taqman showed similar results, with no significant change in the level of contribution of the MDS1 RIS over time in granulocytes or mononuclear cells (MNCs).
Retroviral integration at the MDS1/EVI1 locus. (A) Location of the 14 independent RISs identified within the Mds1/Evi1 locus. Black arrows indicate RISs with the provirus in the same orientation as transcription; gray arrows indicate RISs with the provirus in the reverse orientation. Locations mapped using July 2003 human genome assembly. (B) RIS-specific PCR using insert-specific 5′ genomic nested forward (F) primers and 3′ LTR-R primers to confirm the presence of the fusion sequence in blood granulocytes from individual animals, shown for insertion nos. 1, 2, 3, 5, and 9 (see Table 1 for list of inserts and Table S1 for primer sequences). MPT indicates months after transplantation. Expected amplification product size is indicated. (C) Taqman PCR measuring the contributions from individual Mds1 clones over time. Animal RC501 underwent transplantation in June 1998. The top panel shows the level of insert no. 1 in granulocytes. The bottom panel shows the level of insert no. 2 in granulocytes and mononuclear cells. A total of 4 additional inserts analyzed by Taqman showed similar results, with no significant change in the level of contribution of the MDS1 RIS over time in granulocytes or mononuclear cells (MNCs).
Results and discussion
We characterized a total of 702 RISs in blood granulocytes and T cells from rhesus macaques 6 to 92 months after reinfusion of gene-modified CD34+ cells, including 491 previously reported12 and an additional 211.4 A total of 17 genes were identified as having 2 independent intragenic insertions; one gene, the tyrosine kinase receptor Flt3, had 3 insertions; and another, Mds1, had 14 insertions, representing 1.9% of the insertions (Table 1). The number of hits occurring twice within 30 kb predicted by random Monte Carlo generation would be 5.06, and number of 3 hits occurring within 50 kb would be 0.33, not significantly different from the results we obtained. The probability of observing 14 hits within Mds1 by chance is 1.7 × 10-34.13 The 14 Mds1 insertions were all in the first 2 introns of the Mds1/Evi1 gene complex, and were detected in 9 monkeys out of a total of 22 analyzed (Figure 1A and Table 1). An insertion was also found in Prdm16, a homolog of Mds1/Evi1. The RISs all fall within the same region of Mds1/Evi1 reported to be involved in chromosomal translocations in human myeloid leukemias, and upstream of the RISs found activating the Evi1 gene in murine leukemias reported with replication-competent retroviral infection (http://rtcgd.ncifcrf.govs14 ) and shown to immortalize primary murine bone marrow cells.15
Summary of MDS/EVII retroviral vector insertions
Insertion no. . | Animal . | Vector* . | Date of transplantation . | LTR coordinate† . | Orientation‡ . | Sequence, LTR§ . | Lineages∥ . |
|---|---|---|---|---|---|---|---|
| 1 | RC5018 | LNL6/GINa | 5/15/98 | 170,623,356 | + | AATACACAGTAGTTTACCAGTAGTATACTCAAATTCAACTGGAGATGTACTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 2 | RC5018 | LNL6/GINa | 5/15/98 | 170,429,293 | + | CAATGGCACATTAATGCTCCCTCATGTCTCTGCCCAGAAGGTGTTCAACATGAAAGACCC | Identified in Grans; T cells +, B cells + |
| 3 | 96E0257 | LNL6/GINa | 7/30/99 | 170,213,060 | − | GGCAATGATCCTACCACCAACCAGTTCCTTCTGGCACTGTTGATACTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 4 | RC7067 | LNL6/GINa | 1/14/00 | 170,273,870 | + | AGAGCACATTAGACTGAGTGGTTTATTTACAAGTAAAAATCACATTGATGTGAAAGACCC | Identified in DCs; Gran +, T cells −, B cells − |
| 5 | RQ22977 | LNL6/GINa | 7/7/00 | 170,362,372 | − | GACCACAGTCTTTCTGCCCAGGAGGCCACCAAAGGGCCTGAATGTTGCCTTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 6 | RQ22977 | LNL6/GINa | 7/7/00 | 170,351,399 | + | TCCCTAGACCTGCTCCTCCTATTGTCCTCGAATATTCTATTAGGTGCTGATGAAAGACCC | Identified in Grans |
| 7 | RQ23265 | GINa, MPSV-G6PD | 8/12/00 | 170,362,387 | − | GCCCAGGAGGCCACCAAAGGGTCTGAATGTTGCCACTACAGTCCTACCCTTGAAAGACCC | Identified in Grans |
| 8 | RQ23265 | GINa, MPSV-G6PD | 8/12/00 | 170,361,790 | + | TGCATGTCTGAGGTCTAACAGATTGGTCCCAGGATATTGCATGTTTTCCCTGAAAGACCC | Identified in Grans |
| 9 | RQ22776 | LNL6/GINa | 2/9/01 | 170,278,164 | + | TAGGTTTCCCTTGATGTGAACTTTTCAAGCTATTTCTTTTTGATAGCTATTGAAAGACCC | Identified in Grans |
| 10 | RQ22879 | LNL6/GINa, GIPL1 | 3/30/01 | 170,349,678 | + | TTCCCACGGCTTTCGTGTTTGGATATTACTGTATCTTCTCAATCAGATGTTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 11 | RQ22879 | LNL6/GINa, GIPL1 | 3/30/01 | 170,273,453 | + | AGTGGCAAAGAAAGCATAAGTATTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 12 | RQ22879 | LNL6/GINa, GIPL1 | 3/30/01 | 170,373,081 | − | AATTAACAGATATGCAAAACAAGGTAAAGCACCGGACATTATTAGCTCATTCTGAAAGACCC | Identified in Grans; T cells +, B cells − |
| 13 | RQ24288 | LNL6/GINa | 3/15/02 | 170,699,486 | + | TGGGTAATAAAGGAAATCATACTAAAACTACAGATTTTTGTATTTTAAAGTGAAAGACCC | Identified in Grans |
| 14 | RC9036 | L6/GINa | 7/10/02 | 170,393,825 | − | TGTCGGGAAAAAAGGAAGCCACTTAATGGCTCACAATAGATCTTGAAAGACCC | Identified in T cells |
Insertion no. . | Animal . | Vector* . | Date of transplantation . | LTR coordinate† . | Orientation‡ . | Sequence, LTR§ . | Lineages∥ . |
|---|---|---|---|---|---|---|---|
| 1 | RC5018 | LNL6/GINa | 5/15/98 | 170,623,356 | + | AATACACAGTAGTTTACCAGTAGTATACTCAAATTCAACTGGAGATGTACTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 2 | RC5018 | LNL6/GINa | 5/15/98 | 170,429,293 | + | CAATGGCACATTAATGCTCCCTCATGTCTCTGCCCAGAAGGTGTTCAACATGAAAGACCC | Identified in Grans; T cells +, B cells + |
| 3 | 96E0257 | LNL6/GINa | 7/30/99 | 170,213,060 | − | GGCAATGATCCTACCACCAACCAGTTCCTTCTGGCACTGTTGATACTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 4 | RC7067 | LNL6/GINa | 1/14/00 | 170,273,870 | + | AGAGCACATTAGACTGAGTGGTTTATTTACAAGTAAAAATCACATTGATGTGAAAGACCC | Identified in DCs; Gran +, T cells −, B cells − |
| 5 | RQ22977 | LNL6/GINa | 7/7/00 | 170,362,372 | − | GACCACAGTCTTTCTGCCCAGGAGGCCACCAAAGGGCCTGAATGTTGCCTTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 6 | RQ22977 | LNL6/GINa | 7/7/00 | 170,351,399 | + | TCCCTAGACCTGCTCCTCCTATTGTCCTCGAATATTCTATTAGGTGCTGATGAAAGACCC | Identified in Grans |
| 7 | RQ23265 | GINa, MPSV-G6PD | 8/12/00 | 170,362,387 | − | GCCCAGGAGGCCACCAAAGGGTCTGAATGTTGCCACTACAGTCCTACCCTTGAAAGACCC | Identified in Grans |
| 8 | RQ23265 | GINa, MPSV-G6PD | 8/12/00 | 170,361,790 | + | TGCATGTCTGAGGTCTAACAGATTGGTCCCAGGATATTGCATGTTTTCCCTGAAAGACCC | Identified in Grans |
| 9 | RQ22776 | LNL6/GINa | 2/9/01 | 170,278,164 | + | TAGGTTTCCCTTGATGTGAACTTTTCAAGCTATTTCTTTTTGATAGCTATTGAAAGACCC | Identified in Grans |
| 10 | RQ22879 | LNL6/GINa, GIPL1 | 3/30/01 | 170,349,678 | + | TTCCCACGGCTTTCGTGTTTGGATATTACTGTATCTTCTCAATCAGATGTTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 11 | RQ22879 | LNL6/GINa, GIPL1 | 3/30/01 | 170,273,453 | + | AGTGGCAAAGAAAGCATAAGTATTGAAAGACCC | Identified in Grans; T cells −, B cells − |
| 12 | RQ22879 | LNL6/GINa, GIPL1 | 3/30/01 | 170,373,081 | − | AATTAACAGATATGCAAAACAAGGTAAAGCACCGGACATTATTAGCTCATTCTGAAAGACCC | Identified in Grans; T cells +, B cells − |
| 13 | RQ24288 | LNL6/GINa | 3/15/02 | 170,699,486 | + | TGGGTAATAAAGGAAATCATACTAAAACTACAGATTTTTGTATTTTAAAGTGAAAGACCC | Identified in Grans |
| 14 | RC9036 | L6/GINa | 7/10/02 | 170,393,825 | − | TGTCGGGAAAAAAGGAAGCCACTTAATGGCTCACAATAGATCTTGAAAGACCC | Identified in T cells |
−indicates below limit of detection or more than 2 logs lower than granulocyte lineage signal; + indicates present.
Vector or vectors used to transduce autologous CD34+ cells.
Site of vector insertion, based on the humen July 2003 assembly.
Orientation of the inserted provirus in relation to the Mds1/Evi1 genes: + indicates provirus in same orientation as the genes; − indicates provirus in opposite orientation as the genes.
Junction of genomic DNA (roman typeface) and the proviral 5′ LTR (bold italic typeface).
Lineage in which Mds1/Evi1 insertion initially identified via LAM-PCR and sequencing given in first line, lineage analysis via allele-specific primers given on second line.
Insertion-specific primers confirmed that the fusion between the genomic Mds1 locus and the 5′-LTR of the vector were present (Figure 1B and data not shown), first detectable 2 to 3 months after transplantation, and then continuously thereafter, as we have previously reported for most marked clones, with early marking derived primarily from a much greater number of short-term repopulating cells.10 We used quantitative PCR to assess the level of contribution to hematopoiesis from clones with Mds1/Evi1 RISs. While there were variable levels found in granulocytes, there was no consistent pattern of clonal expansion in any animal up to 7 years after transplantation (Figure 1C). All but one Mds1 insertions were initially identified by LAM-PCR from myeloid cells at more than 6 months after transplantation. Allele-specific primers and Taqman analysis demonstrated that 6 of 8 clones analyzed could only be detected contributing to myeloid lineages, compared with 2 of 8 contributing to both myeloid and lymphoid populations (Table 1).
All 9 animals have normal blood counts, with no evidence of leukemia, and polyclonal hematopoiesis by LAM-PCR (Figure S1) at a median of 66 months (range, 30-92 months) since transplantation. These findings imply that although insertion into the Mds1/Evi1 locus as a single event impacted on engraftment or survival of primitive progenitors, it did not result in abnormal proliferation, differentiation, or clonal dominance.
Expression from the Mds1/Evi1 locus results in multiple isoforms of Mds1, Evi1, and Mds1-Evi1, consisting of the first 2 exons of Mds1 spliced to the second exon of Evi1.16,17 All 3 gene products are transcription factors, and the PR (positive regulatory) domain, a negative regulator of tumorigenesis, is present in Mds1-Evi1 but missing in the shorter Evi1 gene product.18 Overexpression of Evi1 in murine hematopoietic stem cells (HSCs) results in a myelodysplastic/myeloproliferative syndrome, and high EVI1 expression but not MDS1/EVI1 expression is associated with poor survival in human acute myeloid leukemia (AML).19,20 Either loss of the MDS1-EVI1 gene product containing the PR domain or unapposed gain of the EVI1 gene product may impact on the behavior of primitive hematopoietic cells.
The presence of both orientations for inserted proviruses in the locus argues against vector-gene fusion transcripts being critical. Theoretically, the RISs via vector control sequences could up-regulate either Mds1, Mds1-Evi1, or Evi1 transcription, or interfere with splicing and down-regulate Mds1/Evi1 production. These transcripts are expressed endogenously only in primitive hematopoietic cells. We did not detect Mds1, Mds1-Evi1, or Evi1 transcripts in granulocytes from 5 animals (data not shown). The Mds1/Evi1 clones represent only 1 to 2 of up to 50 to 100 total RISs in each animal, making detection of overexpression in circulating mature cells unlikely. To increase sensitivity, neomycin-resistant bone marrow colony-forming units (CFUs) were grown from 2 animals and 11 of 290 CFUs screened had Mds1 RIS. However, none had detectable Mds1-Evi1 or Evi1 expression by reverse transcriptase (RT)-PCR.
The cluster of RISs at the Mds1/Evi1 locus in the progeny of transduced CD34+ cells cannot be due to chance, and suggests either a marked integration hotspot, or perturbation of the hematopoietic potential of CD34+ cells following MLV integration at this locus. Our data, in combination with that reported by Du and Copeland,15 implicate dysregulation of the Mds1/Evi1 locus as a hematopoietic immortalization event. The high frequency of MDS1/Evi1 RISs in our animals long term suggests that these events increased the probability of engraftment or survival of transduced CD34+ cells. Each animal received about 10 million transduced CD34+ cells. If a gene locus is hit at a frequency of 1 in 105,21 even random integration would result in infusion of up to 100 cells with integrations at the Mds1/Evi1 locus. However, the estimate of the frequency of repopulating stem cells within CD34+ cells is only 5 per 105 cells in large animals10,22 or only 500 HSCs per animal in these grafts; therefore, it is unlikely that our findings result from effects primarily on true HSCs in the graft. Instead, we suggest that otherwise-committed progenitor cells, including some with only myeloid potential, gain engraftment potential long term or even become immortalized via vector insertion at this locus.
The lack of progression toward monoclonality or leukemia 3 to 7 years after transplantation in these animals is partially reassuring for clinical use of MLV vectors. Development of clonal dominance or actual malignancy likely depends on occurrence of additional mutations or on cooperating effects of the transgene, for instance the IL2Rγ in SCID-X1 vector-related leukemias, or the truncated nerve growth factor receptor (NGFR) in a murine leukemia.1,23 Continued follow-up in this relevant preclinical model will provide critical information for risk assessment in the development of viral gene therapies.
Prepublished online as Blood First Edition Paper, June 2, 2005; DOI 10.1182/blood-2005-03-1115.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Neal Copeland for helpful discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal