Abstract
Patients with paroxysmal nocturnal hemoglobinuria (PNH) have a large clonal population of blood cells deriving from hematopoietic stem cells (HSCs) deficient in glycosylphosphatidylinositol (GPI)-anchored surface molecules. A current model postulates that PNH arises through negative selection against normal HSCs exerted by autoreactive T cells, whereas PNH HSCs escape damage. We have investigated the inhibitory receptor superfamily (IRS) system in 13 patients with PNH. We found a slight increase in the proportion of T cells expressing IRS. In contrast to what applies to healthy donors, the engagement of IRS molecules on T cells from patients with PNH elicited a powerful cytolytic activity in a redirected killing assay, indicating that these IRSs belong to the activating type. This was confirmed by clonal analysis: 50% of IRS+ T-cell clones in patients with PNH were of the activating type, while only 5% were of the activating type in healthy donors. Moreover, the ligation of IRS induces (1) production of tumor necrosis factor α (TNF-α) and interferon γ (IFN-γ) and (2) brisk cytolytic activity against cells bearing appropriate IRS counter-ligands. In addition, these IRS+ T cells show natural killer (NK)-like cytolytic activity to which GPI- cells were less sensitive than GPI+ cells. Thus, T cells with NK-like features, expressing the activating isoforms of IRS, may include effector cells involved in the pathogenesis of PNH.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is a clonal disorder of the hematopoietic stem cell (HSC)1 characterized by an acquired somatic mutation in the PIG-A gene2,3 ; this results in a deficiency on the cell membrane of all proteins anchored by the glycosylphosphatidylinositol (GPI) molecules.4,5 PNH is closely related to idiopathic aplastic anemia (IAA),6,7 which in turn is thought to result from an auto immune attack against HSCs.8 Indeed, thorough analysis of the T-cell repertoire has revealed an increased frequency of expanded T-cell clones in both PNH9,10 and IAA.10,11 This set of facts could be interpreted as suggesting that in PNH autoreactive T cells cause the selective destruction of normal HSCs, whereas HSCs with a PIG-A mutation and consequently with a GPI- phenotype can escape T-cell-mediated damage, thus being able to survive and expand.12-14 The identity of the putative autoreactive T cells and of their molecular targets remain unknown.
Recent studies have identified a subset of peripheral-blood T lymphocytes expressing on their surface molecules that are members of the inhibitory receptor superfamily (IRS), including killer immunoglobulin (Ig)-like receptor (KIR, CD158) or C-lectin type inhibitory receptors (CLIRs, CD94/NKG2).15-17 Upon interaction with HLA-I expressed on autologous cells, these IRSs can deliver an inhibitory signal leading to the reduction of cytolytic activity as well as to the inhibition of cytokine secretion.18-20 This inhibitory effect is mediated by the association of IRS with tyrosine phosphatases, such as Src homology 2-containing protein tyrosine phosphatase-1 (SHP-1), which in turn block the positive signal delivered by activating T-cell surface molecules.15-17 In addition to the inhibitory isoforms of IRS, activating isoforms also exist.15-17 The engagement of activating IRS leads to activation of effector-cell functions including cytolytic activity, cytokine production, and proliferation.15-24 In healthy donors, IRS+ T lymphocytes express surface markers characteristic of memory T cells.25,26 Thus, it has been proposed that this T-cell subset may be expanded in the course of long-term stimulation (eg, in certain viral infections and autoimmune processes).27 Interestingly, it has been reported that T cells bearing activating isoforms of IRS are increased in the sinovial fluid of patients with rheumatoid arthritis, suggesting that these cells may play a role in the pathogenesis of this disease.24 Indeed, T cells bearing activating IRS may actually recognize self-major histocompatibility complex (MHC) alleles and thus activate autoimmune reaction.
We have characterized IRS+ T cells in patients with PNH, and we have found distinctive qualitative abnormalities. The engagement of IRS molecules on cells belonging to this T-cell subset induced triggering of cytolytic activity and production of cytokines. This indicates that the IRS+ T-cell population found in patients with PNH, in contrast to healthy donors, tends to bear IRS of activating type, which may be involved in the pathogenesis of PNH.
Patients, materials, and methods
Subjects
Blood samples from 13 patients with hemolytic PNH (median age, 45 years; range, 24-55 years) and from 38 healthy individuals (median age, 42 years; range, 28-50 years) were collected after informed consent was obtained according to National Institute for Cancer Research (IST-Genoa) institutional procedure. We included only patients with primary classic PNH who had a large PNH population (GPI- granulocytes over 40%), florid hemoglobinuria, and no severe cytopenias (Table 1). No patient was receiving any immunosuppressive drug at the time of the study. Few patients have been studied during the follow-up with blood sampling about 3 months apart.
Clinical and hematologic features of patients investigated
. | . | . | . | . | . | . | . | . | Size of PNH-cell population* . | . | . | . | . | . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients with PNH . | Sex/age, y . | Time since diagnosis, y . | Hb, g/L . | WBC, x10–9/L . | ANC, x10–9/L . | Lymph, x10–9/L . | Platelets, x10–9/L . | Retics, % . | RBCs, % . | PMN, % . | Hemoglobinuria . | Thrombosis . | Blood transfusion . | Other treatment . | |
| 1 | M/53 | 1 | 102 | 4.4 | 3.2 | 0.9 | 249 | 6.0 | 21 | 81 | Yes | No | Occasional | Supportive | |
| 2 | M/45 | 2 | 94 | 2.9 | 1.6 | 1.1 | 97 | 4.8 | 20 | 55 | Yes | No | Never | Supportive | |
| 3 | M/34 | 2 | 101 | 4.7 | 2.5 | 1.7 | 178 | 10.2 | 50 | 80 | Yes | No | Occasional | Supportive† | |
| 4 | F/54 | 10 | 81 | 3.3 | 2.4 | 0.7 | 57 | 7.1 | 36 | 98 | Yes | Hepato-portal | Never | Supportive | |
| 5 | F/54 | 10 | 77 | 3.8 | 1.8 | 1.5 | 193 | 6.4 | 30 | 93 | Yes | No‡ | Never | Supportive | |
| 6 | M/33 | 5 | 123 | 2.1 | 0.9 | 0.9 | 87 | 2.2 | 22 | 69 | Yes | No | Never | Supportive | |
| 7 | F/25 | 8 | 67 | 2.5 | 1.1 | 1.2 | 83 | 14.5 | 23 | 82 | Yes | No | Regular | Supportive§ | |
| 8 | F/53 | 7 | 121 | 3.6 | 1.6 | 1.7 | 64 | 2.1 | 51 | 96 | Yes | Budd-Chiari | Occasional | Supportive | |
| 9 | F/25 | 1 | 109 | 4.5 | 2.4 | 1.8 | 323 | 9.8 | 26 | 80 | Yes | No‡ | Never | Supportive | |
| 10 | F/45 | 5 | 86 | 8.7 | 7.5 | 1.0 | 143 | 17.0 | 75 | 99 | Yes | No | Occasional | Supportive | |
| 11 | M/33 | 5 | 101 | 5.6 | 3.3 | 1.8 | 262 | 6.8 | 37 | 97 | Yes | No | Occasional | Supportive§ | |
| 12 | F/55 | 23 | 84 | 3.9 | 2.5 | 1.0 | 185 | 10.1 | 54 | 90 | Yes | No | Regular | Supportive | |
| 13 | F/32 | 3 | 78 | 4.0 | 2.2 | 1.3 | 193 | 2.4 | 91 | 69 | Yes | No | Occasional | Supportive§ | |
. | . | . | . | . | . | . | . | . | Size of PNH-cell population* . | . | . | . | . | . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients with PNH . | Sex/age, y . | Time since diagnosis, y . | Hb, g/L . | WBC, x10–9/L . | ANC, x10–9/L . | Lymph, x10–9/L . | Platelets, x10–9/L . | Retics, % . | RBCs, % . | PMN, % . | Hemoglobinuria . | Thrombosis . | Blood transfusion . | Other treatment . | |
| 1 | M/53 | 1 | 102 | 4.4 | 3.2 | 0.9 | 249 | 6.0 | 21 | 81 | Yes | No | Occasional | Supportive | |
| 2 | M/45 | 2 | 94 | 2.9 | 1.6 | 1.1 | 97 | 4.8 | 20 | 55 | Yes | No | Never | Supportive | |
| 3 | M/34 | 2 | 101 | 4.7 | 2.5 | 1.7 | 178 | 10.2 | 50 | 80 | Yes | No | Occasional | Supportive† | |
| 4 | F/54 | 10 | 81 | 3.3 | 2.4 | 0.7 | 57 | 7.1 | 36 | 98 | Yes | Hepato-portal | Never | Supportive | |
| 5 | F/54 | 10 | 77 | 3.8 | 1.8 | 1.5 | 193 | 6.4 | 30 | 93 | Yes | No‡ | Never | Supportive | |
| 6 | M/33 | 5 | 123 | 2.1 | 0.9 | 0.9 | 87 | 2.2 | 22 | 69 | Yes | No | Never | Supportive | |
| 7 | F/25 | 8 | 67 | 2.5 | 1.1 | 1.2 | 83 | 14.5 | 23 | 82 | Yes | No | Regular | Supportive§ | |
| 8 | F/53 | 7 | 121 | 3.6 | 1.6 | 1.7 | 64 | 2.1 | 51 | 96 | Yes | Budd-Chiari | Occasional | Supportive | |
| 9 | F/25 | 1 | 109 | 4.5 | 2.4 | 1.8 | 323 | 9.8 | 26 | 80 | Yes | No‡ | Never | Supportive | |
| 10 | F/45 | 5 | 86 | 8.7 | 7.5 | 1.0 | 143 | 17.0 | 75 | 99 | Yes | No | Occasional | Supportive | |
| 11 | M/33 | 5 | 101 | 5.6 | 3.3 | 1.8 | 262 | 6.8 | 37 | 97 | Yes | No | Occasional | Supportive§ | |
| 12 | F/55 | 23 | 84 | 3.9 | 2.5 | 1.0 | 185 | 10.1 | 54 | 90 | Yes | No | Regular | Supportive | |
| 13 | F/32 | 3 | 78 | 4.0 | 2.2 | 1.3 | 193 | 2.4 | 91 | 69 | Yes | No | Occasional | Supportive§ | |
Hb indicates hemoglobin; PMN, granulocytes; Lymph, lymphocytes; Retics, reticulocytes; RBCs, red blood cells; Yes, recurrent episodes of macroscopic hemoglobinuria throughout several years of clinical history.
PMN reflects the relative size of the PNH-cell population more accurately than the percentage of PNH RBCs, because the latter will be grossly underestimated as a consequence of selective hemolysis; this may be further complicated by blood transfusion
Previous prednisone treatment
Recurrent attacks of severe abdominal pain but no documented thrombosis
Previous antithymocyte globulin treatment
Cell separation and analysis
Mononuclear peripheral-blood cells (PBMCs) were isolated by density gradient centrifugation (Ficoll Hypaque; Sigma, St Louis, MO) as described28 and used in immunofluorescence and functional assays. Absolute number of CD3+KIR+ or CD3-KIR+ or CD3+CLIR+ or CD3-CLIR+ lymphocytes was calculated on the basis of lymphocyte counts and the percentage of each cell population determined after surface staining with the corresponding antibodies and analysis on a FACSort (Becton Dickinson, Palo Alto, CA).
Monoclonal antibodies (mAbs) and reagents
The anti-CD16 (NK54, IgG1) mAb, the anti-CD56 (TA181H12, IgG2a) mAb, the anti-CD54 (14D12D2, IgG1) mAb, the anti-CD3 (JT3A, IgG2a) mAb, the anti-CD8α (astra102, IgG1) mAb, the pan KIR2D NKVFS1 mAb (recognizing a common epitope of CD158a/h and of CD158b/j), and p50.3 (CD158i) were produced as described.15,28,29 The anti-CD3 (Leu4, IgG1), anti-CD4 (Leu3a, IgG1), and anti-CD8 (Leu2a, IgG1) mAbs were from Becton Dickinson. The anti-CD94 (HP-3B1, IgG2a; or E94, IgG1), anti-CD158a/h (KIR2DL1/S1) (EB6, IgG1), and anti-CD158b/j (KIR2DL2/S2) (GL183, IgG1) mAbs were from Serotec (Kidlington, Oxford, United Kingdom). The affinity-purified goat antimouse (GAM) anti-isotype specific antiserum was from Southern Biotechnology (Birmingham, AL). Purified GAM anti-Ig (H+L) was purchased from Sigma. The EasySep Custom kit was purchased from Stemcell Biotechnologies (Vancouver, BC, Canada). Recombinant interleukin 2 (rIL2) was from Chiron (Proleukin; Chiron Italia, Siena, Italy). Cells were cultured in RPMI 1640 medium with glutamine and pennicillin-streptomycin (Biochrom, Berlin, Germany) supplemented with 10% fetal calf serum (FCS; Sigma).
Indirect immunofluorescence
Immunofluorescence staining was performed as described.28 Briefly, aliquots of 105 cells were stained with the indicated mAbs followed by either the fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-conjugated anti-isotype-specific GAM antiserum or with an unrelated mAb followed by the fluorescent second reagent. Samples were analyzed on a flow cytometer (FACSort; Becton Dickinson) equipped with a 488-nm argon ion laser exciting FITC, PE, or peridinin chlorophyll (PerCP), and results are expressed as Log green (x-axis) versus red (y-axis) mean fluorescence intensity (MFI) in arbitrary units (au). In some experiments triple immunofluorescence staining was performed with anti-CD3-PerCP, anti-CD94-PE (HP-3B1), and Leu7-FITC- or CD8-FITC-conjugated mAbs. Then samples were run on a FACSort by gating either CD3+CD94+ or CD3+CD94- cells.
Generation of T-cell clones expressing IRS
CD3+ IRS+ T cells were isolated from PBMCs of patients with PNH or healthy volunteers by positive selection using anti-CD94 (E94, IgG1) mAb and custom preparation of EasySep (Stemcell Biotechnologies). The resulting cell population was 60% to 90% CD94+ and 50% to 95% CD3+ depending on the donor. Purified CD94+ cells were stimulated with 10μg/mL phytohemagglutinin (PHA) and cultured in 96-well U-bottomed microplates (Becton Dickinson) with complete medium in the presence of 100 U/mL rIL2 in a final volume of 200 μL/well in the presence of 105/well irradiated allogeneic PBMCs and 5 × 103/well 721.221 lymphoblastoid cell line transfected with HLA-G.30 CD3+ T-cell clones were obtained by culturing purified CD94+ cells under limiting dilution conditions as previously reported.29,30 Briefly, CD94+ cells were cultured in U-bottomed plates at different numbers of cells per well (25/well, 10/well, 5/well, 1/well, and 0.5/well) in the presence of 105 irradiated PBMCs and 103 irradiated 721.221 HLA-I-negative lymphoblastoid cell line with 1 μg/mL PHA and 100 IU/mL IL2. After 5 days medium was changed and cell proliferation was detected from day 10 of culture. Cloning efficiency ranged from 10% to 30%, calculated as described.31 All cell populations were analyzed for the expression of CD3, CD94/NKG2, CD16, CD56, KIR2DL1/S1 with EB6 mAb, and KIR2DL2/S2 with GL183 mAb. For the expression of KIR2DS4, cells reacted with NKVFS1 mAb but not with EB6 and GL183 mAbs. Each clone was analyzed in a redirected killing assay using the FcγR+ P815 murine mastocytoma cell line in the presence of mAbs recognizing either KIR (anti-CD158 mAb, NKVFS1) or CLIR (anti-CD94 mAb, E94) at the effector-to-target (E/T) ratio of 20:1 or 2:1 to identify clones with inhibiting or activating forms of these HLA-I receptors, respectively.30 Indeed, when the engagement of either KIR or CLIR triggered cytolysis of P815 at a 2:1 E/T ratio, that clone was assigned to the activating group. On the other hand, when cytolysis of P815 at a 20:1 E/T ratio was inhibited by the addition of either anti-KIR or anti-CLIR mAb, that clone expressed IRS of the inhibiting type.30
Determination of IFN-γ and TNF-α in culture supernatants
Interferon γ (IFN-γ) and tumor necrosis factor α (TNF-α) released in culture supernatant from T-cell clones derived from PBMCs of patients with PNH upon incubation for 24 hours with medium alone, or in the presence of anti-IRS or anti-CD3 mAbs followed by GAM to obtain optimal cross-linking of the corresponding surface molecule, as indicated in “Results,” was evaluated by enzyme-linked immunosorbent assay (ELISA; PeproTech, London, United Kingdom).32
Cytolytic assays
Cytolytic activity of either ex vivo-isolated PBMCs from patients with PNH or healthy donors or from T-cell clones was tested in a 4-hour 51 Cr-release assay as previously described.28,29 T-cell clones were selected for the expression of activating forms of either KIR and/or CLIR, and were used as effector cells with the FcγR+ murine mastocytoma cell line P815 in the presence of mAb directed against activating receptor for HLA, at an E/T ratio of 2:1, in a final volume of 200 μL RPMI 1640 medium in V-bottomed microwells.28,29 In order to confirm that killing of target cells by an IRS+ T-cell population or by a T-cell clone is mediated by the ligation of an activating type IRS to its cognate HLA ligand, we used the HLA-I-negative lymphoblastoid cell line 721.221. These cells were transfected either with HLA-G (to induce expression of HLA-E with HLA-G peptides, as appropriate for the CD94/NKG2 complex of activating type); or with HLA-Cw3 (appropriate for CD158h [KIR2D2S1]); or with HLA-Cw4 (appropriate for CD158j [KIR2DS2]). These cells were then used without and with the addition of either anti-HLA-I or anti-IRS mAb (5 μg/mL). In some experiments, to study the spontaneous cytolytic activity of IRS+ T cells, GPI+ K562 erythroleukemia cell line (K562wt) and the parental K562 cell line lacking GPI-linked surface molecules (KCRN) generated by Finberg and collaborators33 were used in cytolytic assays with IRS+ T cells from patients with PNH. To evaluate the contribution of GPI-linked molecules to the interaction between effector and K562 target cells, the experiments were carried out also in the presence of saturating amounts of anti-LFA1 mAb (5 μg/mL).
Analysis of KIR genotype and HLA-I typing in patients with PNH
High-molecular-weight DNA was extracted from peripheral-blood samples by a standard method. KIR genotyping was performed by using a multiplex polymerase chain reaction (PCR) method developed by Sun et al.34 PCR amplification was performed with hot-start DNA polymerase, AmpliTaq Gold (Applied Biosystems), by using the described PCR conditions34 and the mixtures of KIR-allele-specific primers kindly provided by D. Senitzer (City of Hope National Medical Center, Duarte, CA). The presence of selected alleles was confirmed by using primers and PCR conditions described by Hiby et al.35 As a quality control of our KIR genotyping procedures, we used cell lines with known KIR genotypes (NK/KIR Phase II QC Reference Panel, International Histocompatibility Working Group, Seattle, WA). HLA-I typing was determined by PCR using the commercial kit from One Lambda (Dynal Biotech, Oslo, Norway).
Statistical analysis
Data are expressed as the mean plus or minus the standard deviation (SD). Nonparametric Mann-Whitney and Fisher exact tests have been used when appropriate. Statistical significance was accepted for P values below .05.
Results
Immune-phenotypic analysis of circulating T lymphocytes bearing molecules belonging to the IRS in patients with PNH
In order to identify cells with potential autoreactivity that may be responsible for the depletion of GPI+ cells in PNH, we first stained PBMCs from patients with PNH (Table 1) with a panel of monoclonal antibodies directed against different cell-surface markers, including CD3, CD4, CD8, and CD45. No significant differences were found in this respect between patients with PNH and healthy donors (data not shown). Since it has been recently suggested that in autoimmune diseases CD3+IRS+ T cells may be a subset of lymphocytes that is expanded,13-15,22,25 we next analyzed the expression of some members of IRS such as CLIR (CD94) and KIR2D (CD158a/h and CD158b/j) together with CD3 (Figure 1A).
In patients with PNH we found a slight but not statistically significant increase (Mann Whitney, P < .09) in the percentage of T cells expressing IRS (CD3+IRS+). The absolute number of T CD3+IRS+ cells in most patients with PNH was similar to that in healthy donors (Mann Whitney, P > .4; Table 2). On the other hand, the absolute number of NK cells (CD3-IRS+) was markedly reduced in patients with PNH (Table 2; Mann-Whitney, P < .002 for CD3-KIR2D+ and P < .006 for CD3-CLIR+).
Quantitative analysis of the expression of molecules belonging to the IRS on T and NK lymphocytes from patients with PNH
Patient no. . | CD3+ KIR2D+, cell/μL . | CD3– KIR2D+, cell/μL . | CD3+ KIR2D+, % of T cells . | CD3+CLIR+, cell/μL . | CD3–CLIR+, cell/μL . | CD3+CLIR+, % of T cells . |
|---|---|---|---|---|---|---|
| PNH1 | ||||||
| Sample 5 | 98 | 91.6 | 2.9 | – | – | – |
| Sample 3 | – | – | – | 31.3 | 27.3 | 3.1 |
| PNH2 | ||||||
| Sample 2 | 195 | 69 | 20.6 | – | – | – |
| PNH3 | ||||||
| Sample 5 | 51 | 30.6 | 3.7 | – | – | – |
| Sample 3 | – | – | – | 64 | 42 | 4.8 |
| PNH4 | ||||||
| Sample 1 | 55 | 10 | 9.2 | – | – | – |
| PNH5 | ||||||
| Sample 2 | 52.5 | 206 | 3.8 | – | – | – |
| PNH6 | ||||||
| Sample 4 | 70.2 | 10 | 8.3 | – | – | – |
| Sample 2 | – | – | – | 115.5 | 9.5 | 12.7 |
| PNH7 | ||||||
| Sample 4 | 61.8 | 80.3 | 5.8 | – | – | – |
| Sample 2 | – | – | – | 112.5 | 87 | 8.4 |
| PNH8 | ||||||
| Sample 2 | 57.5 | 69.5 | 3.8 | – | – | – |
| PNH9 | ||||||
| Sample 3 | 71.3 | 174.7 | 5 | – | – | – |
| Sample 1 | – | – | – | 87 | 204 | 6.9 |
| PNH10 | ||||||
| Sample 1 | 29 | 10 | 2.9 | – | – | – |
| PNH11 | ||||||
| Sample 4 | 151.3 | 111.8 | 15.3 | – | – | – |
| Sample 3 | – | – | – | 177.3 | 73.0 | 17.0 |
| PNH12 | ||||||
| Sample 1 | 87 | 34 | 9.4 | 67 | 24 | 7.2 |
| PNH13 | ||||||
| Sample 1 | 31 | 27 | 2.6 | 94 | 56 | 7.9 |
| Mean ± SD (range) | 81.8 ± 47.0 (31-195) | 76.2 ± 62.7 (10-206) | 7.5 ± 5.5 (2.6-20.6) | 96.3 ± 43.7 (31.3-177.3) | 65.4 ± 61.7 (9.5-204.0) | 8.5 ± 4.4 (3.1-17.0) |
| Healthy controls* | ||||||
| Mean ± SD (range) | 63 ± 36 (19-171) | 192 ± 155 (8-690) | 4.3 ± 2.4 (1.3-11.4) | 102 ± 71 (15-309) | 214 ± 155 (9-666) | 6.8 ± 4.7 (1.0-20.6) |
Patient no. . | CD3+ KIR2D+, cell/μL . | CD3– KIR2D+, cell/μL . | CD3+ KIR2D+, % of T cells . | CD3+CLIR+, cell/μL . | CD3–CLIR+, cell/μL . | CD3+CLIR+, % of T cells . |
|---|---|---|---|---|---|---|
| PNH1 | ||||||
| Sample 5 | 98 | 91.6 | 2.9 | – | – | – |
| Sample 3 | – | – | – | 31.3 | 27.3 | 3.1 |
| PNH2 | ||||||
| Sample 2 | 195 | 69 | 20.6 | – | – | – |
| PNH3 | ||||||
| Sample 5 | 51 | 30.6 | 3.7 | – | – | – |
| Sample 3 | – | – | – | 64 | 42 | 4.8 |
| PNH4 | ||||||
| Sample 1 | 55 | 10 | 9.2 | – | – | – |
| PNH5 | ||||||
| Sample 2 | 52.5 | 206 | 3.8 | – | – | – |
| PNH6 | ||||||
| Sample 4 | 70.2 | 10 | 8.3 | – | – | – |
| Sample 2 | – | – | – | 115.5 | 9.5 | 12.7 |
| PNH7 | ||||||
| Sample 4 | 61.8 | 80.3 | 5.8 | – | – | – |
| Sample 2 | – | – | – | 112.5 | 87 | 8.4 |
| PNH8 | ||||||
| Sample 2 | 57.5 | 69.5 | 3.8 | – | – | – |
| PNH9 | ||||||
| Sample 3 | 71.3 | 174.7 | 5 | – | – | – |
| Sample 1 | – | – | – | 87 | 204 | 6.9 |
| PNH10 | ||||||
| Sample 1 | 29 | 10 | 2.9 | – | – | – |
| PNH11 | ||||||
| Sample 4 | 151.3 | 111.8 | 15.3 | – | – | – |
| Sample 3 | – | – | – | 177.3 | 73.0 | 17.0 |
| PNH12 | ||||||
| Sample 1 | 87 | 34 | 9.4 | 67 | 24 | 7.2 |
| PNH13 | ||||||
| Sample 1 | 31 | 27 | 2.6 | 94 | 56 | 7.9 |
| Mean ± SD (range) | 81.8 ± 47.0 (31-195) | 76.2 ± 62.7 (10-206) | 7.5 ± 5.5 (2.6-20.6) | 96.3 ± 43.7 (31.3-177.3) | 65.4 ± 61.7 (9.5-204.0) | 8.5 ± 4.4 (3.1-17.0) |
| Healthy controls* | ||||||
| Mean ± SD (range) | 63 ± 36 (19-171) | 192 ± 155 (8-690) | 4.3 ± 2.4 (1.3-11.4) | 102 ± 71 (15-309) | 214 ± 155 (9-666) | 6.8 ± 4.7 (1.0-20.6) |
Peripheral-blood lymphocytes were isolated from the indicated patients. Samples of blood were collected at about 3 months from one to another during the follow-up; the sample number indicates the number of the blood collection. Samples were stained with anti-CD3 mAb and anti-panKIR2D mAb (NKVFS1) or anti-CLIR mAb followed by isotype-specific antimouse antiserum. At least 10 000 events for each sample were analyzed on a FACSort (Becton Dickinson). Percent of CD3+ T lymphocytes that are KIR2D+ or CLIR+ is also shown.
– indicates that the sample was not stained with the indicated antibody combination.
For healthy controls, n = 38 for CD3/KIR2D staining, and n = 17 for CD3/CLIR staining
The number of lymphocytes that fail to express the GPI-anchored protein CD59 is much lower in these patients than the number of CD3+IRS+ cells (not shown). We infer that they do not belong to the PNH clone.
Further characterization of CD3+IRS+ cells from patients with PNH demonstrates that they expressed CD8 and CD57 (Figure 1B). This finding suggests that CD3+IRS+ cells may have a cytolytic function. Indeed, CD8+CD57+ T cells with morphologic features of large granular lymphocytes (LGLs) have been reported in patients with PNH.36-38 The granules of LGLs are known to contain perforins, and we have confirmed that in patients with PNH, CD3+IRS+ T cells express perforins in their cytoplasm (not shown). In addition, by using antibodies specific for individual VβTCR families, we have found considerable heterogeneity of IRS+ T cells within individual patients and among different patients (data not shown).
Immune phenotypic analysis of circulating lymphocytes in PNH patients. (A) Expression of IRS members on peripheral-blood T lymphocytes from a patient with PNH. Peripheral-blood mononuclear cells isolated from patient PNH5 (Ai-v) or healthy donor HD3 (Avi-x) were stained with either anti-pan-KIR2D mAb (Aii, Avii), anti-CD158a/h mAb (Aiii, Aviii), anti-CD158b/j mAb (Aiv, Aix), or anti-CLIR (CD94) mAb (Av, Ax) and anti-CD3 mAb (Aii-Av, Avii-Ax) followed by anti-isotype specific goat antimouse antiserum PE-conjugated (for anti-IRS mAbs) or FITC-conjugated (for anti-CD3 mAb). Samples were run on a FACSort and at least 10 000 events were analyzed with the CellQuest computer program (Becton Dickinson). Panels Ai and Avi were stained with an unrelated mAb followed by the second reagent. Results are expressed as Log green fluorescence intensity versus Log red fluorescence intensity in arbitrary units (au). (B) IRS+ T cells in patients with PNH have features of cytotoxic T cells. Peripheral-blood mononuclear cells (PNH5) were stained with PE-conjugated anti-CD94 mAb, PerCP-conjugated anti-CD3 (Bi), and either FITC-conjugated anti-CD57 (Bii, Biv) or anti-CD8 (Biv, Biii) mAb. Samples were run on a FACSort and analyzed with the CellQuest program. R1 and R2 regions (Bi), corresponding to CD3+CD94+ and CD3+CD94- cells respectively, were depicted to analyze the third color (CD57- or CD8-FITC-conjugated; Bii-Bv). Results are expressed as Log PerCP fluorescence intensity versus Log red (PE) fluorescence intensity in arbitrary units (au) (Bi) or Log green (FITC) fluorescence intensity versus number of cells (Bii-Bv). Similar proportions of the different T-cell subsets were found in 3 additional patients with PNH analyzed.
Immune phenotypic analysis of circulating lymphocytes in PNH patients. (A) Expression of IRS members on peripheral-blood T lymphocytes from a patient with PNH. Peripheral-blood mononuclear cells isolated from patient PNH5 (Ai-v) or healthy donor HD3 (Avi-x) were stained with either anti-pan-KIR2D mAb (Aii, Avii), anti-CD158a/h mAb (Aiii, Aviii), anti-CD158b/j mAb (Aiv, Aix), or anti-CLIR (CD94) mAb (Av, Ax) and anti-CD3 mAb (Aii-Av, Avii-Ax) followed by anti-isotype specific goat antimouse antiserum PE-conjugated (for anti-IRS mAbs) or FITC-conjugated (for anti-CD3 mAb). Samples were run on a FACSort and at least 10 000 events were analyzed with the CellQuest computer program (Becton Dickinson). Panels Ai and Avi were stained with an unrelated mAb followed by the second reagent. Results are expressed as Log green fluorescence intensity versus Log red fluorescence intensity in arbitrary units (au). (B) IRS+ T cells in patients with PNH have features of cytotoxic T cells. Peripheral-blood mononuclear cells (PNH5) were stained with PE-conjugated anti-CD94 mAb, PerCP-conjugated anti-CD3 (Bi), and either FITC-conjugated anti-CD57 (Bii, Biv) or anti-CD8 (Biv, Biii) mAb. Samples were run on a FACSort and analyzed with the CellQuest program. R1 and R2 regions (Bi), corresponding to CD3+CD94+ and CD3+CD94- cells respectively, were depicted to analyze the third color (CD57- or CD8-FITC-conjugated; Bii-Bv). Results are expressed as Log PerCP fluorescence intensity versus Log red (PE) fluorescence intensity in arbitrary units (au) (Bi) or Log green (FITC) fluorescence intensity versus number of cells (Bii-Bv). Similar proportions of the different T-cell subsets were found in 3 additional patients with PNH analyzed.
The engagement of IRS molecules on IRS+ T cells from patients with PNH triggers a powerful cytolytic activity
Lymphocytes can express 2 different isoforms of IRS, one of which is of the activating type and the other of the inhibiting type with respect to cytolytic function.15-18,39 Thus, we analyzed the function of IRS in T cells from patients with PNH. To address this point, we used a redirected killing assay. In this assay, the engagement of the corresponding molecule by an appropriate (anti-IRS) mAb acts both as a bridge between the Fcγ receptor of P815 cells and a surface molecule on T cells and as a cross-linking device that triggers the function of T cells themselves. In general, among PBMCs, the CD3-CD16+ NK cells, on which IRSs have been first described, are the most potent cytolytic effector cells.15-17 In order to minimize interference in this assay by NK cells, we selected those samples with a low number of NK cells. We found that ex vivo-isolated PBMCs from 2 patients with PNH, incubated with either anti-KIR or anti-CLIR mAbs in a redirected killing assay, displayed a strong lytic effect against the target cells, indicating that their IRSs are predominantly of the activating type (Figure 2).
When we used the same assay to test highly purified IRS+ T cells activated by IL2 from patients with PNH (n = 4), we observed that either anti-KIR or anti-CLIR mAbs induced a strong lytic activity (data not shown). Taken together, these findings indicate that, in patients with PNH, T cells are equipped with activating IRS.18-20
T-cell clones from patients with PNH mainly express activating isoforms of IRS molecules
In order to determine the relative proportions of activating versus inhibiting isoforms of IRS present on T cells in patients with PNH, we proceeded to isolate T-cell clones. This was done by purifying CLIR+ cells from patients with PNH and healthy donors by positive immune-magnetic selection using anti-CLIR mAb. CLIR+ cells were cloned by limiting dilution and cultured with IL2.28,29 The percentage of CD3+ cells among newly isolated CLIR+ cells ranged from 80% to 95%. The cloning efficiency was lower (25%) in patients with PNH than that found with CD3+CD8+ or CD3+CD4+ T cells from healthy donors (70% and 95%, respectively), but it was similar to that of CLIR+ cells from healthy donors (27%; data not shown). As expected, the large majority of these T-cell clones, whether derived from patients with PNH or from healthy donors, expressed CLIR, whereas only a minority expressed KIR molecules (not shown).
In a redirected killing assay, a given clone was classified as being of the activating type when anti-KIR or anti-CLIR mAb triggered cytolysis of P815 at an E/T ratio of 2:1, whereas a clone was classified as being of the inhibiting type if it inhibited lysis of P815 at an E/T ratio of 20:1.
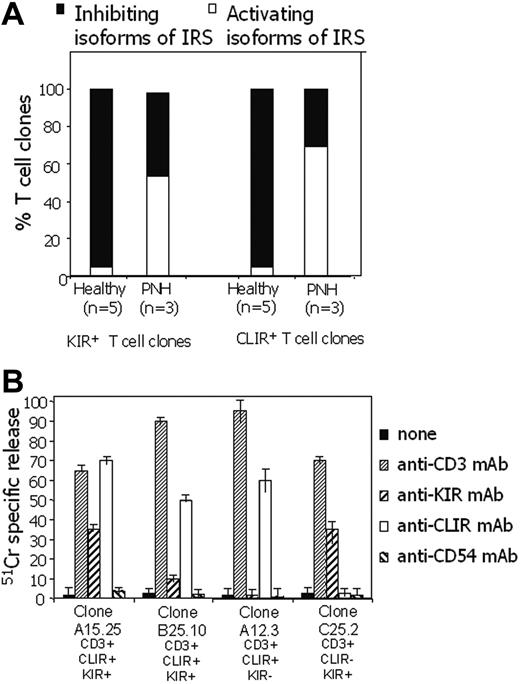
We found that the large majority of CD3+IRS+ T-cell clones from patients with PNH expressed activating isoforms of IRS (Figure 3). In some CD3+IRS+ T-cell clones, both KIR and CLIR were of the activating type, whereas other T-cell clones bore activating CLIR but inhibiting KIR or vice versa (not shown). Twenty-four of 45 of the T-cell clones derived from patients with PNH bore only KIR of the activating type (from PNH5, 12 CD158h [KIR2DS1] and 2 CD158j [KIR2DS2] of 23 KIR+ clones; from PNH6, 5 CD158h [KIR2DS1] of 10 KIR+ clones; from PNH7, 5 CD158j [KIR2DS2] of 12 KIR+ clones). Forty-five of 65 T-cell clones bore CLIR of the activating type (17 from PNH5, 13 from PNH6, and 15 from PNH7). By contrast, in healthy donors T-cell clones bearing the activating isoforms of these 2 IRSs were very rare (only 1 of 21 and 2 of 43, respectively, bore either KIR or CLIR of the activating type; Figure 4A; Fisher exact test, P < .001).
The majority (about 90%) of clones from patients with PNH bearing activating IRS were TCRαβ and a few were TCRγδ. These findings indicate that T-cell clones expressing activating IRS are present with high frequency in patients with PNH, whereas they are an exception in healthy donors.13-15
The engagement of activating IRS on IRS+ T-cell clones from patients with PNH induces production of high levels of IFN-γ and TNF-α
Given the expression of activating IRS on T-cell clones derived from patients with PNH, we further analyzed whether the ligation of these IRSs lead to the production of cytokines potentially involved in the regulation of hematopoiesis. Indeed, it has been shown that IFN-γ and TNF-α may play a key role in suppressing hematopoiesis in AA.40-44 To analyze whether T-cell clones bearing activating IRS were able to produce IFN-γ and TNF-α, we stained these clones with either anti-CLIR or anti-KIR mAbs; thus, these clones were incubated for 24 hours at 37°C on GAM-coated plates to achieve the cross-linking of the corresponding IRS. As shown in Figure 5, both IFN-γ and TNF-α were produced by IRS+ T-cell clones upon the engagement of either CLIR or KIR on the cell surface. It is of note that the amount of IFN-γ or TNF-α released in culture supernatant was comparable to that elicited by the engagement of the CD3/TCR complex achieved by using anti-CD3 mAb, in the same experimental system. T-cell clones that bore only one member of IRS of the activating type were able to produce the IFN-γ and TNF-A cytokines only when the corresponding receptor was engaged by the specific mAb (data not shown). These findings suggest that activating IRS, when appropriately engaged by the corresponding natural ligand, can induce not only activation of the cytolytic machinery, but also the release of cytokines, which may play a role in regulating hematopoiesis.
T cells expressing activating IRS lyse target cells bearing the cognate IRS counter-receptors
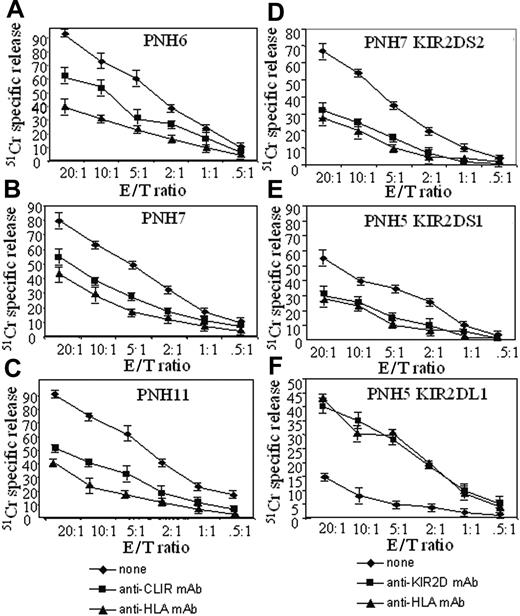
721.221 cells do not express HLA class I molecules but, once transfected with HLA-G, they will express HLA-E, the counter-ligand for CD94/NKG2-presenting HLA-G-derived peptides. When we exposed 721.221 lymphoblastoid cells transfected with HLA-G to T-cell populations from patients PNH6, PNH7, and PNH11, who expressed the CLIR CD94/NKG2 of the activating type, we obtained lysis in all 3 cases (Figure 5). Addition of either anti-HLA class I or anti-CD94 mAb to the cytolytic assay strongly inhibited lysis (Figure 5A-C). To determine that also activating KIR-expressing T cells lyse cells bearing KIR ligand, we selected T-cell clones from patients with PNH that express only one activating KIR. Clone B25.5 CD158j+ (KIR2DS2+) from patient PNH7 killed 721.221 cells transfected with the counter-ligand HLA-Cw3 allele (Figure 5D); and clone C15.10 CD158h+ (KIR2DS1+) from patient PNH5 killed 721.221 cells transfected with the corresponding ligand HLA-Cw4 (Figure 5E). Again, the addition of anti-HLA-I or anti-pan KIR mAbs inhibited by 50% to 60% the lysis of target cells bearing the KIR counter-ligands (Figure 5D-E). As a negative control, we found that clone A14.25, expressing the inhibiting KIR2DL1, did not lyse 721.221 Cw4-transfected cells, unless the inhibiting signal delivered via KIR2DL1 was impaired by the addition of anti-HLA-I or anti-KIR mAb.
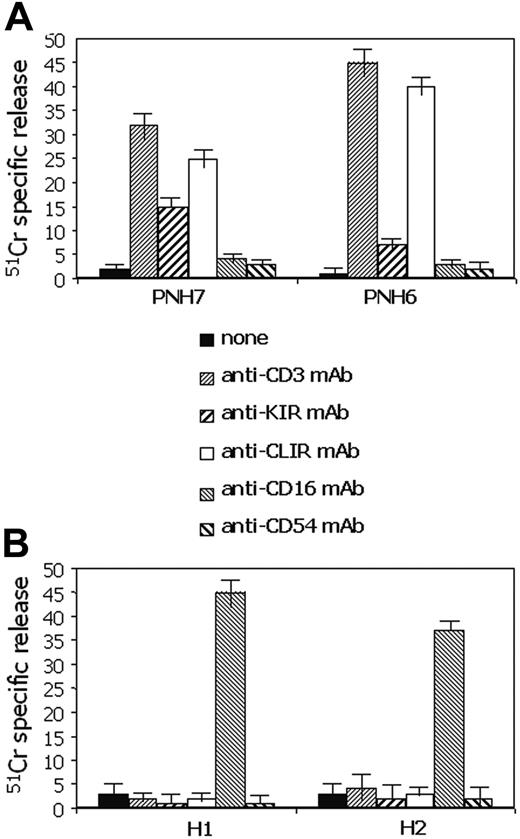
Peripheral-blood mononuclear cells from patients with PNH have strong cytolytic activity. Peripheral-blood mononuclear cells were isolated from either patients with PNH (A) or healthy donors (B) and their cytolytic activity was analyzed in a 4-hour redirected killing assay using the FcγR+ murine cell line P815 in the absence (none) or in the presence of mAbs recognizing the indicated surface molecules. Anti-CD54 mAb was used as an isotype-matched control mAb. The PNH donor samples (PNH6 and PNH7) used in this experiment had a low number of CD16+ NK cells (9 cells/μL in PNH6 and 24 cells/μL in PNH7) but sizeable proportions of CD3+IRS+ cells (109 cells/μL in PNH6 and 112 cells/μL in PNH7). Cytolytic activity of peripheral-blood lymphocytes from 2 representative healthy donors (H1 and H2) is shown for comparison (B). Results are expressed as % 51 Cr-specific release at an E/T ratio of 10:1 mean of triplicate samples; bars indicate standard deviation of triplicate samples. A comparable difference between PBMCs of patients with PNH and healthy donors was found also at 20:1 and at 40:1 E/T ratios.
Peripheral-blood mononuclear cells from patients with PNH have strong cytolytic activity. Peripheral-blood mononuclear cells were isolated from either patients with PNH (A) or healthy donors (B) and their cytolytic activity was analyzed in a 4-hour redirected killing assay using the FcγR+ murine cell line P815 in the absence (none) or in the presence of mAbs recognizing the indicated surface molecules. Anti-CD54 mAb was used as an isotype-matched control mAb. The PNH donor samples (PNH6 and PNH7) used in this experiment had a low number of CD16+ NK cells (9 cells/μL in PNH6 and 24 cells/μL in PNH7) but sizeable proportions of CD3+IRS+ cells (109 cells/μL in PNH6 and 112 cells/μL in PNH7). Cytolytic activity of peripheral-blood lymphocytes from 2 representative healthy donors (H1 and H2) is shown for comparison (B). Results are expressed as % 51 Cr-specific release at an E/T ratio of 10:1 mean of triplicate samples; bars indicate standard deviation of triplicate samples. A comparable difference between PBMCs of patients with PNH and healthy donors was found also at 20:1 and at 40:1 E/T ratios.
Matching activating KIR genotypes and HLA-C genotypes in patients with PNH
IRS+ molecules can exert their action only when they find the appropriate counter-ligands on target cells. Both the CLIR gene and its counter-ligands (ie, HLA-G and HLA-E) are not known to be polymorphic: therefore, a match for CLIR is always potentially available. In contrast, the genes encoding KIR molecules as well as those encoding their counter-ligands (HLA-C) are polymorphic. In fact, each KIR allele can exert its effect only if it finds on the target cell at least one HLA-C molecule that is a specific match for it. Therefore, we proceeded to determine both the activating KIR genotypes and the HLA-C genotypes of patients with PNH (Table 3). In patients PNH4, PNH5, PNH6, and PNH9, we found KIR2DS1 and 1 or 2 of its counter-ligands (HLA-Cw4, HLA-Cw5, Cw6). In patients PNH3, PNH5, PNH7, and PNH9, we found KIR2DS2 and 1 or 2 of its counter-ligands (HLA-Cw1, HLA-Cw7, HLA-Cw8). In patients PNH1, PNH2, PNH4, and PNH12, we found the KIR2DS4 gene and its counter-ligand HLA-Cw4.45 Thus, in 9 of 11 patients with PNH we found both the activating KIR gene and the appropriate HLA-C allele, so that target cells bearing the latter would be susceptible to KIR-mediated killing. We note that the 2 remaining patients (PNH11 and PNH13) share the KIR2DS4 gene, for which a possible alternative (non-MHC) counter-ligand has been recently suggested.46
The genotype of activating KIR and of HLA-C in PNH patients
Patient no. . | Activating KIR genotype . | HLA-C allele . |
|---|---|---|
| PNH1 | 2DS4 | 04,‡16 |
| PNH2 | 2DS4 | 04,‡07 |
| PNH3 | 2DS2,†2DS4 | 08,†12 |
| PNH4 | 2DS1,*2DS2, 2DS3, 2DS4,‡2DS5, 3DS1 | 04,‡06 |
| PNH5 | 2DS1,*2DS2,†2DS3, 3DS1 | 04,*07† |
| PNH6 | 2DS1,*2DS3, 2DS4, 3DS1 | 05,*15 |
| PNH7 | 2DS2,†2DS3, 2DS4 | 01,†07† |
| PNH9 | 2DS1,*2DS2,†2DS3, 2DS4, 2DS5, 3DS1 | 06,*07† |
| PNH11 | 2DS4 | 07, 07 |
| PNH12 | 2DS4‡ | 03, 04‡ |
| PNH13 | 2DS4 | 06, 15 |
Patient no. . | Activating KIR genotype . | HLA-C allele . |
|---|---|---|
| PNH1 | 2DS4 | 04,‡16 |
| PNH2 | 2DS4 | 04,‡07 |
| PNH3 | 2DS2,†2DS4 | 08,†12 |
| PNH4 | 2DS1,*2DS2, 2DS3, 2DS4,‡2DS5, 3DS1 | 04,‡06 |
| PNH5 | 2DS1,*2DS2,†2DS3, 3DS1 | 04,*07† |
| PNH6 | 2DS1,*2DS3, 2DS4, 3DS1 | 05,*15 |
| PNH7 | 2DS2,†2DS3, 2DS4 | 01,†07† |
| PNH9 | 2DS1,*2DS2,†2DS3, 2DS4, 2DS5, 3DS1 | 06,*07† |
| PNH11 | 2DS4 | 07, 07 |
| PNH12 | 2DS4‡ | 03, 04‡ |
| PNH13 | 2DS4 | 06, 15 |
Patients as in Table 1. The ligands of KIR2DS1 are the HLA-CLys80 (w2, w4, w5, w6); the ligands of KIR2DS2 are the HLA-CAsn80 (w1, w3, w7, w8); the ligand of KIR2DS4 is the HLA-Cw4; the ligands of KIR2DS3 and KIR2DS5 are still unknown.
Positive matches for KIRDS1
Matches for KIR2DS2
Different susceptibility to lysis of GPI+ versus GPI- cells
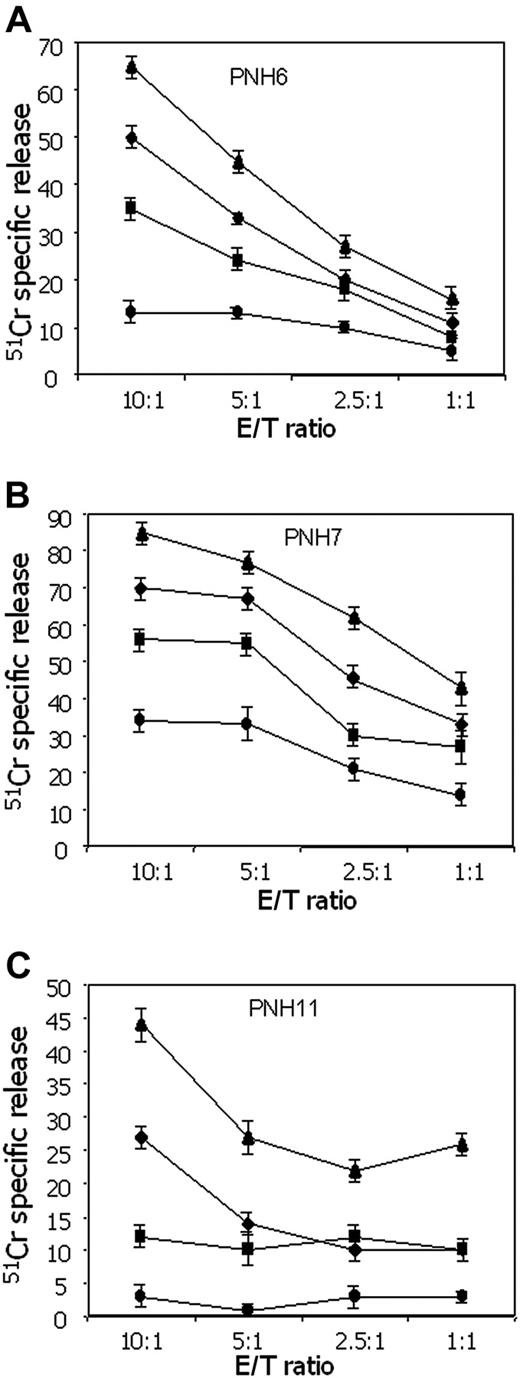
In patients with PNH there is an expansion of hematopoietic cells lacking GPI-linked molecules.47-50 This is thought to result not from an intrinsic growth advantage of GPI- precursors, but rather from negative selection against GPI+ precursors by autoreactive cells.47-52 Therefore, we next tested whether IRS+ T cells from patients with PNH would exert a different spontaneous cytolytic activity against GPI+ versus GPI- K562 cells. As shown in Figure 6, the cytolytic activity exerted by IRS+ T cells from 3 different patients with PNH against GPI+ K562 was consistently higher (ranging from 15% to 55%), at different E/T ratios, than that observed using GPI- K562 cell line. It is well known that the binding between LFA-1 on effector lymphocytes and intercellular adhesion molecule-1 (ICAM-1) expressed on target cells plays a key role in triggering cytolytic activity.53-55 Pretreatment of cells with lymphocyte function-associated antigen 1 (LFA-1)-specific mAbs strongly reduced lysis of both GPI+ and GPI- K562 cell lines, confirming that LFA-1/ICAM-1 binding is involved in effector-target interaction. However, in the presence of anti-LFA-1 mAb, IRS+ T cells from patient PNH11 were still able to lyse K562 cells, but only if they were GPI+ (Figure 6C).
Most IRS+ T-cell clones from patients with PNH bear IRS of the activating type. T-cell clones expressing IRS were derived from 3 different patients with PNH (PNH5, PNH6, PNH7) and from 5 healthy donors (HD3, HD8, HD9, HD15, HD19). (A) Each clone was analyzed in redirected killing assay using P815 target cells in the presence of anti-CLIR or anti-KIR mAbs at (1) an E/T ratio of 20:1 to identify T-cell clones bearing inhibiting IRS isoforms (▪) and (2) an E/T ratio of 2:1 to identify T-cell clones bearing activating IRS isoforms (▪). (B) Cytolysis by T-cell clones obtained from patients with PNH upon the engagement of activating IRS. Cytolytic activity of T-cell clones derived from patients with PNH was analyzed in a 4-hour redirected killing assay in the absence (none) or in the presence of anti-CLIR or anti-KIR mAb or anti-CD3 mAb. Anti-CD54 mAb was used as an isotype-matched control mAb. The clone A15.25 (PNH5) was CLIR+KIR2DS1+ (CD158h+) and B25.10 (PNH7) was CLIR+KIR2DS2+ (CD158j+) while the clones from PNH6 A12.3 expressed only CLIR (CD94) and the clone C25.2 was CLIR-KIR2DS1+. All the clones were selected for the expression of activating isoforms of either CLIR and/or KIR. These clones are representative of 65 clones analyzed. Results are expressed as % of 51 Cr release, mean of triplicate samples; bars indicate standard deviation of triplicate samples.
Most IRS+ T-cell clones from patients with PNH bear IRS of the activating type. T-cell clones expressing IRS were derived from 3 different patients with PNH (PNH5, PNH6, PNH7) and from 5 healthy donors (HD3, HD8, HD9, HD15, HD19). (A) Each clone was analyzed in redirected killing assay using P815 target cells in the presence of anti-CLIR or anti-KIR mAbs at (1) an E/T ratio of 20:1 to identify T-cell clones bearing inhibiting IRS isoforms (▪) and (2) an E/T ratio of 2:1 to identify T-cell clones bearing activating IRS isoforms (▪). (B) Cytolysis by T-cell clones obtained from patients with PNH upon the engagement of activating IRS. Cytolytic activity of T-cell clones derived from patients with PNH was analyzed in a 4-hour redirected killing assay in the absence (none) or in the presence of anti-CLIR or anti-KIR mAb or anti-CD3 mAb. Anti-CD54 mAb was used as an isotype-matched control mAb. The clone A15.25 (PNH5) was CLIR+KIR2DS1+ (CD158h+) and B25.10 (PNH7) was CLIR+KIR2DS2+ (CD158j+) while the clones from PNH6 A12.3 expressed only CLIR (CD94) and the clone C25.2 was CLIR-KIR2DS1+. All the clones were selected for the expression of activating isoforms of either CLIR and/or KIR. These clones are representative of 65 clones analyzed. Results are expressed as % of 51 Cr release, mean of triplicate samples; bars indicate standard deviation of triplicate samples.
IRS+ T-cell clones produce a high amount of IFN-γ and TNF-α. T-cell clones expressing IRS activating isoforms (CD3+KIR+CLIR+ clones: C3.50 [KIR2DS2+, from PNH5], ▪; and A1.25 [KIR2DS1+, from PNH6], □) were stained with either anti-CD3, anti-KIR, anti-CLIR, or anti-CD54 (isotype-matched control mAb) mAbs and incubated in plates precoated with goat antimouse antiserum. After 24 hours, culture supernatants were harvested and analyzed by ELISA for the presence of either IFN-γ (A) or TNF-α (B). “None” represents the release of either IFN-γ or TNF-α in the absence of any mAb. Results are expressed as mean of triplicate samples; bars indicate standard deviation of triplicate samples.
IRS+ T-cell clones produce a high amount of IFN-γ and TNF-α. T-cell clones expressing IRS activating isoforms (CD3+KIR+CLIR+ clones: C3.50 [KIR2DS2+, from PNH5], ▪; and A1.25 [KIR2DS1+, from PNH6], □) were stained with either anti-CD3, anti-KIR, anti-CLIR, or anti-CD54 (isotype-matched control mAb) mAbs and incubated in plates precoated with goat antimouse antiserum. After 24 hours, culture supernatants were harvested and analyzed by ELISA for the presence of either IFN-γ (A) or TNF-α (B). “None” represents the release of either IFN-γ or TNF-α in the absence of any mAb. Results are expressed as mean of triplicate samples; bars indicate standard deviation of triplicate samples.
These results indicate that lack of surface GPI molecules renders cells less susceptible to lysis mediated by cytolytic effector T lymphocytes, suggesting that GPI-linked molecules have a critical role in effector-target ligation.
Discussion
The classic clinical triad of PNH consists of intravascular hemolysis, thrombosis, and bone marrow failure.52,53 At the molecular level, PNH is characterized by the deficiency on the surface of blood cells of all proteins anchored to the membrane by the glycosylphosphatidylinositol (GPI) molecule. The absence of GPI-linked molecules, such as CD59 and CD55, explains hemoglobinuria,56,57 and probably also the tendency to thrombosis.58 On the other hand, there is no obvious explanation as to why deficiency of GPI-linked proteins by itself ought to also cause bone marrow failure. A plausible model has been, for some time, that an autoimmune process is at work in PNH, as it is in idiopathic aplastic anemia (IAA).8,40 If the auto reactive T cells involved in this process recognize a GPI-linked protein, or the GPI molecule itself, then GPI+ cells, including GPI+ HSCs, would be damaged selectively; whereas GPI- HSCs, if they exist, would escape damage and be able to proliferate.59 This model has been corroborated by the finding that small GPI- (ie, PNH-like) clones exist in the peripheral blood of healthy subjects48 ; therefore, in PNH it is the expansion rather than the presence of GPI- cells that we need to explain. Based on this notion, it was natural to seek evidence for auto reactive T cells. In fact, it has emerged that skewing of the T-cell repertoire is common in PNH,9,10 and the coexistence of LGL leukemia and PNH in the same patient38 suggests that the T cells responsible for bone marrow damage in PNH may belong to the CD3+CD8+CD57+ cell population displaying NK-like cytolytic activity.15-17
NK-like T cells can express IRS molecules. In terms of their structure, IRSs are of 2 types: the immunoglobulin (Ig) superfamily inhibitory receptors (ISIRs), which include killer Ig-like receptors (KIR, CD158a, b, and p50); and C-type lectin inhibitory receptors (CLIRs),15-17 such as the CD94/NKG2 complex. An increase of T cells expressing IRS has been reported in a range of chronic disorders, in which they may serve to maintain memory T cells alive, as well as in a case of pure red-cell aplasia.60
For this reason, we have systematically studied in patients with PNH those NK-like T cells that carry IRS molecules. We have observed in these patients a slight but not statistically significant increase of the proportion of T cells bearing IRS molecules, whereas CD3-CD16+ NK cells were decreased, as previously reported.61,62 This shortage of NK cells might be part of the bone marrow failure syndrome present in patients with PNH.63,64 On the other hand, in most patients with PNH the absolute number of IRS+ T cells was similar to that found in healthy controls. These findings were in keeping with a qualitative rather than with a quantitative abnormality of IRS+ T cells in PNH; therefore, we proceeded to their functional characterization.
CD3+IRS+ T cells lyse target cells bearing the appropriate HLA-I counter ligand. Polyclonal (A: PNH6; B: PNH7; C: PNH11) bulk populations homogeneously expressing CD3 and CLIR (CD94+) antigens were challenged in a 4-hour 51 Cr release assay with 721.221 target cells bearing HLA-E with G peptides as ligand for CD94/NKG2 complex of the activating type. Anti-HLA-I mAb or anti-CD94 mAb (5 μg/mL) was added to the cytolytic assay to determine whether the masking of CD94/NKG2-HLA-I interaction leads to inhibition of target lysis. (D-F) The CD3+KIR+CLIR- T-cell clones KIR2DS2+ (D; from PNH7) or KIR2DS1+ (E; from PNH5) were used as effector cells with the 721.221 transfected with HLA-Cw3 or HLA-Cw4 (ligands of KIR2DS2 or KIR2DS1, respectively). E/T ratio indicates effector-target ratio. Anti-HLA-I mAb or anti-panKIR2D mAb (5 μg/mL) was added to the cytolytic assay to determine whether the masking of KIR2D-HLA-I interaction leads to inhibition of target lysis. The CD3+KIR+ T-cell clone expressing KIR2DL1, the inhibiting form of KIR2D recognizing HLA-Cw4, was challenged with 721.221 target cells HLA-Cw4 transfected. In this instance, the addition of anti-HLA-I mAb or anti-panKIR2D mAb (5 μg/mL) led to triggering of lysis as the impairment of KIR2D-HLA-I binding does not protect the target cells any more. Results are expressed as % 51 Cr-specific release, mean of triplicate samples plus or minus SD.
CD3+IRS+ T cells lyse target cells bearing the appropriate HLA-I counter ligand. Polyclonal (A: PNH6; B: PNH7; C: PNH11) bulk populations homogeneously expressing CD3 and CLIR (CD94+) antigens were challenged in a 4-hour 51 Cr release assay with 721.221 target cells bearing HLA-E with G peptides as ligand for CD94/NKG2 complex of the activating type. Anti-HLA-I mAb or anti-CD94 mAb (5 μg/mL) was added to the cytolytic assay to determine whether the masking of CD94/NKG2-HLA-I interaction leads to inhibition of target lysis. (D-F) The CD3+KIR+CLIR- T-cell clones KIR2DS2+ (D; from PNH7) or KIR2DS1+ (E; from PNH5) were used as effector cells with the 721.221 transfected with HLA-Cw3 or HLA-Cw4 (ligands of KIR2DS2 or KIR2DS1, respectively). E/T ratio indicates effector-target ratio. Anti-HLA-I mAb or anti-panKIR2D mAb (5 μg/mL) was added to the cytolytic assay to determine whether the masking of KIR2D-HLA-I interaction leads to inhibition of target lysis. The CD3+KIR+ T-cell clone expressing KIR2DL1, the inhibiting form of KIR2D recognizing HLA-Cw4, was challenged with 721.221 target cells HLA-Cw4 transfected. In this instance, the addition of anti-HLA-I mAb or anti-panKIR2D mAb (5 μg/mL) led to triggering of lysis as the impairment of KIR2D-HLA-I binding does not protect the target cells any more. Results are expressed as % 51 Cr-specific release, mean of triplicate samples plus or minus SD.
IRS molecules exist in 2 functionally different isoforms. In healthy donors, most IRS+ T cells bear isoforms of IRS molecules with inhibiting properties. In fact, their engagement inhibits strongly cytolytic activity and cytokine production triggered via activating receptors (ie, the CD3/TCR complex).25,26 Nevertheless, a small proportion of IRS+ T cells bear isoforms of IRS molecules with activating properties. The physiologic role of the activating isoforms of IRS is incompletely understood, although several hypotheses have been put forward to explain their presence on different lymphocyte subsets.15-17,39 Expansion of these cells has been reported in rheumatoid arthritis,24 suggesting that they may play a role in some autoimmune process.
Unlike in healthy subjects, in patients with PNH the ligation of IRS molecules either on freshly isolated PBMCs (Figure 3) or on highly purified IL2-activated IRS+ T cells triggered cytolysis in a redirected killing assay. This indicates that in patients with PNH there is a marked increase in IRS+ T cells bearing activating isoforms of IRS. This was corroborated by generating T-cell clones. Indeed, from healthy subjects more than 95% of these clones were of the inhibiting type. In contrast, under the same experimental conditions, in patients with PNH more than 50% of IRS+ T-cell clones in our redirected killing assay were of the activating type (Figure 4A). This activating function has been more conclusively confirmed by the finding that the engagement of the activating IRS with the appropriate HLA counter-ligands (HLA-E with G peptide for CLIR; HLA-Cw4 for KIR2DS1; HLA-CW3 for KIR2DS2) triggered the cytolytic activity of IRS+ T cells (Figure 5). The counter-ligand of the activating CLIR is potentially present in all individuals, whereas activating KIR alleles recognize specific HLA-C alleles. In this respect, our analysis has shown that in most patients with PNH, activating KIR isoforms are properly matched with their HLA-C counter-ligands (Table 3). Although we cannot positively state that HLA-C-matched activating KIR alleles are expressed on IRS+ T cells, the genotype data suggest that the cytotoxic ability of the IRS+ T cells could occur in vivo.
In principle, we cannot say whether the increase in IRS+ T cells bearing activating IRS isoforms in PNH is a secondary or a primary phenomenon. Earlier reports from other groups and from our group have not revealed GPI- cells to be less sensitive than GPI+ cells to T-cell65,66 or NK-mediated lysis.33,66 Now we have found that a specific T-cell subset (ie, IRS+ T cells obtained from patients with PNH) shows NK-like cytolytic activity to which GPI+ K562 cells are more sensitive than GPI- K562 cells. If this difference applies to HSCs as well, this is one plausible mechanism whereby GPI deficiency would constitute a selective advantage for GPI- HSCs, explaining the expansion of PNH hematopoiesis in the context of the bone marrow failure present in patients with PNH. This bone marrow failure might be determined also by IFN-γ and TNF-α, cytokines that suppress hematopoiesis,40-44 produced by IRS+ T-cell clones. Thus, the IRS+ T cells we have characterized are good candidates for being able to damage GPI+ HSCs in PNH.
IRS+ T cells from patients with PNH kill more efficiently GPI+ than GPI- cells. Cytolytic activity of IRS+ T cells from 3 different patients with PNH (A: PNH6; B: PNH7; C: PNH11) against GPI+ K562 cell line or its GPI- counterpart (KCRN) was analyzed in a 4-hour 51 Cr release assay at the indicated effector-target (E/T) ratios. Anti-LFA-1 mAb (5 μg/mL) was added to the cytotoxic assay in order to block the contribution of LFA-1/ICAM-1 interaction to the activation of T cells. As the 2 K562 cell lines differ from each other for the expression of GPI-linked molecules, this shows the relative contribution of GPI molecules to target-cell lysis. Results are expressed as % of 51 Cr release, mean of triplicate samples plus or minus SD.♦ indicates KCRN (K562 GPI-); •, KCRN (K562 GPI-) plus anti-LFA-1 mAb; ▴, K562 (GPI+); ▪, K562 (GPI+) plus anti-LFA-1 mAb.
IRS+ T cells from patients with PNH kill more efficiently GPI+ than GPI- cells. Cytolytic activity of IRS+ T cells from 3 different patients with PNH (A: PNH6; B: PNH7; C: PNH11) against GPI+ K562 cell line or its GPI- counterpart (KCRN) was analyzed in a 4-hour 51 Cr release assay at the indicated effector-target (E/T) ratios. Anti-LFA-1 mAb (5 μg/mL) was added to the cytotoxic assay in order to block the contribution of LFA-1/ICAM-1 interaction to the activation of T cells. As the 2 K562 cell lines differ from each other for the expression of GPI-linked molecules, this shows the relative contribution of GPI molecules to target-cell lysis. Results are expressed as % of 51 Cr release, mean of triplicate samples plus or minus SD.♦ indicates KCRN (K562 GPI-); •, KCRN (K562 GPI-) plus anti-LFA-1 mAb; ▴, K562 (GPI+); ▪, K562 (GPI+) plus anti-LFA-1 mAb.
Prepublished online Blood First Edition Paper, June 14, 2005; DOI 10.1182/blood-2004-11-4315.
Supported in part by the Associazione Italiana Ricerca Cancro (A.P., M.R.Z., L.L., and R.N.) and by the Fondo Investimenti Ricerca di Base-MIUR (L.L. and R.N.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Robert Finberg (Boston, MA) for providing the GPI+ and GPI- K562 cell lines and David Sanitzer (Duarte, CA) for providing the primers for KIR genotyping. We are grateful to Ji-Yao Sun (Duarte, CA) for the helpful technical suggestion for KIR genotyping and Martina Serra for help in the Laboratory of Human Genetics (LST, Genoa).


![Figure 4. IRS+ T-cell clones produce a high amount of IFN-γ and TNF-α. T-cell clones expressing IRS activating isoforms (CD3+KIR+CLIR+ clones: C3.50 [KIR2DS2+, from PNH5], ▪; and A1.25 [KIR2DS1+, from PNH6], □) were stained with either anti-CD3, anti-KIR, anti-CLIR, or anti-CD54 (isotype-matched control mAb) mAbs and incubated in plates precoated with goat antimouse antiserum. After 24 hours, culture supernatants were harvested and analyzed by ELISA for the presence of either IFN-γ (A) or TNF-α (B). “None” represents the release of either IFN-γ or TNF-α in the absence of any mAb. Results are expressed as mean of triplicate samples; bars indicate standard deviation of triplicate samples.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/7/10.1182_blood-2004-11-4315/6/m_zh80190584930004.jpeg?Expires=1767762404&Signature=xpBJ4oLu71p4EeCFMPyMy~h7Z0rp7mrqVURNhPgVIo~fsaefgTfvLgBIZf8aYxMIoOqNRhCpV1MnH3yPpgzD09akQrWMHgI-lYUwxtUkX8e~g4Kn1I0Ww5BgJqDjUw53JYquWuHJao~epamCYS1OIJxxeZP5KNG-kg~qOy2iK1kv1Ysf9KX3xFGSJovNcfjpCTPJmlLrWi5LPsyq4X9a3HZICKXHuSUGtLPOvZgGtRQVZHYFISeZqgpNjX-tF5bK7he6Kh0wCcL2hJCecNfasjzWiw4WUeHT1D7iGCQpNAJimnJYfHpEoA59buw2Az7nKPKTIiJRNQBJNtfcPHB9Cw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal