Abstract
Enucleation, a rare feature of mammalian differentiation, occurs in 3 cell types: erythroblasts, lens epithelium, and keratinocytes. Previous investigations suggest that caspase activation functions in lens epithelial and keratinocyte enucleation, as well as in early erythropoiesis encompassing erythroid burst-forming unit (BFU-E) differentiation to proerythroblast. To determine whether caspase activation contributes to later erythropoiesis and whether nuclear substructures other than chromatin reorganize, we analyzed distributions of nuclear subcompartment proteins and assayed for caspase-induced cleavage of subcompartmental target proteins in mouse erythroblasts. We found that patterns of lamin B in the filamentous network interacting with both the nuclear envelope and DNA, nuclear matrix protein NuMA (Nuclear mitotic apparatus), and splicing factors Sm and SC35 persisted during nuclear condensation, consistent with effective transcription of genes expressed late in differentiation. Thus, nuclear reorganization prior to enucleation is selective, allowing maintenance of critical transcriptional processes independent of extensive chromosomal reorganization. Consistent with these data, we found no evidence for caspase-induced cleavage of major nuclear subcompartment proteins during late erythropoiesis, in contrast to what has been observed in early erythropoiesis and in lens epithelial and keratinocyte differentiation. These findings imply that nuclear condensation and extrusion during terminal erythroid differentiation involve novel mechanisms that do not entail major activation of apoptotic machinery. (Blood. 2005;106:2200-2205)
Introduction
Enucleation is an extremely rare feature in mammalian tissue differentiation with only 3 cell types, erythroblasts, lens epithelial cells, and keratinocytes, undergoing this process. Prior to erythroblast enucleation, nuclear chromatin condenses, and the nucleus reduces in size and moves adjacent to the plasma membrane.1-11 Accompanying alterations in chromatin higher-order folding and nuclear shrinkage, DNA replication, and RNA transcription are largely inactivated. Despite these marked perturbations select nuclear processes remain robust, such as transcription of genes encoding α and β globins,12 structural protein 4.1R,13-15 B-cell leukemia XL (Bcl-XL),16 embryonic epithelial gene 1 proteins (EEG-1 proteins),17 and p21.18 We therefore speculated that to protect the nuclear microenvironment and enable functional transcription and nucleoplasmic transit, some nuclear domains must remain structurally intact.
The eukaryotic nucleus is demarcated from cytoplasm by a double membrane that is fused at nuclear pores, elaborate membrane machines composed of approximately 50 proteins which function to regulate trafficking of proteins and RNA between the cytoplasm and the nucleoplasm. The inner layer of the nuclear membrane is lined with fibrous lamin proteins, forming a 2-dimensional network capable of binding DNA during interphase but rapidly disassembling early in cell division. Within the nucleus, DNA replication, RNA transcription, and ribosomal processes are spatially and temporally regulated. Factors involved in replication, transcription, and RNA splicing are concentrated at unique functional subcompartments identifiable by immunofluorescence.
In probing mechanisms for enucleation, 2 studies suggested that caspase activation plays a role in both lens epithelial19 and keratinocyte20 enucleation. Caspases, a family of cysteine proteases, are critical for programmed cell death (as reviewed in Budihardjo et al21 and Fadeel et al22 ). During in vivo rodent lens epithelial-cell differentiation cleavage of poly-(ADP [adenosine diphosphate]-ribose) polymerase (PARP), a well-recognized substrate for caspase-3 during apoptosis, suggested that either caspase-3 or a related caspase was activated preceding enucleation.19 In addition, a pan-caspase inhibitor, z-Val-Ala-DL-Asp-fluoromethyl-ketone (zVAD-fmk), inhibited both PARP cleavage and enucleation in lens cells differentiating in culture. Similarly, caspase inhibitors blocked nuclear extrusion during keratinocyte differentiation in culture, and a cleaved activated form of caspase-3 was detected by Western blot analysis.20 These findings raise the intriguing possibility that in lens epithelium and keratinocytes some of the biochemical machinery of apoptosis may participate in normal terminal differentiation. Several, more recent studies suggest that caspase activation may also function in erythroid differentiation. One investigation of CD34+CD36+ human hematopoietic progenitor cells reported that in the presence of pan caspase inhibitors, cells did not enucleate and, in fact, did not differentiate beyond the blast/basophilic stage.23 A second study showed caspase-3 activation in differentiating human erythroblast populations expressing erythroid burst-forming unit (BFU-E), erythroid colony-forming unit (CFU-E), and proerythroblast surface markers.24 Treatment with caspase-3 small interfering RNA (siRNA) blocked cells at the proerythroblast stage. Considered together, the data on erythroid differentiation appear to indicate that caspase activation plays a role in the stages of erythropoiesis encompassing BFU-E differentiation to proerythroblast.
In light of these observations, we wanted to determine the potential contribution of caspase activation during the proerythroblast to enucleation phase of erythropoiesis. An in vitro model system of terminal differentiation exists that is appropriately suited to address this question: erythroid-cell populations that are predominantly proerythroblasts (0% BFU-E, 10%-20% CFU-E) can be obtained from spleens of mice infected with anemia-inducing strain of Friend erythroleukemia virus (FVA cells).12,25 When cultured with erythropoietin over 43 hours, these proerythroblasts proliferate and differentiate to orthochromatic normoblasts, of which the majority enucleate.26 This well-studied model system of terminal erythroid differentiation closely mimics erythropoiesis in vivo.
To investigate whether nuclear substructures, other than chromatin, reorganize extensively late in erythropoiesis and whether caspase activation plays a role in preparing nuclei for extrusion, we performed studies in erythroid progenitors analyzing selected, nuclear subcompartment protein distributions and assaying for caspase-induced cleavage of subcompartmental target proteins. We found that despite marked chromatin condensation, there were few major changes in distribution of selected nuclear structural proteins and splicing factors, consistent with on-going effective transcription of genes expressed late in differentiation. Consistent with these data, we found no evidence for caspase-induced cleavage of target proteins within these nuclear subcompartments during late-stage erythropoiesis, in contrast to what has been reported in early-stage erythropoiesis and in differentiation of lens epithelium and keratinocytes. These findings imply that nuclear condensation and extrusion during late-stage terminal erythroid differentiation involve novel mechanisms that do not entail major activation of apoptotic machinery.
Materials and methods
Reagents
Anti-Lamin B clone M-20 (Santa Cruz Biotechnology, Santa Cruz, CA); anti-NuMA (Nuclear mitotic apparatus) clone 204-41 (Oncogene Research Products, Boston, MA); anti-nuclear pore 62 (Nup62) clone Mab414 (Babco, Richmond, CA); anti-nuclear pore 153 (Nup53), anti-nuclear pore 358 (Nup358), and anti-tetratricopeptide repeat (Tpr; gifts from Dr Volker Cordes, Zentrum für Molekulare Biologie Heidelberg [ZMBH], Heidelberg, Germany); anti-splicing factor SC35 clone SC35 (Sigma, Saint Louis, MO); anti-Smith antigen (SM) clone Y12 (Neomarkers, Fremont, CA). Secondary antibody reagents coupled to horseradish peroxidase (HRP), fluorescein isothiocyanate (FITC), or Texas Red were obtained from Jackson Immuno Research, Westgrove, PA. Phycoerythrin (PE)-conjugated Ter119 was obtained from Ebiosciences, San Diego, CA.
Mouse erythroblasts
Erythroblasts were isolated and cultured from mice infected with anemia-inducing strain of Friend erythroleukemia virus (FVA cells) using methods established by Koury et al.12,25 Briefly, CD2F1 mice were infected with 1 × 104 spleen focus-forming units. Cells were harvested from spleens 2 weeks later and separated by velocity sedimentation at unit gravity (pooled cells sedimenting at approximately 6 mm/h or greater). Cell separation yielded approximately 5 × 108 cells per spleen, which were predominantly proerythroblasts (0% BFU-E, 10%-20% CFU-E). When cultured with 1.0 U/mL recombinant erythropoietin over 43 hours, proerythroblasts proliferated and differentiated to yield approximately 2 × 109 late-stage erythroblasts per spleen, with about two thirds undergoing enucleation.
Erythroblasts from native bone marrow were also studied. For these experiments bone marrow cells were flushed from mouse femurs and tibiae with a 25-gauge needle into Iscoves modified Dulbecco medium (IMDM; Gibco BRL Products, Rockville, MD) and then washed by centrifugation in IMDM. For immunofluorescence microscopy, erythroblasts were identified by staining with erythroid-specific antibody TER119 against glycophorin A.27
Western blotting
FVA cells were solubilized in sample buffer containing 3% sodium dodecyl sulfate (SDS) and 10% dithiothreitol and then boiled for 10 minutes. Lysate from 1 million FVA cells were loaded per lane and separated on a 4% to 12% SDS-polyacrylamide gel (Biorad, Hercules, CA). Proteins were then transferred by semidry blotting onto nitrocellulose membrane (Biorad) or polyvinylidene difluoride (PVDF) membrane (Perkin-Elmer Life Sciences, Boston, MA) for 1 hour at 8 mA/cm2. The membrane was blocked for 1 hour in phosphate-buffered saline (PBS) containing 5% nonfat dry milk and 0.1% Tween-20, then probed for 1 hour with primary antibody diluted in PBS and 0.1% Tween-20. After several washes, blots were incubated with secondary antibody coupled to HRP diluted in PBS and 0.1% Tween-20, washed, and developed on X-Omat Blue film (Perkin-Elmer Life Sciences) using the Renaissance chemiluminescence detection kit (Perkin-Elmer Life Sciences).
Immunofluorescence microscopy
Bone marrow erythroblasts or FVA cells were washed 3 times by centrifugation in PBS or IMDM (Gibco BRL Products), respectively, and allowed to adhere for 1 hour to slides coated with Cell-Tak (BD Pharmingen, San Diego, CA). The cells were washed 3 times in PBS; fixed with 4% paraformaldehyde, 0.2% Triton X-100 in PBS at room temperature for 10 minutes; further permeablized by 1% Triton X-100 in PBS for 3 minutes; and then rinsed with PBS. After incubation in 10% donkey serum (Jackson Immuno Research) and 10 mg/mL bovine serum albumin (BSA; Sigma) in PBS for 1 hour to block nonspecific protein binding, fixed cells were treated with primary antibodies diluted in 10 mg/mL BSA/PBS for 1 hour at room temperature. Cells were washed for 5-minute intervals 6 times with gentle shaking, incubated for 1 hour with secondary antibodies at 1:100 in 10 mg/mL BSA/PBS, washed 3 times in PBS; then mounted using Vectashield with DAPI (4′, 6-diamino-2-phenylindole) (Vector Laboratories, Burlingame, CA). Fluorescence was imaged using a Zeiss Axiovert 135 microscope with a 63×/1.25 oil immersion objective or a Nikon TE2000U microscope with a 60×/1.4 oil immersion objective, equipped with a CCD camera. Images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA) and Image Pro (Microsoft, Redman, WA).
Measurement of nuclear surface area and volume
DNA in early (0 hour) and late (43 hours) FVA erythroblasts and in early- and late-stage erythroblasts from freshly harvested mouse bone marrow were stained with Hoescht 33342 DNA stain (Sigma) at 100 μg/mL for 10 minutes. Live cells were visualized with a Nikon TE2000U microscope using a 60×/1.4 oil immersion objective. Measurements of nuclear diameter were determined with a micrometer and used in calculating surface area and volume.
Results
Lamin remains intact during terminal differentiation
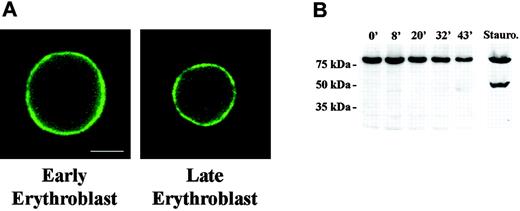
Lamin proteins form a filamentous network underlying the inner layer of the nuclear envelope. This lamin network interacts with both the nuclear envelope and DNA during interphase, thus linking chromatin and the nuclear membrane. It was therefore pertinent to determine whether lamins reorganize during the period of increasing chromatin condensation and decreasing nuclear size. For this analysis we studied lamin B distribution in murine erythroblasts from freshly obtained bone marrow. Using triple label fluorescence microscopy we compared the staining pattern of large early erythroblasts to that of small late erythroblasts with condensed chromatin. Bone marrow cells were paraformaldehyde-fixed and stained with anti-lamin B and DAPI to image DNA. To distinguish erythroblasts from other hematopoietic cells, TER119 was used to specifically label cells expressing glycophorin A.27 We observed that large, early erythroblasts exhibited strong continuous staining around the rim of the nucleus (Figure 1A). This pattern was unchanged in small late erythroblasts, strongly suggesting that lamin B is not reorganized late in erythropoiesis.
Distribution and Western blot analysis of Lamin B in erythroid progenitors. (A) Fluorescence micrograph of early and late mouse erythroblasts from freshly harvested bone marrow labeled with antilamin. The pattern of rim staining was similar in early and late erythroblasts. Bar is 2.5 μm. (B) Western blot analysis of FVA-cell lysates obtained from cells cultured from 0 hour to 43 hours. Gel lanes were loaded with equivalent cell numbers and blot probed with anti-lamin B. Immunoreactive bands of lamin B were observed at 69 kDa; no apoptotic products were detected. A staurosporine-treated sample of FVA cells, serving as positive control for apoptosis, showed the classic apoptotic 49-kDa lamin B cleavage product.
Distribution and Western blot analysis of Lamin B in erythroid progenitors. (A) Fluorescence micrograph of early and late mouse erythroblasts from freshly harvested bone marrow labeled with antilamin. The pattern of rim staining was similar in early and late erythroblasts. Bar is 2.5 μm. (B) Western blot analysis of FVA-cell lysates obtained from cells cultured from 0 hour to 43 hours. Gel lanes were loaded with equivalent cell numbers and blot probed with anti-lamin B. Immunoreactive bands of lamin B were observed at 69 kDa; no apoptotic products were detected. A staurosporine-treated sample of FVA cells, serving as positive control for apoptosis, showed the classic apoptotic 49-kDa lamin B cleavage product.
Lamin B undergoes caspase 3-induced cleavage early in apoptosis.28 As such, it is a relevant indicator of activation of apoptotic pathways. To determine whether lamin B is cleaved during erythropoiesis we performed Western blot analysis on FVA cells over a 43-hour time course of terminal differentiation. We used the FVA model system of erythroid differentiation because it faithfully mimics in vivo development and provides adequate numbers of cells at specific developmental stages for study. Proteins from an equivalent number of erythroblasts obtained at 5 time points, from proerythroblast (0 hour) to enucleating late-stage erythroblast (43 hours), were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and Western blots were probed with antibody to lamin B. Immunoreactive bands migrating at approximately 69 kDa were observed at each time point of the FVA time course, and no degradation products were detected (Figure 1B). To determine whether one reason for the apparent decreased intensity of the 69-kDa band during the 43-hour time course was related to a change in nuclear volume and surface area, these parameters were calculated from measurements of nuclear diameter of 0-hour and 43-hour live FVA cells labeled with Hoechst DNA stain. Surface area and volume of 0-hour cells were 205.4 μm2 and 276.8 μm3, respectively, while surface area and volume of 43-hour cells were 91.6 μm2 and 82.5 μm3, respectively. Hence, surface area and volume decreased 2- to 3-fold between 0 and 43 hours. These data are consistent with electron microscopic measurements showing that approximately 2-fold decrease in nuclear surface area occurred in FVA cells between 2 and 45 hours of culture.29 We also measured nuclear diameters of erythroblasts freshly harvested from mouse bone marrow and determined that, as with differentiating FVA cells, surface area and volume decreased 2- to 3-fold between early- and late-stage erythroblasts. Thus, the decrease in nuclear surface area could account for the observed decrease in lamin B on a per erythroblast basis, as detected by Western blot.
As a positive control for caspase-specific degradation, we included an FVA sample exposed to 0.25 mM staurosporine, which activates apoptotic cascades and generates a 49-kDa product. The classic lamin B caspase cleavage product was present in the staurosporine-treated sample but was not evident in any of the untreated samples during the entire time course of erythroid differentiation (Figure 1B). Taken together, these data clearly show that lamin B is neither relocalized nor degraded by an apoptotic or nonapoptotic mechanism during terminal differentiation of FVA cells, supporting the conclusion that a signature event in the classic apoptotic pathway is not operative prior to enucleation.
Nuclear pore proteins redistribute in late bone marrow erythroblasts
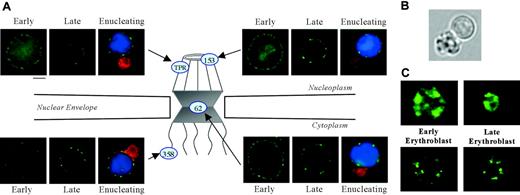
Nuclear pores, complex multiprotein channels that regulate RNA and protein movement between nucleoplasm and cytoplasm, are composed of 3 major regions: a nucleoplasmic basket, a central transporter region, and cytoplasmic fibrils (Figure 2A). To analyze these critical nuclear membrane substructures during terminal differentiation, we studied the distribution of nuclear pore protein, Nup62, localized within the central transporter region. Using triple label fluorescence microscopy we compared the staining pattern of large early erythroblasts to that of small late erythroblasts with condensed chromatin in cells from freshly obtained mouse bone marrow. Paraformaldehyde-fixed cells were stained with a monoclonal antibody Mab414, which detects Nup62,30 erythroid specific antibody TER119, and DAPI. We observed that early erythroblasts exhibited strong punctate Nup62 staining around the nuclear rim. In marked contrast, small late erythroblasts with condensed DNA, as well as enucleating erythroblasts, had an irregular, clustered pattern (Figure 2A). In enucleating cells, Nup62 segregated to the extruding nucleus and not to the nascent reticulocyte.
To explore whether proteins in other regions of the pore structure also relocalized during erythropoiesis, we probed erythroblasts from bone marrow with antibodies specific for 2 proteins in the nucleoplasmic basket, Nup153 and Tpr, and 1 protein in the cytoplasmic fibrils, Nup358. Interestingly, Nup153, Tpr, and Nup358 each exhibited a change in staining pattern in late erythroblasts and enucleating erythroblasts similar to Nup62 (Figure 2A). These findings clearly show that nuclear pore proteins from all regions of the pore redistribute at the nuclear periphery in late bone marrow erythroblasts.
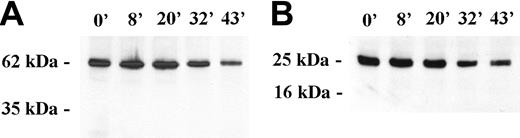
To investigate whether the redistribution of nuclear pore components was indicative of protein degradation we analyzed Western blots of FVA cells over a 43-hour time course of terminal differentiation. Proteins from an equivalent number of erythroblasts from each time point were separated by SDS-PAGE, and the blots were probed with Mab414 to detect Nup62. An immunoreactive band migrating at approximately 62 kDa was observed at each time point, and no degradation products were detected (Figure 3A). We conclude that Nup62 is not detectably degraded during terminal differentiation of FVA cells and, therefore, that the changes in pore pattern observed by immunofluorescence appear to reflect redistribution of intact proteins.
Immunofluorescent localization of various nuclear pore and spliceosome proteins in erythroid progenitors. (A) Diagram illustrating localization within nuclear pore structure of proteins probed with specific fluorescent antibodies: Nup153 and Tpr (nucleoplasmic basket), Nup62 (central transporter), and Nup358 (cytoplasmic fibrils). Fluorescence micrographs show early and late mouse erythroblasts from freshly harvested bone marrow labeled with antibody to Nup153, Tpr, Nup62, or Nup358 (green). Micrographs of enucleating erythroblasts show cells stained for erythroid-specific marker Glycophorin A (GPA) (Ter119, red), nuclear pore (green), and DNA (Hoechst, blue). Nuclear pore proteins exhibit a more uniform rim fluorescence in early erythroblasts, but then they redistribute to form an irregular punctuate pattern in late and enucleating erythroblasts. Bar is 2.5 μm. (B) Bright-field micrograph of a live enucleating erythroblast, freshly harvested from mouse bone marrow, that was identified by positive staining for erythroid-specific marker GPA and DNA. In the paraformaldehyde-fixed antibody and Hoechst-stained enucleating cells shown in panel A, reticulocytes are smaller than extruding nuclei because of a decrease in reticulocyte volume secondary to fixation. In the live-cell image, the nascent multilobulated reticulocyte and extruding nucleus appear similar in size. Because the reticulocyte is multilobulated, a visual comparison in size to the spherical nucleus can be misleading. In an earlier report we determined the surface area of multilobular young reticulocytes and extruding nuclei to be 83 μm2 and 60 μm2, respectively.45 (C) Micrographs of early and late erythroblasts probed with anti-Sm (top panels; green) and anti-SC35 (bottom panels; green) show that these spliceosome components maintain their irregular nucleoplasmic cluster patterns in late-stage erythroblasts. Bar is 2.5 μm.
Immunofluorescent localization of various nuclear pore and spliceosome proteins in erythroid progenitors. (A) Diagram illustrating localization within nuclear pore structure of proteins probed with specific fluorescent antibodies: Nup153 and Tpr (nucleoplasmic basket), Nup62 (central transporter), and Nup358 (cytoplasmic fibrils). Fluorescence micrographs show early and late mouse erythroblasts from freshly harvested bone marrow labeled with antibody to Nup153, Tpr, Nup62, or Nup358 (green). Micrographs of enucleating erythroblasts show cells stained for erythroid-specific marker Glycophorin A (GPA) (Ter119, red), nuclear pore (green), and DNA (Hoechst, blue). Nuclear pore proteins exhibit a more uniform rim fluorescence in early erythroblasts, but then they redistribute to form an irregular punctuate pattern in late and enucleating erythroblasts. Bar is 2.5 μm. (B) Bright-field micrograph of a live enucleating erythroblast, freshly harvested from mouse bone marrow, that was identified by positive staining for erythroid-specific marker GPA and DNA. In the paraformaldehyde-fixed antibody and Hoechst-stained enucleating cells shown in panel A, reticulocytes are smaller than extruding nuclei because of a decrease in reticulocyte volume secondary to fixation. In the live-cell image, the nascent multilobulated reticulocyte and extruding nucleus appear similar in size. Because the reticulocyte is multilobulated, a visual comparison in size to the spherical nucleus can be misleading. In an earlier report we determined the surface area of multilobular young reticulocytes and extruding nuclei to be 83 μm2 and 60 μm2, respectively.45 (C) Micrographs of early and late erythroblasts probed with anti-Sm (top panels; green) and anti-SC35 (bottom panels; green) show that these spliceosome components maintain their irregular nucleoplasmic cluster patterns in late-stage erythroblasts. Bar is 2.5 μm.
Western blot analysis of Nup62 and Sm proteins during erythropoiesis. Western blot analysis of FVA-cell lysates obtained from cells cultured from 0 hour to 43 hours. Gel lanes were loaded with equivalent cell numbers and blot probed with (A) mAb414 against Nup62 and (B) anti-Sm. Immunoreactive bands of Nup62 and Sm were observed at 62 kDa and 29 kDa, respectively; no apoptotic cleavage products were detected.
Western blot analysis of Nup62 and Sm proteins during erythropoiesis. Western blot analysis of FVA-cell lysates obtained from cells cultured from 0 hour to 43 hours. Gel lanes were loaded with equivalent cell numbers and blot probed with (A) mAb414 against Nup62 and (B) anti-Sm. Immunoreactive bands of Nup62 and Sm were observed at 62 kDa and 29 kDa, respectively; no apoptotic cleavage products were detected.
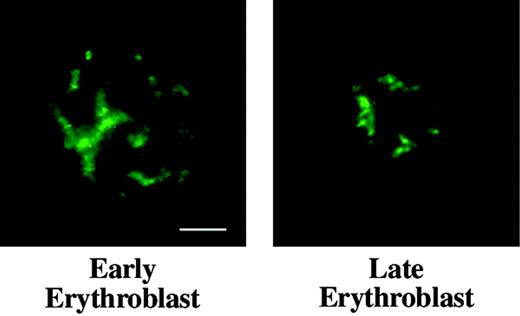
Distribution of nuclear matrix protein NuMA in erythroid progenitors. Early and late mouse erythroblasts from freshly harvested bone marrow labeled with anti-NuMA (green) show similar staining patterns, demonstrating that NuMA does not undergo detectable redistribution during later phases of terminal differentiation. Bar is 2.5 μm.
Distribution of nuclear matrix protein NuMA in erythroid progenitors. Early and late mouse erythroblasts from freshly harvested bone marrow labeled with anti-NuMA (green) show similar staining patterns, demonstrating that NuMA does not undergo detectable redistribution during later phases of terminal differentiation. Bar is 2.5 μm.
Splicing machinery does not undergo major rearrangement during terminal differentiation
Our earlier investigations14,15 and those of others13 convincingly show that protein 4.1R pre-RNA splicing continues to occur in late erythroblasts. These data argue strongly that at least a portion of RNA splicing machinery maintains function in late-stage nuclei despite marked chromatin condensation. To explore whether spliceosomes rearrange within late stage, largely heterochromatic nuclei, we probed bone marrow erythroblasts with anti-Sm, specific for spliceosome subunits of small ribonucleoproteins (snRNPs) U1, U2, U4/6, and U5. Both large early erythroblasts and small late-stage erythroblasts exhibited similar staining patterns of irregular nucleoplasmic clusters (Figure 2C). We also examined the distribution of the non-snRNP splicing protein, SC35. As observed with anti-Sm, large early erythroblasts and late-stage small erythroblasts probed with anti-SC35 exhibited similar staining patterns of irregular nucleoplasmic clusters (Figure 2C). The data obtained with the 2 spliceosome-specific probes strongly suggest that splicing machinery is retained late in erythropoiesis, despite marked chromatin condensation. Furthermore, by Western blot analysis, Sm protein remained intact throughout the 43-hour time course of FVA-cell terminal differentiation (Figure 3B).
Nuclear matrix protein, NuMA, does not redistribute in late erythropoiesis
NuMA is a component of nuclear matrix or skeleton, an insoluble nonchromatin network intimately involved during interphase in regulation of DNA replication and RNA transcription. NuMA appears to play an important architectural role in nuclei of most cells and silencing of NuMA by RNAi results in apoptosis in HeLa cells.31 As a probe for the presence of a nuclear matrix component during terminal differentiation, we compared the distribution of NuMA in interphasic early- and late-stage bone marrow erythroblasts. By immunofluorescence microscopy the staining pattern of NuMA did not differ markedly in early and late cells (Figure 4), suggesting that a NuMA-containing nuclear skeleton survives chromatin condensation and nuclear shrinkage during erythroid differentiation.
Discussion
Intriguing prior studies presented evidence that caspase activation plays a role in enucleation of lens epithelia and keratinocytes, the only other mammalian cell types besides erythroblasts that expel nuclei during differentiation. These findings stimulated us to investigate whether caspase activation is instrumental in preparing erythroblast nuclei for extrusion. We therefore performed studies in erythroid progenitors from the proerythroblast to enucleation phase of erythropoiesis looking for evidence of caspase-induced cleavage of specific nuclear target proteins. A major finding of the current investigation was that there was no evidence of widespread activation of classic apoptotic machinery during the later stages of terminal erythroid differentiation. Immunochemical analysis of lamin B, Nup62, and Sm clearly showed that these nuclear target proteins did not undergo caspase-specific cleavage. Hence, in 3 distinct nuclear substructures, fibrous lamina, nuclear pores, and spliceosomes, no caspase-dependent cleavage was detected.
Interestingly, these findings contrast with observations of caspase activation in an earlier phase of erythropoiesis. Carlile et al24 detected the active form of caspase-3 in cultures of normal human CD34+ progenitor cells from days 0 to 8. At day 0 these cells were predominantly BFU-E by surface marker expression. By day 8 the BFU-E marker was expressed on 45% of cells, the CFU-E marker on 75%, and 45% of cells expressed GPA, indicative of proerythroblasts. Transfection of siRNA directed to caspase-3 blocked cells at the proerythroblast stage, suggesting that caspase-3 functions in early erythroid development. Another laboratory reported that CD34+CD36+ human progenitor cells treated with caspase inhibitors did not differentiate beyond the blast/basophilic stage and did not enucleate.23 Unfortunately, the concentration of caspase inhibitors used by those investigators resulted in an approximate 86% loss in cell number, leaving for further study an uncharacterized residual blast/basophilic-cell population. Although these nondifferentiating residual cells were most likely members of the erythroid lineage, they were not definitely identified as such; hence, the conclusion regarding the importance of caspase activation in early erythropoiesis is not as strongly supported as in the report of Carlile et al.24 However, considered together, these 2 studies suggest that caspase activation plays a role in the stages of erythropoiesis encompassing BFU-E differentiation to proerythroblast.
Our conclusion that caspase activation is not instrumental in the later stages of terminal erythroid differentiation is bolstered by a number of other investigations. Knockouts of proapoptotic proteins do not support a role for caspase activation in enucleation: phenotypic abnormalities in erythropoiesis or red cells were not observed in knockouts of caspases-1, -2, -3, -9 (as reviewed in Wang and Lenardo32 ), although subtle pathology may have been missed. Caspase-8 null mice do have abnormal hematopoiesis, but the effect is seen much earlier than the final stages of erythropoiesis.33 It has also been reported that a critical function of erythropoietin and its receptor is to protect erythroblasts from apoptosis.34,35 Proapoptotic caspases as well as antiapoptotic B-cell lymphoma-2 (Bcl-2) family members, including Bcl-XL, have been characterized in committed erythroid progenitors.16,36 In the presence of erythropoietin, antiapoptotic Bcl-XL expression markedly increases at the end of terminal differentiation16,37 and may, in fact, be critical in preventing apoptosis38 in cooperation with transcription factor GATA-1.39
In contrast, erythropoietin deprivation activates caspases resulting in apoptosis.36 Investigations of Stat5a-/-5b-/- mice suggest that Stat5 (signal transducer and transcriptional activator 5) may function in the antiapoptotic signaling erythropoietin receptor (EpoR) by mediating the induction of Bcl-XL.40 Immature erythroblasts express death receptors CD95, Fas, and tumor necrosis factor (TNF) and exposure of erythroid progenitors to death receptor ligands appears to negatively regulate erythropoiesis by inducing caspase-mediated cleavage of GATA-1.41
Another striking finding of our current studies was that, despite the marked decrease in nuclear size and chromatin condensation occurring prior to enucleation, several important nuclear subcompartments appear to remain intact when analyzed by immunofluorescent microscopy. Lamin B, a filamentous protein linking the inner layer of the nuclear envelope to DNA, exhibited continuous nuclear rim staining in both early and late freshly harvested mouse erythroblasts. This may be particularly significant because transcription continues up until erythroblast enucleation, and lamins have been proposed to be involved in mRNA synthesis. Disruption of lamin organization inhibits RNA polymerase II (pol II) transcription and alters the distribution of TATA-binding protein (TBP) transcription factor.42 Moreover, we note that in Fas-induced apoptosis in HeLa cells, lamin B shows a folded staining pattern,43 markedly different from the smooth staining of late erythroblasts.
Further, NuMA, a component of the nuclear matrix structural network, was distributed throughout multiple areas of nuclei in both early and late erythroblasts. This is in contrast to reorganization of NuMA into large intranuclear foci during differentiation of breast cancer cells into acini when grown in 3D cultures.44 Finally, snRNPs and SC35 components of spliceosomes both exhibited similar distributions throughout late-stage differentiation. These observations are consistent with maintenance of a suitable nuclear microenvironment that could enable selected genes, such as α and β globin and protein 4.1R, to continue to be transcribed despite major replicational and transcriptional shut down during terminal erythroid differentiation.
In contrast to our data on components residing within nuclei, we did observe differentiation stage-specific rearrangement of 4 proteins within the nuclear pore complex. Two nuclear pore proteins in the nucleoplasmic basket (Nup153 and Tpr), 1 component in the central transporter region (Nup62), and 1 Nup in the cytoplasmic fibrils (Nup358) exhibited punctate nuclear rim staining in early erythroblasts but an irregular, clustered pattern in late stage and enucleating erythroblasts. Because there was no evidence of apoptotic or nonspecific cleavage of Nup62 and 4 distinct Nups gave similar relocalization patterns, we propose that nuclear pores remain intact but redistribute late in differentiation. Retention of functional, albeit redistributed, nuclear pores is consistent with other evidence clearly showing that certain specific RNAs, such as protein 4.1R and α and β globin, continue to be effectively translated during reticulocyte maturation. This would suggest that these RNAs are successfully trafficked through functional pores late in differentiation, prior to enucleation.
Although marked chromatin condensation and decreased nuclear size have long been recognized as hallmarks of the final stages of mammalian erythropoiesis, little is known about whether perturbations in nuclear subcompartments accompany these dramatic changes. The current investigations provide clear evidence that nuclear subcompartments containing lamin B, NuMA, Sm, and SC35 persist during nuclear condensation and decreasing nuclear volume. Thus, nuclear reorganization of erythroblasts prior to enucleation is selective, allowing maintenance of critical transcriptional processes independent of gross chromosomal reorganization and cessation of DNA replication and cell cycling. DNA isolated from extruded nuclei of FVA cells cultured for 42 hours migrates as intact, high-molecular-weight DNA.34 Whether replication and transcription processes that have been largely shut down in terminal nuclei could be reactivated, similar to what has been demonstrated for avian nuclei in mature nucleated avian erythrocytes, is a future area to be investigated.
The current investigations also provide clear evidence that apoptotic biochemical machinery is not widely activated during the later stages of terminal erythroid differentiation. Hence, activated caspase function in committed differentiating erythroblasts appears to be limited to the early stage of development, where it may play a role in maintaining an adequate pool of CFU-Es for acute stress erythropoiesis. Erythroblast enucleation also occurs without evidence of major activation of apoptotic proteins, in contrast to what has been reported for the only other 2 cell types that undergo enucleation during their normal differentiation: lens epithelial cells and keratinocytes. Hence, erythroblasts appear to be unique among enucleating mammalian cells in their late terminal differentiation program.
Prepublished online as Blood First Edition Paper, June 2, 2005; DOI 10.1182/blood-2005-04-1357.
Supported in part by the National Institutes of Health (grants DK32094, DK56267, DK059079, and HL79565), by a Merit Review Award from the Department of Veterans Affairs, and by the Office of Health and Environment Research Division, US Department of Energy (contract DE-AC03-76SF00098).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Mr Prapaporn Kopsombut (VA Tennessee Valley Healthcare System, Nashville, TN) for help in the culture and processing of FVA cells and Dr Volker Cordes (ZMBH, Heidelberg, Germany) for his kind gift of antibodies directed against nuclear pore proteins.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal