Abstract
Hereditary hemochromatosis (HH), an iron overload disease associated with mutations in the HFE gene, is characterized by increased intestinal iron absorption and consequent deposition of excess iron, primarily in the liver. Patients with HH and Hfe-deficient (Hfe-/-) mice manifest inappropriate expression of the iron absorption regulator hepcidin, a peptide hormone produced by the liver in response to iron loading. In this study, we investigated the contribution of Hfe expression in macrophages to the regulation of liver hepcidin levels and iron loading. We used bone marrow transplantation to generate wild-type (wt) and Hfe-/- mice chimeric for macrophage Hfe gene expression. Reconstitution of Hfe-deficient mice with wt bone marrow resulted in augmented capacity of the spleen to store iron and in significantly decreased liver iron loading, accompanied by a significant increase of hepatic hepcidin mRNA levels. Conversely, wt mice reconstituted with Hfe-deficient bone marrow had a diminished capacity to store iron in the spleen but no significant alterations of liver iron stores or hepcidin mRNA levels. Our results suggest that macrophage Hfe participates in the regulation of splenic and liver iron concentrations and liver hepcidin expression. (Blood. 2005;106:2189-2195)
Introduction
Hereditary hemochromatosis (HH) type 1, an autosomal recessive disease of iron overload, is one of the most common inherited disorders. It is characterized by failure in the regulation of duodenal iron absorption, leading to iron overloading that can eventually impair organ systems and cause cirrhosis, diabetes, and cardiomyopathy.1
HH is caused by mutations in the HFE gene encoding a major histocompatibility complex (MHC) class 1-like protein that requires β2-microglobulin (B2m) for cell surface expression.2 A link between HFE and cellular iron metabolism is suggested by the observation that wild-type (wt) HFE-β2m molecules form a stable complex with transferrin receptor 1 (TfR1).3
Most patients with HH are homozygous for a missense mutation in the HFE gene that results in cysteine-to-tyrosine substitution at amino acid 282 of HFE protein (C282Y).2 The mutation disrupts a critical disulfide bond in the α3 domain of HFE protein and abrogates binding of the mutant HFE protein to B2m, leading to impaired HFE protein intracellular trafficking, incorporation into the cell membrane, and association with TfR1.3
Studies in mice with targeted inactivation of the Hfe or B2m gene have confirmed the critical role of HFE-B2m complexes in iron metabolism.4-6 As do humans with HH, Hfe-deficient mice develop iron overloading with an accumulation of excess iron, primarily in liver parenchymal cells, as opposed to reticuloendothelial (RE) cell storage characteristic of secondary iron overload. Thus, abnormal regulation of iron metabolism in RE cells seems to take place in HH because these cells are relatively iron deficient compared with surrounding parenchymal hepatocytes.7 Importantly, RE cells—monocytes and tissue macrophages—have a central role in regulating iron homeostasis because they recognize and phagocytose senescent and damaged erythrocytes, process iron from heme, and return it to the circulation for reuse by red cell precursors during erythropoiesis.8
In a previous study, we showed that the reconstitution of B2m-/- mice with normal hematopoietic cells leads to cellular redistribution of iron stored in the liver,6 suggesting an important role of Hfe expression in hematopoietic-derived cells in iron homeostasis. This was based on a histologic investigation into iron distribution in the liver. In this model, attempts to reconstitute wt-type mice with B2m-/- hematopoietic cells failed because of rejection of the transplanted MHC class 1-negative cells by host natural killer (NK) cells.9
There is now considerable evidence that Hfe-deficiency results in inappropriate expression of the putative iron regulator hepcidin in animal models10,11 and in patients with HH.12 Hepcidin, the product of the Hamp gene, is a β-defensin-like antimicrobial peptide13 whose liver expression is modulated by iron stores, anemia/hypoxia, and inflammation.14,15 Further convincing evidence of an essential role for hepcidin in iron metabolism and in the regulation of intestinal iron uptake is provided by findings that hepcidin knockout mice develop severe iron overload,16 whereas, conversely, transgenic mice overexpressing hepcidin incur severe anemia.17
In the present study, we investigated the functional role of Hfe in macrophages and the contribution of this cell type to the regulation of liver hepcidin levels and iron loading by using bone marrow transplantation (BMT). We used the Hfe-/- mouse model for BMT to generate mice expressing Hfe in RE macrophages and mice with selected inactivation of Hfe in RE macrophages.
Materials and methods
Animals
All procedures were performed in accordance with Canadian Council on Animal Care guidelines after approval by the Institutional Animal Care Committee of the Centre hospitalier de l'Université de Montréal (CHUM). Hfe-/- mice were kindly provided by Dr Nancy C. Andrews (Howard Hughes Medical Institute and Harvard Medical School, Children's Hospital, Boston, MA).5 The Hfe-/- and wt mice in these experiments, females from the 129/SvEvTac background, were permanently housed under specific pathogen-free conditions. Green fluorescence protein (GFP) transgenic mice (GFP+wt) expressing GFP from the chicken Actb promoter (strain Tg[Actb-EGFP]) were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA) and were backcrossed 5 times with 129/SvEvTac mice in our animal facility. GFP+Hfe-/- transgenic mice were obtained by crossing Hfe-/- mice with GFP+wt mice.
Cell preparations
Spleens were mashed through a 40-μm nylon cell strainer, and erythrocytes in splenocyte suspensions were lysed with Puregene RBC Lysis Solution (Gentra Systems, Minneapolis, MN). Livers were perfused with liver digest media (Gibco/BRL Life Technologies, Burlington, ON, Canada), and the resultant suspension was centrifuged at 70g for 5 minutes to separate hepatocytes (pellet) and nonparenchymal cells (NPCs; supernatant). Mononuclear cells (MNCs) were obtained by centrifugation of NPC suspensions on Lympholyte-M (Cedarlane, Hornby, ON, Canada). B lymphocytes, T lymphocytes, NK cells, and macrophages were isolated from splenocyte and NPC suspensions by positive magnetic cell separation, with EasySep kits (Stem Cell Technologies, Vancouver, BC, Canada). The following antibodies (PharMingen, San Diego, CA) were used: CD45R/B220 (B cells), CD90/Thy-1 (T cells), panNK-PE (NK cells), and F4/80 (macrophages and Kupffer cells). Enrichment of the recovered B220+, CD90+, NK+, and F4/80+ cells was confirmed by flow cytometric analysis (Coulter Epics Elite counter; Coulter, Hialeah, FL) and was routinely greater than 95%.
Bone marrow transplantation
Bone marrow cells were harvested by flushing the femurs and tibias of wt and Hfe-/- mice with medium. Six-week-old female wt mice and Hfe-/- mice were lethally irradiated (9.5 Gy) and served as recipients. Irradiated mice intravenously received 10 to 15 × 106 bone marrow cells from wt or Hfe-/- mice and were killed 14 weeks after BMT. In experiments in which bone marrow cells were harvested from GFP+wt and GFP+Hfe-/- mice, wt or Hfe-/- mice that underwent transplantation were screened by flow cytometry for GFP+ leukocytes in the liver. B cells, T cells, and macrophages in nonparenchymal cells of the liver were identified by fluorescence-activated cell sorter (FACS) staining with anti-B220, anti-T-cell receptor (TCR), and anti-F4/80, respectively.
Quantitative and conventional reverse transcription-polymerase chain reaction
Total RNA was isolated with Trizol reagent (Invitrogen, Burlington, ON, Canada), and reverse transcription (RT) was performed with the Thermoscript RT-PCR system (Invitrogen). Levels of Hfe, Actb, hepcidin (Hamp), and glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA were measured by real-time PCR in a Rotor Gene 3000 Real Time DNA Detection System (Montreal Biotech Inc, Kirkland, QC, Canada) with the QuantiTect SYBRGreen I PCR kit (Qiagen, Mississauga, ON, Canada). All primers were designed with Primer3 software to include at least one intron. For each primer pair, the amplified cDNA fragments were verified in agarose gel to confirm the absence of the intron on the amplified fragment and the absence of nonspecific products. The primers used were: wt-type Hfe (wtHfe), 5′GAATGGGACGAGCACAAGAT3′ and 5′TGATGTTCTGGGGGAAGAAG3′; mutated Hfe (neoHfe), AGTTGGGAGTGGTGTCCGA3′ and CTAGCTTCGGCCGTGACG; Actb, 5′-TGTTACCAACTGGGACGACA-3′ and 5′-GGTGTTGAAGGTCTCAAA-3′; Hamp, 5′AGAGCTGCAGCCTTTGCAC3′ and 5′GAAGATGCAGATGGGGAAGT3′; and Gapdh, 5′TCAAGAAGGTGGTGAAGCAG3′ and 5′TGGGAGTTGCTGTTGAAGTC3′. Relative quantitation was performed using standard curves constructed from serial dilutions of PCR products. All standard curves generated were found to have excellent PCR amplification efficiency (90%-96%; 100% indicates that after each cycle the amount of template is doubled), as determined by their slopes. mRNA expression for each gene was determined by direct comparison with the standard curve of the specific target generated in each PCR run. Expression levels were normalized to the housekeeping gene Gapdh or to Actb.
For conventional PCR used in Hfe expression studies, the primers were: Hfe, 5′AGTTGGGAGTGGTGTCCGA3′ and 5′CCTCCAAGTCTTTGGCTGAG3′; and Actb, 5′-AGCCATGTACGTAGCCATCC-3′ and 5′-TTTGATGTCACGCACGATTT-3′. Amplified RT-PCR products were visualized by ethidium bromide staining in 1.0% agarose gels.
Hematologic measurements and transferrin saturation
Red blood cell (RBC) count, hemoglobin (Hb) level, hematocrit (HCT), and mean corpuscular volume (MCV) were measured with an automated cell counter calibrated for murine samples (ABC vet counter; ABX hématologie, Montpellier, France). Serum iron, total iron-binding capacity (TIBC), and transferrin saturation were assessed by colorimetric assay18 with the Kodak Ektachem DT60 system (Johnson & Johnson, Ortho-Clinical Diagnostics, Mississauga, ON, Canada).
Measurement of tissue iron concentration
Liver and spleen iron concentrations were assessed by acid digestion of tissue samples, followed by iron quantification with atomic absorption spectroscopy.18
Statistical analysis
All values in the figures are expressed as mean plus or minus SD. Student t test (unpaired, 2-tailed) was used for comparison between 2 groups. Multiple comparisons were statistically evaluated by one-way analysis of variance (ANOVA) followed by the Tukey test.
Results
qRT-PCR analysis of Hfe expression in various mouse organs and in isolated cell fractions
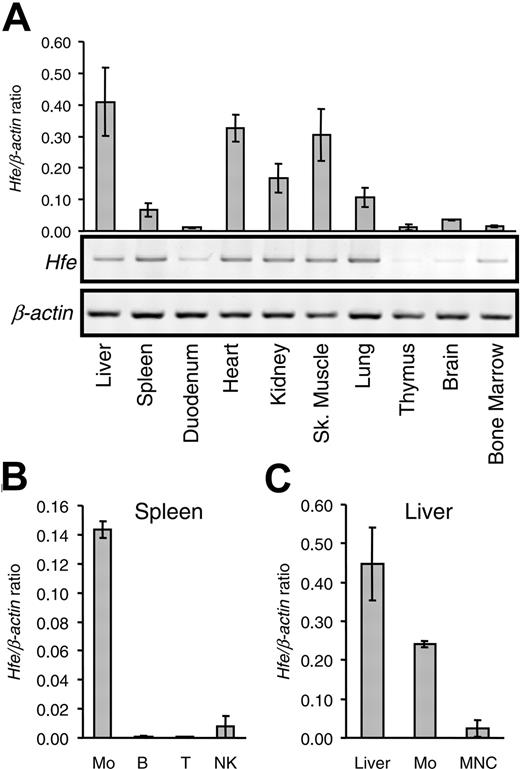
Previous reports using immunocytochemistry showed that human HFE protein is abundantly expressed in the small intestine, particularly in duodenal crypt cells,19 whereas in the liver, HFE is found in Kupffer cells.20 Recent studies with RT-PCR analysis, in situ hybridization, and Western blot analysis demonstrated HFE expression also in rat hepatocytes.21,22 We examined the pattern of Hfe expression in mice by quantitative RT-PCR (qRT-PCR). As shown in Figure 1A, we found that the mouse Hfe gene is expressed in most organs, with the highest expression levels in the liver, heart, and skeletal muscle, followed by the kidneys, lungs, and spleen. The lowest mRNA levels were found in the duodenum, thymus, brain, and bone marrow. These results are consistent with previous work on the Hfe expression pattern in mice by Northern blot analysis.23
To determine which hematopoietic cell lineages express Hfe, we performed qRT-PCR analysis in purified splenic cell populations (Figure 1B). Hfe was highly expressed in macrophages (F4/80+ cells) but was barely detectable in B and T lymphocytes or in NK cells.
Analysis of Hfe gene expression.Hfe expression was analyzed in various organs and in isolated cell fractions from spleen and liver. (A) mRNA was isolated from various organs, as indicated, and was used to synthesize cDNA. Hfe mRNA expression was quantified by qRT-PCR and normalized to Actb (graph). The Hfe/Actb × 103 ratio is shown. Conventional RT-PCR products were visualized by ethidium bromide staining in 1.0% agarose gels. (B) Hfe mRNA expression in positively selected splenic macrophages (Mo), B lymphocytes (B), T lymphocytes (T), NK cells, and (C) whole liver (liver), liver macrophages (Mo), and mononuclear cells (MNC) assessed by qRT-PCR and normalized to Actb. The Hfe/Actb × 103 ratio is shown. Results are presented as mean ± SD (n = 3 mice per organ or cell isolate).
Analysis of Hfe gene expression.Hfe expression was analyzed in various organs and in isolated cell fractions from spleen and liver. (A) mRNA was isolated from various organs, as indicated, and was used to synthesize cDNA. Hfe mRNA expression was quantified by qRT-PCR and normalized to Actb (graph). The Hfe/Actb × 103 ratio is shown. Conventional RT-PCR products were visualized by ethidium bromide staining in 1.0% agarose gels. (B) Hfe mRNA expression in positively selected splenic macrophages (Mo), B lymphocytes (B), T lymphocytes (T), NK cells, and (C) whole liver (liver), liver macrophages (Mo), and mononuclear cells (MNC) assessed by qRT-PCR and normalized to Actb. The Hfe/Actb × 103 ratio is shown. Results are presented as mean ± SD (n = 3 mice per organ or cell isolate).
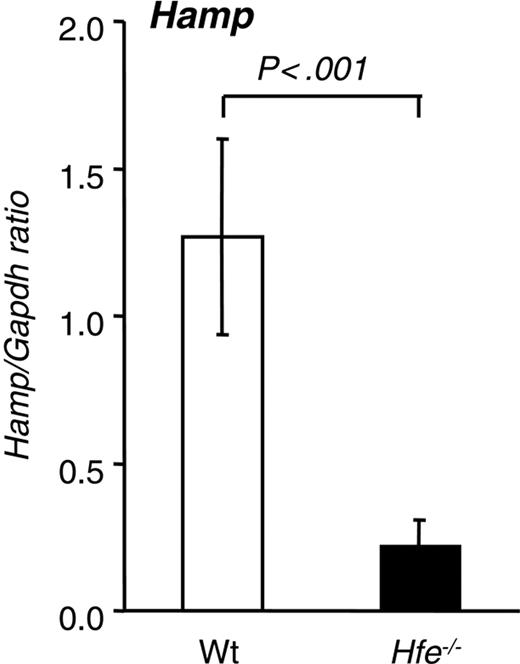
Hepatic hepcidin expression in 10-week-old Hfe-/- mice from the 129/SvEvTac background.Hamp mRNA expression was quantified by qRT-PCR and normalized to Gapdh. The Hamp/Gapdh ratio is shown. Results are presented as mean ± SD (n = 6 mice per group). Statistical analysis was performed by Student t test.
Hepatic hepcidin expression in 10-week-old Hfe-/- mice from the 129/SvEvTac background.Hamp mRNA expression was quantified by qRT-PCR and normalized to Gapdh. The Hamp/Gapdh ratio is shown. Results are presented as mean ± SD (n = 6 mice per group). Statistical analysis was performed by Student t test.
Similarly, in the liver, macrophages expressed high Hfe mRNA levels, whereas considerably lower levels were observed in intrahepatic MNC isolates, which consisted mostly of T and B lymphocytes with 4% to 8% F4/80+ cells.
Iron misregulation in Hfe-/- mice
The severity of iron overloading and the expression levels of liver hepcidin in HH animal models are known to depend on genetic background, age, and sex.24-28 Thus, we assessed the degree of iron loading in 2 Hfe-deficient mouse strains, 129/SvEvTac and C57Bl/6, by measuring serum iron levels, transferrin saturation, and iron concentrations in the liver and spleen. We used qRT-PCR analysis to quantify hepatic Hamp mRNA levels in wt and Hfe-/- mice.
As expected, serum iron levels and transferrin saturation were significantly elevated in Hfe-deficient 10-week-old females from the 129/SvEvTac background (Table 1). Hepatic iron concentration was 5-fold higher in Hfe-/- mice, whereas splenic iron levels were not significantly different from those of wt mice (Table 1). Resistance to iron loading in the spleens of Hfe-deficient mice has been observed previously4,29 and is believed to be caused by defective handling of iron in macrophages.
Iron parameters in 129/SvEvTac Hfe−/− mice
Parameter . | WT . | Hfe−/− . | P . | |||
|---|---|---|---|---|---|---|
| Serum iron, μM | 32 ± 7 | 44 ± 5 | <.01 | |||
| Transferrin saturation, % | 57 ± 4 | 110 ± 12 | <.001 | |||
| Iron concentration, μg iron/g dry weight | ||||||
| Liver | 322 ± 50 | 1675 ± 249 | <.001 | |||
| Spleen | 2451 ± 671 | 1791 ± 257 | NS | |||
| Data are presented as mean ± SD. | ||||||
| NS indicates not significant. | ||||||
Parameter . | WT . | Hfe−/− . | P . | |||
|---|---|---|---|---|---|---|
| Serum iron, μM | 32 ± 7 | 44 ± 5 | <.01 | |||
| Transferrin saturation, % | 57 ± 4 | 110 ± 12 | <.001 | |||
| Iron concentration, μg iron/g dry weight | ||||||
| Liver | 322 ± 50 | 1675 ± 249 | <.001 | |||
| Spleen | 2451 ± 671 | 1791 ± 257 | NS | |||
| Data are presented as mean ± SD. | ||||||
| NS indicates not significant. | ||||||
A 6-fold reduction of hepatic Hamp mRNA expression was detected in Hfe-/- compared with wt mice (Figure 2). These results are consistent with previously reported data11 and with the scenario that the lack of Hfe expression leads to inappropriate hepatic Hamp levels and ultimately to augmented duodenal iron absorption in this Hfe-deficient mouse strain. When similar studies were performed in Hfe-deficient mice of the same age and sex that had been backcrossed 11 times onto the C57Bl/6 background, the degree of iron loading was significantly lower (1675 ± 249 μg in 129/SvEvTac vs 739 ± 228 μg iron/g dry weight in C57Bl/6; P < .001; n = 6 per group). In addition, hepatic Hamp mRNA levels, albeit lower, were not significantly different from those found in wt C57Bl/6 mice (data not shown).
Based on our data and to monitor the effects of BMT on iron homeostasis, we tested Hfe-deficient mice from the 129/SvEvTac background in subsequent studies.
BMT studies
To investigate the importance of Hfe expression on RE macrophages in the regulation of iron homeostasis, we transplanted bone marrow from wt mice into lethally irradiated Hfe-/- mice to obtain mice that expressed Hfe in macrophages. Furthermore, we generated mice lacking Hfe expression in macrophages, but not in hepatocytes or in intestinal epithelial cells, by transplanting bone marrow from Hfe-/- mice into wt recipients. To control for the effects of irradiation, respective controls were generated by wt → wt and Hfe-/- → Hfe-/- transplantations.
Given that earlier BMT studies show that bone marrow cells (BMCs) have the potential to differentiate into various cell types in the liver, including hepatocytes30,31 and Kupffer cells,32 we first set out to evaluate the cell fate potential of transplanted BMCs in the liver using a cell-marking method with GFP transgenic mice. To generate a source of GFP+Hfe-/- BMCs, we crossed Hfe mutant mice with transgenic animals that constitutively express GFP in all tissues under control of the chicken Actb promoter.33 Chimeric animals were produced by transplantation of GFP+Hfe-/- or GFP+wt BMCs into GFP-negative, wt, or Hfe-/- irradiated recipients. Nonparenchymal cells from the liver were isolated and analyzed by flow cytometry to determine the degree of reconstitution in animals after transplantation, as described in “Materials and methods.” We found that approximately 33% to 37% of liver macrophages, identified as cells expressing the macrophage-specific marker F4/80, were of donor origin (ie, GFP positive), whereas 63% to 67% of total macrophages were of host origin because they were negative for GFP expression (Table 2). No significant differences were observed between the different BMT groups, indicating that Hfe-/- BMCs have a capacity similar to that of wt BMCs to repopulate irradiated recipients.
Degree of engraftment of donor macrophages in the livers of recipient mice after BMT
Donor → recipient . | n . | GFP+F4/80+ donor macrophages, % . | GFP−F4/80+ host macrophages, % . |
|---|---|---|---|
| GFP+wt → wt | 4 | 35 ± 3 | 65 ± 3 |
| GFP+Hfe−/− → wt | 4 | 37 ± 2 | 63 ± 2 |
| GFP+Hfe−/− → Hfe−/− | 4 | 33 ± 3 | 67 ± 3 |
| GFP+wt → Hfe−/− | 4 | 36 ± 4 | 64 ± 4 |
Donor → recipient . | n . | GFP+F4/80+ donor macrophages, % . | GFP−F4/80+ host macrophages, % . |
|---|---|---|---|
| GFP+wt → wt | 4 | 35 ± 3 | 65 ± 3 |
| GFP+Hfe−/− → wt | 4 | 37 ± 2 | 63 ± 2 |
| GFP+Hfe−/− → Hfe−/− | 4 | 33 ± 3 | 67 ± 3 |
| GFP+wt → Hfe−/− | 4 | 36 ± 4 | 64 ± 4 |
Liver NPC fractions from mice that underwent BMT were analyzed by flow cytometry for the presence of GFP+ donor-derived macrophages. NPCs were stained for the macrophage marker F4/80, and the percentage of GFP+ and GFP− within all F4/80+ cells is shown. Data are presented as mean ± SD.
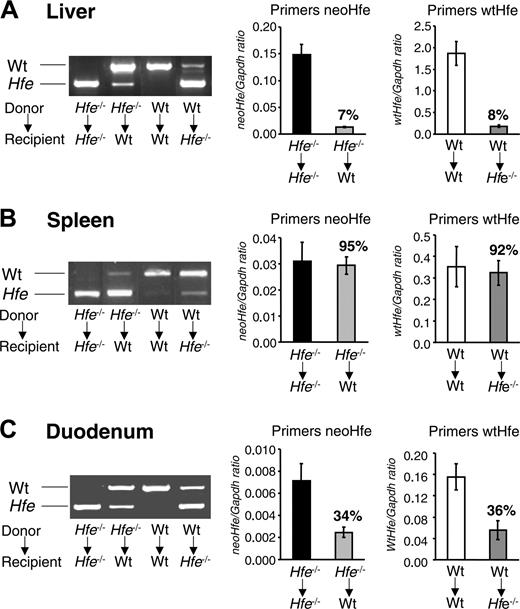
Engraftment of bone marrow-derived cells in the liver, spleen, and duodenum. Total RNA was isolated from the (A) liver, (B) spleen, and (C) duodenum 14 weeks after BMT and was used to synthesize cDNA. (left) cDNA was PCR amplified with one set of primers spanning exons 3 and 6 of the mouse Hfe gene (primers Hfe) and the products were visualized by ethidium bromide staining in 1.0% agarose gels. In Hfe-/- → wt mice, but not in wt → wt mice, the amplification product of donor Hfe-/- mice (Hfe, 499 bp) appears in wt recipient mice (wt, 775 bp) after transplantation. Conversely, in wt → Hfe-/- mice, but not in Hfe-/- → Hfe-/- mice, the amplification product of donor wt mice (wt, 775 bp) appears in Hfe-/- recipient mice (Hfe, 499 bp) after transplantation. One representative mouse is shown per BMT group. (right) Quantification of donor Hfe mRNA by qRT-PCR and normalized to Gapdh. Two distinct sets of primers were used—neoHfe and wtHfe—shown separately in the graphs. neoHfe/Gapdh × 103 and wtHfe/Gapdh × 103 ratios are presented. Results are mean ± SD (n = 6-12 mice per group).
Engraftment of bone marrow-derived cells in the liver, spleen, and duodenum. Total RNA was isolated from the (A) liver, (B) spleen, and (C) duodenum 14 weeks after BMT and was used to synthesize cDNA. (left) cDNA was PCR amplified with one set of primers spanning exons 3 and 6 of the mouse Hfe gene (primers Hfe) and the products were visualized by ethidium bromide staining in 1.0% agarose gels. In Hfe-/- → wt mice, but not in wt → wt mice, the amplification product of donor Hfe-/- mice (Hfe, 499 bp) appears in wt recipient mice (wt, 775 bp) after transplantation. Conversely, in wt → Hfe-/- mice, but not in Hfe-/- → Hfe-/- mice, the amplification product of donor wt mice (wt, 775 bp) appears in Hfe-/- recipient mice (Hfe, 499 bp) after transplantation. One representative mouse is shown per BMT group. (right) Quantification of donor Hfe mRNA by qRT-PCR and normalized to Gapdh. Two distinct sets of primers were used—neoHfe and wtHfe—shown separately in the graphs. neoHfe/Gapdh × 103 and wtHfe/Gapdh × 103 ratios are presented. Results are mean ± SD (n = 6-12 mice per group).
To evaluate the potential of transplanted BMCs to generate hepatocytes, we analyzed liver tissues from engrafted recipients for the presence of GFP+ hepatocytes (GFP+, albumin-positive cells) by fluorescence microscopy. No GFP+ hepatocytes were detected in any of the recipient mice (data not shown).
These results demonstrate that wt and Hfe-/- BMCs can reconstitute approximately one third of liver macrophages but that BMCs contribute little or not at all to the replacement of hepatocytes in the context of iron overload caused by Hfe deficiency. This is in agreement with a number of more recent studies using several models and showing that, under physiologic conditions and without the presence of strong selection pressure, bone marrow-derived hepatocytes are rarely, if at all, generated34-37 (for a review, see Fausto38 ).
Successful engraftment of donor bone marrow cells after 14 weeks was also assessed by conventional RT-PCR analysis of Hfe mRNA from total liver, spleen, and duodenal extracts using primers that span exon 3 to exon 6 of the Hfe gene. Amplification of cDNA obtained from wt and Hfe-/- mice yielded 775-bp and approximately 499-bp products, respectively (Figure 3), because of the deletion of the entire exon 4 (approximately 276 bp) in Hfe-/- mice.29 Expression of disrupted, exon 4-deficient Hfe mRNA was detected in the total liver, spleen, and duodenum of wt mice repopulated with Hfe-/- bone marrow, but not in those that underwent transplantation with control wt bone marrow (Figure 3). The opposite (ie, intact Hfe mRNA) was detected in samples from Hfe-/- mice repopulated with wt bone marrow.
To further quantify donor mRNA in Hfe-/- → wt and wt → Hfe-/- mice, we used qRT-PCR (Figure 3). wt and mutant Hfe were separately amplified with primers wtHfe and neoHfe, as described in “Materials and methods.” In the liver, donor mRNA was approximately 7% to 8% of the level observed in the relevant BMT control group (Figure 3A). Because Kupffer cells represent approximately 20% of the liver, these results suggest approximately 35% to 40% repopulation of liver macrophages from the donor marrow, assuming that Hfe expression in donor macrophages is similar to that in host macrophages. In the spleen, 92% to 95% of mRNA detected in BMT chimeras was of donor origin (Figure 3B).
Overall, engraftment efficiency in our BMT mice was similar to that reported in other studies, which found 20% to 35% replacement of macrophages in the liver 8 weeks after BMT and 80% to 95% replacement of plasma leukocytes 4 to 5 weeks after BMT.39-42 In the duodenum, donor mRNA reached 34% to 36% of the levels encountered in the pertinent BMT control group (Figure 3C).
Given that Hfe is highly expressed in macrophages compared with lymphocytes (Figure 1B-C), these findings are consistent with the accumulation of bone marrow-derived wt macrophages in the liver, spleen, and duodenum of wt → Hfe-/- mice and of Hfe-deficient macrophages in Hfe-/- → wt mice.
Serum iron and transferrin saturation analysis
Next, we examined changes in serum iron (SI) levels and transferrin saturation (TS), the earliest laboratory abnormalities in HH. SI and TS were significantly higher in Hfe-/- → Hfe-/- mice than in wt → wt mice (SI, 50 ± 3 μM vs 25 ± 5 μM, respectively; TS, 115% ± 8% vs 58% ± %15, respectively; P < .001), as expected,29 and were not affected by changes of Hfe expression in macrophages in BMT chimeras (data not shown).
Alterations of iron levels in bone marrow chimeras
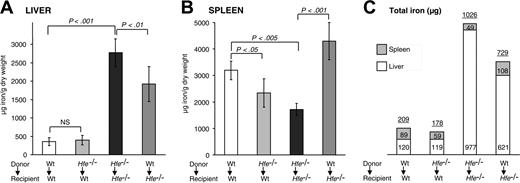
To monitor changes in body iron levels in BMT mice, we measured its concentration and calculated total iron in livers and spleens, the most relevant organs of iron storage. We found that wt mice whose bone marrow had been reconstituted with Hfe-deficient cells (Hfe-/- → wt) did not have significantly altered iron concentrations in the liver. In contrast, Hfe-/- mice reconstituted with wt cells (wt → Hfe-/-) had significantly decreased iron concentrations in the livers compared with control Hfe-/- → Hfe-/- mice (1920 ± 477 μg vs 2774 ± 380 μg iron/g dry weight, respectively) (Figure 4A).
Surprisingly, splenic iron concentrations were reduced by 38% in Hfe-/- → wt mice (Figure 4B). In fact, the capacity to store iron in the spleen was significantly impaired in Hfe-/- → wt mice (89 ± 16 μg in wt → wt vs 59 ± 17 μg total iron in Hfe-/- → wt; P < .005) (Figure 4C). The opposite effect—an improved capacity to store iron in the spleen—developed in Hfe-/- mice reconstituted with wt cells (1711 ± 229 μg in Hfe-/- → Hfe-/- vs 4301 ± 702 μg iron/g dry weight in wt → Hfe-/- mice) (Figure 4B). We observed a 2-fold increase of total iron content in the spleens of wt → Hfe-/- mice (108 ± 24 μg total iron) compared with control Hfe-/- → Hfe-/- mice (49 ± 6 μg total iron; P < .001). These data indicate that iron storage in the spleen is considerably altered in bone marrow chimeras of Hfe expression.
Liver and spleen iron storage in mice 14 weeks after BMT. Iron concentration in the (A) liver and (B) spleen. Results are presented as mean ± SD (n = 6-12). Statistical analysis was performed by one-way ANOVA. (C) Relative amounts of total iron stored in the liver and spleen. Each rectangle represents the absolute proportion of iron, expressed in micrograms, found in the liver (□) and spleen (▦). Numbers on top of the boxes indicate cumulative iron in the liver and spleen.
Liver and spleen iron storage in mice 14 weeks after BMT. Iron concentration in the (A) liver and (B) spleen. Results are presented as mean ± SD (n = 6-12). Statistical analysis was performed by one-way ANOVA. (C) Relative amounts of total iron stored in the liver and spleen. Each rectangle represents the absolute proportion of iron, expressed in micrograms, found in the liver (□) and spleen (▦). Numbers on top of the boxes indicate cumulative iron in the liver and spleen.
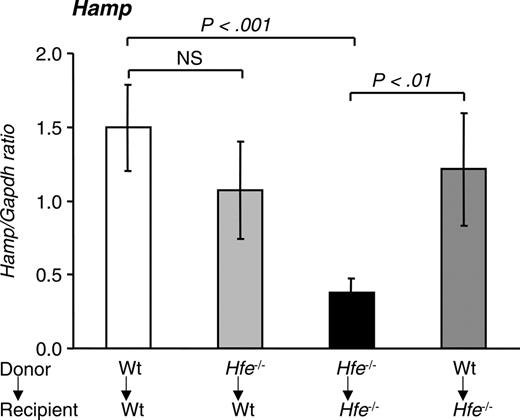
Hepcidin levels in mice 14 weeks after BMT. Quantitative alterations in liver hepcidin (Hamp) mRNA levels were measured by qRT-PCR and normalized to Gapdh. The Hamp/Gapdh ratio is shown. Results are presented as mean ± SD (n = 6-12). Statistical analysis was performed by one-way ANOVA.
Hepcidin levels in mice 14 weeks after BMT. Quantitative alterations in liver hepcidin (Hamp) mRNA levels were measured by qRT-PCR and normalized to Gapdh. The Hamp/Gapdh ratio is shown. Results are presented as mean ± SD (n = 6-12). Statistical analysis was performed by one-way ANOVA.
Augmented iron storage in the spleens of wt → Hfe-/- mice was accompanied by a decrease of approximately one third of total iron in the liver (621 ± 96 μg in wt → Hfe-/- vs 977 ± 135 μg total iron in Hfe-/- → Hfe-/-; P < .05). Reductions in hepatic iron levels in wt → Hfe-/- mice started relatively early after BMT, and it reached statistical significance 6 weeks after BMT (data not shown). Based on calculations of total iron in the livers and spleens in 10-week-old Hfe-/- mice (Table 1), which totaled 565 ± 83 μg iron, 20-week-old Hfe-/- → Hfe-/- mice had gained, on average, an additional 461 μg excess iron (1026 ± 131 μg iron). In comparison, wt → Hfe-/- mice had gained only 296 μg iron (729 ± 98 μg iron; P < .05) in these organs. Our results indicate that iron absorption was partially inhibited in wt → Hfe-/- compared with Hfe-/- → Hfe-/- mice.
Contribution of Hfe gene-expressing macrophages to the regulation of hepatic hepcidin levels
Next, we evaluated the effect of BMT on the expression of the hepatic iron regulator hepcidin. Hfe-/- → Hfe-/- mice expressed 25% of hepcidin mRNA levels encountered in wt → wt mice (P < .001) (Figure 5). When Hfe-/- mice were reconstituted with wt bone marrow, hepcidin levels rose to 81% of those expressed in wt → wt mice (P < .01). Finally, wt mice reconstituted with Hfe-deficient bone marrow cells had a small decrease in hepcidin levels that did not reach statistical significance.
Discussion
Macrophages are central to the maintenance of iron homeostasis because they recycle iron from damaged and senescent red blood cells back into the circulation for reuse by erythropoiesis. In the present study, we explored the role of Hfe expression in macrophages in the maintenance of macrophage iron homeostasis and in the regulation of hepatic hepcidin production and iron loading. We showed that in murine hematopoietic-derived cells, Hfe is expressed predominantly in macrophages and Kupffer cells.
Using BMT, we effectively reconstituted wt mice with Hfe-deficient macrophages and Hfe-/- mice with wt macrophages. The donor phenotype was readily detected in the liver, duodenum, and spleen. Reconstitution in the spleen was highly efficient because 92% to 95% of Hfe mRNA was of donor origin. We found that BMT resulted in dramatic changes in the ability of splenic macrophages to store iron in Hfe chimeras. This was evidenced by diminished iron concentrations in the spleens of wt mice repopulated with Hfe-deficient marrow and, conversely, by the enhanced iron concentration found in the spleens of Hfe-/- mice reconstituted with wt marrow. Considerable evidence indicates that in HH, the lack of appropriately expressed HFE molecules leads to defective iron storage in macrophages and Kupffer cells.43-45 In fact, a hallmark of the HH phenotype is the relative absence of iron in macrophages in spite of body iron overload. HH macrophages were found to have high iron regulatory protein (IRP) activity, which would prevent ferritin mRNA translation, the major cellular iron storage protein, and thus result in insufficient iron storage.7 Convincingly, restoration of HFE expression in monocytes and monocytic cell lines that lack functional HFE by means of transfection with wt HFE increases transferrin-dependent iron uptake46 and inhibits iron efflux,47 resulting in an augmentation of ferritin and cellular iron pools. These observations suggest that HFE mutations can directly affect iron accumulation in HH macrophages, independently of the presence of hepcidin. Our in vivo experiments on BMT chimeras demonstrate that Hfe expression in macrophages clearly determines the capacity of iron storage in the spleen. This could be attributed to a direct effect of Hfe expression in splenic macrophages and/or it can be a consequence of the elevation of hepatic hepcidin levels.
The second important consequence of Hfe deficiency is the higher setting of iron absorption levels in the duodenum. Considerable evidence now indicates that hepcidin plays a crucial role in the pathogenesis of HH and that the regulation of basal hepcidin levels is Hfe dependent. In fact, hepcidin levels are lower than expected in patients with HH12 and in animal models,10,11 particularly taking into account their body iron burden. How iron regulates hepatic hepcidin production remains to be elucidated. However, it is noteworthy that, though iron loading in mice strongly elicits the hepcidin response,14 direct exposure of primary human hepatocyte cultures to iron does not lead to augmented hepcidin expression.48 These observations indicate that hepcidin regulation by iron may rely on cross-communication between hepatocytes and RE macrophages in an Hfe-dependent fashion. Moreover, the fact that HH patients and animal models retain the capacity to appropriately regulate iron absorption in response to variations in iron status and erythroid demand49,50 suggests that Hfe deficiency leads to quantitative rather than qualitative changes in hepcidin expression.
Our experiments, in which Hfe-/- mice were reconstituted with Hfe-expressing macrophages (wt → Hfe-/-), show that restoring Hfe expression in RE cells can significantly inhibit iron loading. In fact, wt → Hfe-/- mice had hepatic iron levels that were approximately one third lower than those in Hfe-/- → Hfe-/- mice. This was observed in conjunction with a 3-fold increase in hepatic hepcidin levels in wt → Hfe-/- compared with Hfe-/- → Hfe-/- mice. These results may be underestimated because macrophages in the liver are replaced at a slower rate and less successfully than macrophages in hematopoietic tissues.39,40 In fact, other studies using similar techniques estimated that donor bone marrow-derived cells repopulate only approximately 35% of liver macrophages within 14 weeks of BMT.41 These values are close to those obtained in the present study using 2 different methods, GFP-marked macrophages and qRT-PCR to quantify donor-derived Hfe mRNA expression. Furthermore, BMT was performed when Hfe-/- mice were 6 weeks of age and when already significant excess iron had accumulated in their livers.
In previous experiments using the B2m-/- mouse, we found that adoptive transfer of wt fetal liver cells into lethally irradiated B2m-/- mice affected iron distribution in the liver in similar ways.6 However, in these studies, no significant differences were detected in terms of liver iron loading and iron absorption between B2m-/- → B2m-/- and wt → B2m-/- mice. Several factors may explain these differences. First, unlike wt → Hfe-/- mice, not all cell lineages are reconstituted in wt → B2m-/- mice. For example, CD8+ lymphocytes, which depend on interactions with MHC class 1 molecules on radioresistant cells in the thymus,51 are not reconstituted in wt → B2m-/- mice (R.J.S., unpublished observations, September 2002, and Ohteki and MacDonald52 ). In turn, lymphocyte numbers have been shown to affect the degree of iron loading in animal models18,53 and in patients with HH.54,55 Second, B2m-/- mice also lack other MHC class 1 molecules, and recent studies indicate that other B2m-dependent MHC class 1 molecules may be involved in iron metabolism. For example, HfeB2m-/- compound mutants deposit more tissue iron than mice lacking Hfe only,5 and mice lacking classic MHC class 1 molecules have higher hepatic iron levels than wt controls.56 Overall, these studies indicate that there are important differences between the Hfe-/- and B2m-/- mouse models. Possibly other factors, including MHC class 1 molecules and lymphocytes, that may contribute to the pathogenesis of iron overload in B2m-/- mice remain unaffected by the adoptive transfer of hematopoietic cells. Third, the animals used in these studies were from different genetic backgrounds; B2m-/- mice were from the C57Bl/6 background and had a milder phenotype than Hfe-/- mice from the 129/SvEvTac background.
In reciprocal experiments in which wt mice were reconstituted with Hfe-deficient marrow, we found that liver iron content—and, hence, iron absorption levels—remained utterly unaffected. In fact, Hfe-/- → wt mice did not develop hemochromatosis, and hepcidin levels, albeit slightly decreased, did not reach statistical significance. Presumably, the remaining host Kupffer cells, expressing wt Hfe, were sufficient to maintain appropriate control of hepcidin and iron absorption levels. In fact, our experiments using GFP-marked BMCs showed that approximately 62% to 67% of hepatic F4/80+ macrophages remain GFP negative and are thus of host origin (Table 2).
Our interpretation is that Hfe expression by part of the Kupffer cells is sufficient to increase basal hepcidin production to such levels as to reduce duodenal iron absorption and, consequently, significantly inhibit iron loading in Hfe-/- mice and that it is enough to maintain appropriate iron homeostasis in wt mice. This suggests a central role for hepcidin rather than Hfe expression in the duodenum for the regulation of intestinal iron absorption and thus the control of iron loading. Supporting evidence for this interpretation is provided by studies on Hfe-/- mice crossed with transgenic mice overexpressing hepcidin under a liver-specific promoter demonstrating that constitutive hepcidin expression in the liver is sufficient to prevent iron accumulation normally observed in Hfe-/- mice.57
A clue to how hepcidin, in turn, may regulate iron absorption is provided by the recent finding that it can bind to Fp1, the major cellular iron exporter protein, inducing Fp1 internalization and degradation.58 This presumably results in reduced iron efflux from hepatocytes, macrophages, and duodenal enterocytes.
In summary, our data show that the regulation of iron handling in macrophages is dependent on Hfe expression in these cells. They also show that Hfe expression by part of the Kupffer cells is sufficient to significantly increase basal hepcidin production in the liver and partially prevent iron loading in a background of total Hfe deficiency, and to maintain appropriate iron homeostasis in a wt background. This suggests an important contribution of Hfe expression in Kupffer cells to the regulation of hepatic hepcidin production.
Prepublished online as Blood First Edition Paper, May 24, 2005; DOI 10.1182/blood-2005-02-0629.
Supported by grant MOP44045 from the Canadian Institutes of Health Research (CIHR) and grant CBO/33485/99-00 from the Fundação para a Ciência e a Tecnologia. M.M.S. is the recipient of a CIHR New Investigator award.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Christian Dallaire for his work with atomic absorption spectroscopy, Cristina Escrevente for her help with cell isolation, and Ovid Da Silva for his editorial assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal