Abstract
Two major genetic categories of multiple myeloma (MM) exist. Hyperdiploid MM (48 to 74 chromosomes, median 53 chromosomes) is associated with trisomies especially of chromosomes 3, 7, 9, 11, 15, and 19, whereas the nonhyperdiploid (< 48 chromosomes or more than 74 chromosomes) MM is associated with primary translocations such as t(11;14), t(4;14), and t(14;16). Whether this dichotomy exists in monoclonal gammopathy of undetermined significance (MGUS) is uncertain due to limitations of current methods in the study of ploidy. This is especially true in MGUS where the number of clonal plasma cells is small. In this study, we derived a fluorescent in situ hybridization (FISH)-based trisomy index from pooled cytogenetic data (karyotype analysis) from 2 large cohorts of patients with MM with abnormal karyotype, and then validated it in 2 independent cohorts of patients who had known ploidy status either by karyotyping or DNA content measurement using flow cytometry. Using the criteria of 2 or more trisomies from a 3-chromosome combination, hyperdiploid myeloma can be detected with high specificity. Applying this index on 28 patients with smoldering multiple myeloma (SMM) or MGUS (11 SMM, 17 MGUS) who had normal karyotype, 11 cases of hyperdiploid SMM/MGUS were detected. This percentage (40%) is remarkably similar to the percentage of hyperdiploid MM reported in the literature, suggesting that hyperdiploid MM may originate early during disease evolution. (Blood. 2005;106: 2156-2161)
Introduction
Multiple myeloma (MM) is characterized by complex cytogenetics and aneuploidy in almost all cases when techniques that do not require metaphases like flow cytometry and interphase fluorescent in situ hybridization (FISH) are used.1-5 Recent independent studies have identified subgroups that cluster closely based on genetic abnormalities: the hyperdiploid group (48 to 74 chromosomes) is associated with recurrent trisomies, particularly of chromosomes 3, 5, 7, 9, 11, 15, and 19; and the nonhyperdiploid group is composed of cases with hypodiploid, pseudodiploid, or near tetraploid chromosome number (fewer than 48 or more than 74 chromosomes) and structural abnormalities, particularly primary translocations such as t(11;14) and t(4;14) and other unknown translocations involving the immunoglobulin heavy chain (IgH) locus at 14q32.6-10 This grouping also provides powerful prognostic information independent of 13q status within each group.7,11 Whereas hypodiploid myeloma probably arises as a result of oncogene activation due to IgH translocations, the oncogenic events of the hyperdiploid variants are largely unknown. Monoclonal gammopathy of undetermined significance (MGUS) is the precursor state for MM and the study of this dichotomy in the early stages of the disease is critical for the full understanding of clonal pathways of evolution.12
Current available methods for the study of ploidy in MM include conventional cytogenetics and flow cytometry. The use of conventional cytogenetics in MM is hampered by detection of abnormal karyotypes in only 20% to 50% of cases, failure to obtain metaphases in most patients,13-16 and predominance of normal karyotypes probably originating from the normal hematopoietic component.17 Flow cytometry measurement of DNA content may be an improvement over conventional cytogenetics but is complicated by technical issues. These technical problems have led to a large variability in reported results.18 Furthermore, ploidy results by flow cytometry have never been validated against conventional cytogenetics,5 tend to underestimate hypodiploid myeloma,19 and have conflicting information on prognosis.20-23 These problems are compounded in MGUS due to the small percentage of clonal cells present. The ability of FISH to detect genetic abnormalities in interphase cells lends itself to the study of genetic abnormalities in MGUS. Indeed, FISH studies in MGUS have shown that aneuploidy can be detected in a large fraction of patients.24-26 Furthermore, many of the genetic abnormalities detected in MM, such as t(4;14), t(11;14), and 13q deletions, have also been found in MGUS using FISH.27-31 Therefore a FISH-based technique to identify ploidy subtypes may facilitate the study of ploidy in MGUS.
In this study, we derived and validated a FISH-based trisomy index (TI) that is highly specific for hyperdiploid myeloma. Using this index, we detected a significant proportion of hyperdiploid MGUS in an unselected cohort of patients with MGUS with normal karyotype.
Patients, materials, and methods
Bone marrow (BM) samples were obtained following informed consent before the time of routine procurement for clinical samples. These studies were conducted under approval from the Mayo Foundation institutional review board (Rochester, MN) and following the Declaration of Helsinki guidelines for research with human subjects. The diagnostic criteria were consistent with those published in the literature.
Derivation cohorts for TI
The TI was derived from pooled complete cytogenetic karyotypes of 2 large published cohorts of patients with MM with abnormal karyotypes, comprising 254 patients from the Mayo clinical cytogenetic database7 and 138 patients previously reported by Smadja et al11 (“Derivation group”). Of these, 153 patients (39%) were classified as hyperdiploid.
Validation cohorts
The index was validated on 2 independent patient cohorts using interphase FISH. The first validation cohort came from patients enrolled in the Eastern Cooperative Oncology Group (ECOG) E9486 and its associated correlative laboratory clinical study E9487 (“ECOG validation group”). These patients all had newly diagnosed MM and have been described in detail elsewhere.32 Of these patients, 97 underwent DNA content determination by flow cytometry and had stored slides for FISH studies.
The second cohort contained 82 Mayo Clinic patients diagnosed with MGUS, smoldering multiple myeloma (SMM), MM, and plasma-cell leukemia (PCL) (“Mayo validation group”). These patients had karyotype information and had stored materials for FISH studies. Testing was done on 26 of the patients with MM/SMM/PCL with abnormal karyotypes. The TI was also applied to the other 56 patients with MM/SMM/MGUS with normal karyotypes to see if the hyperdiploid state could be detected.
Derivation of index
From the pooled karyotype data, a combination of the 5 most commonly trisomic chromosomes (chromosomes 3, 9, 11, 15, and 19) was selected. These chromosomes were found to be trisomic in more than 50% of patients with hyperdiploid myeloma in the derivation cohort. Next, using these chromosomes, we determined how many chromosomes needed to be trisomic for a sample to be considered hyperdiploid, and whether the number of chromosomes used in the combination could be reduced. To accomplish this, using standard formulas, we calculated the sensitivity, specificity, positive, and negative predictive values of all possible chromosome combinations from 5 down to the predetermined cut-off number of chromosomes. The combination with the best compromise between sensitivity and specificity was selected as the TI for validation.
Validation of index
The derived index was then validated in 2 independent cohorts of patients using centromeric FISH probes for the respective chromosomes against the 2 currently established methods for ploidy determination in MM: conventional cytogenetics (the Mayo validation group) and flow cytometric DNA content analysis (the ECOG validation group).
Karyotype analysis
The karyotypes were obtained with the use of both short-term and long-term cultures and processed by conventional cytogenetic techniques. We used the following modal chromosome number to define ploidy categories: hyperdiploid group (48 to 74 chromosomes) and nonhyperdiploid group comprising hypodiploid (up to 44 chromosomes), pseudodiploid group (45 to 47 chromosomes), and near-tetraploid (75 or more chromosomes) group. This categorization has been used in our previous study.6
Cytoplasmic immunoglobulin (cIg)-FISH studies
Interphase FISH was performed as previously described.33 To screen for aneuploidy we used a commercially available direct-labeled chromosome enumeration probe (CEP) for chromosomes 3, 9, 11, and 15 (Vysis, Downers Grove, IL) and a direct-labeled bacterial artificial chromosome (BAC) clone for the p-arm of chromosome 19 (RPCI 11 88I12; BACPAC Resources, Children's Hospital Oakland Research Institute, Oakland, CA). Noncommercial probes were labeled using standard nick translation with SpectrumOrange or SpectrumGreen (Vysis). The normal signal pattern for each CEP probe should be 2 signals of the same color in each interphase cell, depending on the color of the label. Three signals would be trisomy and more than 3 signals would be tetrasomy or more.
These probes were tested on normal cells to establish the incidence of nuclei with false-positive signal patterns: the upper limit of normal calculated from mean plus 3 standard deviations (SDs)34 for 3 and 4 or more signals is different for the different CEP probes, but never exceeded 7%. To improve the stringency of our criteria, we arbitrarily chose higher cut-offs of 10% for 3 signals and 4 or more signals as the upper limit of normal.
Only cIg-positive plasma cells (PCs) were scored using either a Zeiss Axioplan 2 microscope (Carl Zeiss, Gottingen, Germany) or a Leitz Epifluorescence microscope (Leitz, Wetzlar, Germany) with fluoroisothiocyanate, Texas red, and 4′,6-diamidino-2-phenylindole (DAPI) ultraviolet filters (Chromotechnology, Brattleboro, VT). We aimed to score 100 cells per sample, but at least 20 PCs were scored for each sample.
DNA content analysis
The total DNA content of the ECOG patient samples was analyzed by dual-channel flow cytometric analysis using propidium iodide to measure the DNA content and kappa and lambda light-chain antisera to identify the clonal cells, as previously described.20 The following DNA index criteria were used for the determination of ploidy: less than 0.95 hypodiploid, 0.95 to 1.05 pseudodiploid, between 1.06 and 1.74 hyperdiploid, and more than 1.75 tetraploid.
Statistics
Sensitivity was calculated as the percentage of hyperdiploid MM (true positives) by the trisomy index among those with hyperdiploidy MM by the gold standard method (karyotype or flow cytometry). Specificity was calculated as a percentage of nonhyperdiploid MM by the trisomy index (true negatives) among those with nonhyperdiploid MM by the gold standard method. The positive predictive value (PPV) was calculated as the percentage of “true positives” among all patients identified as hyperdiploid by TI, and the negative predictive value (NPV) was calculated as the percentage of “true negatives” among all patients identified as nonhyperdiploid by TI.
The distributions for overall survival (OS) and progression-free survival (PFS) in the ECOG cohort were estimated using the method of Kaplan and Meier. To test for the difference in OS and PFS between patients with hyperdiploid and nonhyperdiploid MM as classified by the DNA index or TI, a log-rank test was used.
Results
Trisomy clustering and derivation of trisomy index
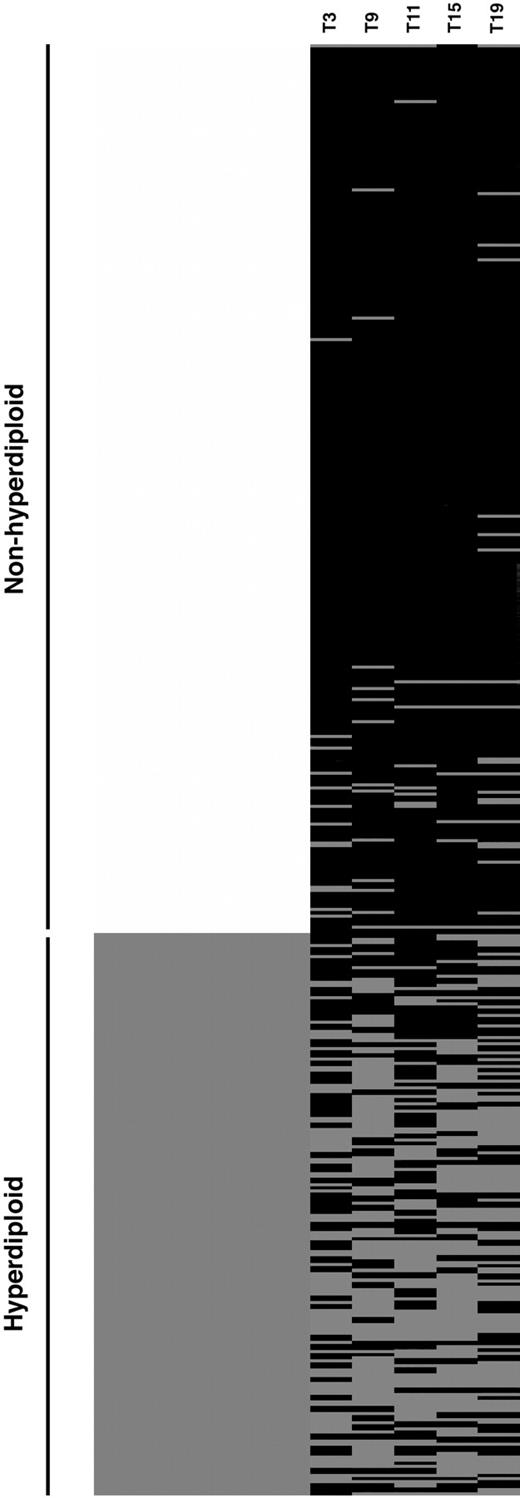
When the trisomy status for the 5 chromosomes selected (chromosome 3, 9, 11, 15, and 19) is presented as a cluster diagram for the derivation group, a striking paucity of trisomy is seen in the nonhyperdiploid group (Figure 1) and is consistent with our previous observations. We also observe that very few of the nonhyperdiploid MMs have more than 2 trisomies and very few of the hyperdiploid MMs have less than 2 trisomies. When we assess whether a minimum of 2 or 3 trisomies within this chromosome combination will give the best discrimination between hyper and nonhyperdiploid MM, we find that the presence of at least 2 trisomies can identify hyperdiploid MM with high sensitivity (88%) and specificity (94%). The use of 3 trisomies or more as the cut-off will severely compromise sensitivity (reduced to 65%) without much improvement in specificity (increased to 98%). Most of the cases misclassified as hyperdiploidy using this simple index belonged to the near-tetraploid group, in which the distinction between trisomies and a 4N karyotype can be complex. Only one case of hypodiploid MM was misclassified as hyperdiploid MM. This is an unusual patient from the French series who had a modal chromosome number of 30 but also trisomy of chromosomes 3, 7, 9, 11, 18, and 19.11 We therefore used 2 trisomies as the cut-off for identification of hyperdiploid MM.
Trisomy clustering diagram. Each row represents results from a single patient. The left column represents ploidy categories. The lighter color represents patients with nonhyperdiploid myeloma, whereas the darker color represents hyperdiploid myeloma. Each right column represents one of the commonly trisomic chromosomes, chromosomes 3, 9, 11, 15, and 19, with pale and dark bars representing the presence and absence of trisomy, respectively, for each chromosome.
Trisomy clustering diagram. Each row represents results from a single patient. The left column represents ploidy categories. The lighter color represents patients with nonhyperdiploid myeloma, whereas the darker color represents hyperdiploid myeloma. Each right column represents one of the commonly trisomic chromosomes, chromosomes 3, 9, 11, 15, and 19, with pale and dark bars representing the presence and absence of trisomy, respectively, for each chromosome.
Next, we wanted to see if the number of chromosomes used could be reduced from this set of 5. To do this, the sensitivity, specificity, and negative and positive predictive values of all possible combinations from 2 to 5 chromosomes within this chromosome set are determined for the derivation group (Table 1). As fewer chromosomes were used, the sensitivity was reduced while the specificity improved. This is paralleled by the trend for PPV and NPV. Although the use of the 2-chromosome combination can result in near 100% specificity, sensitivity is severely compromised because 5 times as many hyperdiploid myeloma cases will be missed compared with when all the 5 chromosomes are used. What is also clear from this analysis is that not all combinations of similar numbers of chromosomes produce the same sensitivity and specificity. For example, among the 4-chromosome combinations, chromosome combinations 3, 9, 15, and 19 and 9, 11, 15, and 19 appear to be better that the rest. Similarly, certain 3-chromosome combinations are better than others. This suggests that certain combinations of trisomies tend to track together in hyperdiploid patients.
Sensitivity and specificity of different chromosome combinations for identification of hyperdiploid myeloma derived from the validation group
Chromosome combinations . | Sensitivity, % . | Specificity, % . | PPV, % . | NPV, % . |
|---|---|---|---|---|
| 3,9,11,15,19 | 88 | 94 | 90 | 93 |
| 3,9,11,15 | 77 | 96 | 93 | 87 |
| 3,9,11,19 | 78 | 95 | 90 | 87 |
| 3,9,15,19 | 83 | 95 | 91 | 90 |
| 3,11,15,19 | 74 | 95 | 91 | 85 |
| 9,11,15,19 | 82 | 96 | 93 | 90 |
| 3,9,11 | 63 | 98 | 95 | 81 |
| 3,9,15 | 69 | 98 | 95 | 83 |
| 3,9,19 | 67 | 97 | 93 | 82 |
| 3,11,15 | 59 | 97 | 93 | 78 |
| 3,11,19 | 58 | 96 | 90 | 78 |
| 3,15,19 | 69 | 97 | 93 | 83 |
| 9,11,15 | 69 | 98 | 95 | 83 |
| 9,11,19 | 69 | 98 | 95 | 83 |
| 9,15,19 | 75 | 96 | 93 | 86 |
| 11,15,19 | 66 | 97 | 94 | 82 |
| 3,9 | 44 | 99 | 97 | 74 |
| 3,11 | 33 | 99 | 94 | 70 |
| 3,15 | 44 | 100 | 99 | 74 |
| 3,19 | 39 | 98 | 94 | 72 |
| 9,11 | 41 | 99 | 97 | 73 |
| 9,15 | 53 | 99 | 98 | 77 |
| 9,19 | 48 | 98 | 95 | 75 |
| 11,15 | 42 | 99 | 96 | 73 |
| 11,19 | 34 | 98 | 91 | 70 |
| 15,19 | 51 | 98 | 94 | 76 |
Chromosome combinations . | Sensitivity, % . | Specificity, % . | PPV, % . | NPV, % . |
|---|---|---|---|---|
| 3,9,11,15,19 | 88 | 94 | 90 | 93 |
| 3,9,11,15 | 77 | 96 | 93 | 87 |
| 3,9,11,19 | 78 | 95 | 90 | 87 |
| 3,9,15,19 | 83 | 95 | 91 | 90 |
| 3,11,15,19 | 74 | 95 | 91 | 85 |
| 9,11,15,19 | 82 | 96 | 93 | 90 |
| 3,9,11 | 63 | 98 | 95 | 81 |
| 3,9,15 | 69 | 98 | 95 | 83 |
| 3,9,19 | 67 | 97 | 93 | 82 |
| 3,11,15 | 59 | 97 | 93 | 78 |
| 3,11,19 | 58 | 96 | 90 | 78 |
| 3,15,19 | 69 | 97 | 93 | 83 |
| 9,11,15 | 69 | 98 | 95 | 83 |
| 9,11,19 | 69 | 98 | 95 | 83 |
| 9,15,19 | 75 | 96 | 93 | 86 |
| 11,15,19 | 66 | 97 | 94 | 82 |
| 3,9 | 44 | 99 | 97 | 74 |
| 3,11 | 33 | 99 | 94 | 70 |
| 3,15 | 44 | 100 | 99 | 74 |
| 3,19 | 39 | 98 | 94 | 72 |
| 9,11 | 41 | 99 | 97 | 73 |
| 9,15 | 53 | 99 | 98 | 77 |
| 9,19 | 48 | 98 | 95 | 75 |
| 11,15 | 42 | 99 | 96 | 73 |
| 11,19 | 34 | 98 | 91 | 70 |
| 15,19 | 51 | 98 | 94 | 76 |
The 3-chromosome combinations seem to offer good specificity without severely compromising sensitivity. Furthermore, 3 centromeric probes can be used on one slide, resulting in a more efficient use of patients' limited diagnostic materials with consequent cost savings. From our analysis, several 3-chromosome combinations appear similar. We arbitrarily chose to use the combination of chromosomes 9, 11, and 15 for validation. The TI for hyperdiploid MM is therefore any 2 trisomies among chromosomes 9, 11, and 15. In the ECOG validation group, in which there is enough material to stain 2 slides, all 5 chromosomes were used and the diagnostic efficacies of 5-, 4-, and 3-chromosome combinations were further compared.
Validation of trisomy index
Mayo validation group. Of the 26 Mayo Clinic patients with PCL, MM, or SMM selected for the presence of abnormal karyotypes, 25 had materials for FISH on the 3 chromosomes. The index correctly identified all cases of hyperdiploid MM (Table 2). The 3 cases that fit the criteria for hyperdiploidy but actually had near-tetraploid karyotype could also be identified as near-tetraploid as they had 4 or more signals in more than 10% of clonal plasma cells for 2 or more of these chromosomes. Using this additional criterion, hyperdiploid MM can be identified with a sensitivity and specificity of 100% when using the trisomy index in this cohort of patients with abnormal karyotype. This additional criterion is used for subsequent analysis with the TI.
Accuracy of trisomy index in identifying patients with MM and SMM of various ploidy categories who had abnormal karyotypes (Mayo validation group)
Diagnosis . | Ploidy (cyto) . | T9 . | T11 . | T15 . | >T9 . | >T11 . | >T15 . | Ploidy by TI . |
|---|---|---|---|---|---|---|---|---|
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Hypo | − | + | − | − | + | + | TP |
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| SMM | Dip | − | − | − | − | − | − | NH |
| MM | Dip | − | − | − | − | − | − | NH |
| MM | Dip | − | − | + | − | − | − | NH |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | − | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| PCL | TP | + | + | + | + | + | + | TP |
| MM | TP | − | − | + | + | − | + | TP |
| SMM | TP | + | + | + | + | + | + | TP |
Diagnosis . | Ploidy (cyto) . | T9 . | T11 . | T15 . | >T9 . | >T11 . | >T15 . | Ploidy by TI . |
|---|---|---|---|---|---|---|---|---|
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Hypo | − | + | − | − | + | + | TP |
| PCL | Hypo | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| PCL | Dip | − | − | − | − | − | − | NH |
| SMM | Dip | − | − | − | − | − | − | NH |
| MM | Dip | − | − | − | − | − | − | NH |
| MM | Dip | − | − | + | − | − | − | NH |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | − | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| MM | H | + | + | + | − | − | − | H |
| PCL | TP | + | + | + | + | + | + | TP |
| MM | TP | − | − | + | + | − | + | TP |
| SMM | TP | + | + | + | + | + | + | TP |
Hypo indicates hypodiploid; Dip, diploid; H, hyperdiploid; NH, nonhyperdiploid; TP, near-tetraploid; TI, trisomy index.
ECOG validation group. Using ploidy by flow cytometry as the standard for these 97 patients with active multiple myeloma and slides available for FISH, the TI using a 3-chromosome combination identified hyperdiploid MM with a sensitivity of 60% and specificity of 90%. Only 2 nonhyperdiploid cases were identified inappropriately as hyperdiploid (Table 3). As expected, the use of more chromosomes increases the sensitivity of the TI (up to 75% if 5 chromosomes are used) with little change in specificity. Of note, sensitivities for all 3 combinations are 5% to 10% less than the corresponding sensitivities obtained from the derivation group, based on cytogenetics. These means that more cases identified as hyperdiploid by DNA index than karyotyping were classified as nonhyperdiploid by the TI. This discrepancy may be related to the fact that the TI is derived from cases with abnormal karyotypes and may select for more proliferative cases, although the plasma-cell labeling index is not different between flow cytometry-classified hyperdiploid cases that are classified as hyperdiploid or nonhyperdiploid by the TI (data not shown). Therefore, the TI may actually slightly underestimate the total number of hyperdiploid cases present.
Comparison of ploidy status by trisomy index using different chromosome combinations and flow cytometry on ECOG validation group
. | DNA index, N . | . | . | . | . | . | |
|---|---|---|---|---|---|---|---|
| Chromosome combinations . | < 1.06 or > 1.74 . | 1.06 to 1.74 . | Sensitivity, % . | Specificity, % . | PPV, % . | NPV, % . | |
| 9, 11, 15 | - | - | 60 | 90 | 93 | 50 | |
| Nonhyperdiploid by TI | 27 | 27 | - | - | - | - | |
| Hyperdiploid by TI | 3 | 40 | - | - | - | - | |
| 3, 9, 15, 19 | 64 | 90 | 93 | 53 | |||
| Nonhyperdiploid by TI | 27 | 24 | - | - | - | - | |
| Hyperdiploid by TI | 3 | 43 | - | - | - | - | |
| 3, 9, 11, 15, 19 | - | - | 75 | 87 | 93 | 60 | |
| Nonhyperdiploid by TI | 26 | 17 | - | - | - | - | |
| Hyperdiploid by TI | 4 | 50 | - | - | - | - | |
. | DNA index, N . | . | . | . | . | . | |
|---|---|---|---|---|---|---|---|
| Chromosome combinations . | < 1.06 or > 1.74 . | 1.06 to 1.74 . | Sensitivity, % . | Specificity, % . | PPV, % . | NPV, % . | |
| 9, 11, 15 | - | - | 60 | 90 | 93 | 50 | |
| Nonhyperdiploid by TI | 27 | 27 | - | - | - | - | |
| Hyperdiploid by TI | 3 | 40 | - | - | - | - | |
| 3, 9, 15, 19 | 64 | 90 | 93 | 53 | |||
| Nonhyperdiploid by TI | 27 | 24 | - | - | - | - | |
| Hyperdiploid by TI | 3 | 43 | - | - | - | - | |
| 3, 9, 11, 15, 19 | - | - | 75 | 87 | 93 | 60 | |
| Nonhyperdiploid by TI | 26 | 17 | - | - | - | - | |
| Hyperdiploid by TI | 4 | 50 | - | - | - | - | |
- indicates not applicable.
Application of TI on patients from the Mayo validation group with normal karyotype. Seventeen patients with MM with normal karyotypes were identified as hyperdiploid by the TI. Therefore, the percentage of hyperdiploid MM based on the TI in this cohort of patients with MM was 56%, which is similar to other larger studies6,7,11,35 (Table 4).
Comparison of percentage of hyperdiploid MM detected by the trisomy index in the current study with other published studies
Series . | Hyperdiploid, % . | Nonhyperdiploid, % . |
|---|---|---|
| Current, n = 41 | 56 | 44 |
| French, n = 13811 | 55 | 44 |
| Mayo, n = 2547 | 31 | 69 |
| ECOG, n = 1996 | 51 | 49 |
| Mitleman database, n = 78335 | 39 | 61 |
All the patients with MGUS had normal karyotypes, whereas 2 of the patients with SMM had abnormal karyotypes (1 diploid with t(11;14) and 1 near-tetraploid) that were accurately identified as nonhyperdiploid by TI. In addition, 11 patients with SMM or MGUS (6 MGUS and 5 SMM) with normal karyotype were identified as hyperdiploid. Therefore, 40% of 28 patients with MGUS or SMM are hyperdiploid by TI although it is possible that this is a slight underestimation (see “ECOG validation group”).
In this cohort, trisomies of chromosomes 9 and 15 are more common (61% and 60% of all cases, respectively) than trisomy of chromosome 11 (49% of all cases). As would be expected, trisomies are more common in hyperdiploid cases than in nonhyperdiploid cases. Frequency of trisomies appears similar between MM and MGUS or SMM (Table 5). These observations are similar to those previously published using cytogenetics data.7,11 Of note, among the hyperdiploid cases, the percentage of clonal plasma cells with trisomies is similar for MM and MGUS or SMM (median 51% [range, 15%-75%] versus median 45% [range, 20%-68%], Wilcoxon rank sum test P = .09). These percentages are also similar to those previously published using FISH.24,25
Frequency of trisomies according to disease and genetic categories in the Mayo validation group
Disease and ploidy by TI . | Trisomy 9, N (%) . | Trisomy 11, N (%) . | Trisomy 15, N (%) . |
|---|---|---|---|
| MM, total, n = 41 | 26 (63) | 23 (56) | 27 (66) |
| Hyperdiploid, n = 22 | 21 (96) | 20 (91) | 20 (91) |
| Nonhyperdiploid, n = 19 | 5 (26) | 3 (16) | 7 (37) |
| SMM/MGUS, total, n = 28 | 16 (57) | 11 (39) | 14 (50) |
| Hyperdiploid, n = 11 | 11 (100) | 8 (73) | 11 (100) |
| Nonhyperdiploid, n = 17 | 5 (29) | 3 (18) | 3 (18) |
| Total hyperdiploid, n = 33 | 32 (97) | 28 (85) | 31 (94) |
| Total nonhyperdiploid, n = 36 | 10 (28) | 6 (17) | 10 (28) |
| Overall total, n = 69 | 42 (61) | 34 (49) | 41 (60) |
Disease and ploidy by TI . | Trisomy 9, N (%) . | Trisomy 11, N (%) . | Trisomy 15, N (%) . |
|---|---|---|---|
| MM, total, n = 41 | 26 (63) | 23 (56) | 27 (66) |
| Hyperdiploid, n = 22 | 21 (96) | 20 (91) | 20 (91) |
| Nonhyperdiploid, n = 19 | 5 (26) | 3 (16) | 7 (37) |
| SMM/MGUS, total, n = 28 | 16 (57) | 11 (39) | 14 (50) |
| Hyperdiploid, n = 11 | 11 (100) | 8 (73) | 11 (100) |
| Nonhyperdiploid, n = 17 | 5 (29) | 3 (18) | 3 (18) |
| Total hyperdiploid, n = 33 | 32 (97) | 28 (85) | 31 (94) |
| Total nonhyperdiploid, n = 36 | 10 (28) | 6 (17) | 10 (28) |
| Overall total, n = 69 | 42 (61) | 34 (49) | 41 (60) |
Outcome of hyperdiploid patients defined by DNA index and TI. Of the 97 patients in the ECOG validation cohort, 67 were classified as hyperdiploid and 30 as nonhyperdiploid by DNA index, whereas 43 were classified as hyperdiploid and 54 as nonhyperdoploid by the TI. The median OS and PFS of hyperdiploid patients classified by the TI is 54.2 months and 35.2 months, respectively, whereas it is 52.3 months and 31.7 months, respectively, when classified by DNA index. The survival of patients classified by the 2 methods is therefore similar. There were 27 patients classified as hyperdiploid by DNA index who were classified as nonhyperdiploid by the TI. The median OS and PFS of these patients are worse than those of the patients classified as hyperdiploid by the 2 methods. Conversely, the 3 cases classified as nonhyperdiploid by flow cytometry but as hyperdiploid by the TI have better OS and PFS compared with those patients classified as nonhyperdiploid by both methods. In fact, 2 of the 3 cases were the ones with the best survival among the cases classified as nonhyperdiploid by flow cytometry. However, these differences did not reach statistical differences probably due to the small number of cases within each subset (Table 6). The TI may therefore provide additional information on the survival of patients with MM.
Comparison of patient survival based on different methods of ploidy classification (TI versus DNA index)
. | . | Ploidy classification method . | . | . | |
|---|---|---|---|---|---|
| Median, months (range) . | N . | TI . | DNA index . | Log-rank P . | |
| Overall survival | |||||
| 30.7 (22.0-48.7) | 27 | NH | NH | 0.46 | |
| 73.1 (51.7-94.4) | 3 | H | NH | — | |
| 46.7 (35.6-78.5) | 27 | NH | H | 0.58 | |
| 54.2 (40.5-60.9) | 40 | H | H | — | |
| Progression-free survival | |||||
| 20.0 (12.7-25.3) | 27 | NH | NH | 0.40 | |
| 41.8 (36.9-46.7) | 3 | H | NH | — | |
| 29.2 (24.3-35.1) | 27 | NH | H | 0.61 | |
| 35.1 (30.7-42.5) | 40 | H | H | — | |
. | . | Ploidy classification method . | . | . | |
|---|---|---|---|---|---|
| Median, months (range) . | N . | TI . | DNA index . | Log-rank P . | |
| Overall survival | |||||
| 30.7 (22.0-48.7) | 27 | NH | NH | 0.46 | |
| 73.1 (51.7-94.4) | 3 | H | NH | — | |
| 46.7 (35.6-78.5) | 27 | NH | H | 0.58 | |
| 54.2 (40.5-60.9) | 40 | H | H | — | |
| Progression-free survival | |||||
| 20.0 (12.7-25.3) | 27 | NH | NH | 0.40 | |
| 41.8 (36.9-46.7) | 3 | H | NH | — | |
| 29.2 (24.3-35.1) | 27 | NH | H | 0.61 | |
| 35.1 (30.7-42.5) | 40 | H | H | — | |
H indicates hyperdiploid; NH, nonhyperdiploid; and —, not applicable.
Discussion
In this study, we have developed a FISH-based trisomy index, using a cut-off of 2 trisomies within a 3-chromosome combination (chromosomes 9, 11, and 15) that is highly specific for hyperdiploid MM. This index is easy to perform as it only requires the application of FISH probes for 3 chromosomes. The TI is also robust, having been derived from a large cytogenetic dataset and validated against both karyotyping and flow cytometry in different independent patient cohorts. Furthermore, it is applicable to the full spectrum of plasma-cell neoplasms. When the index is applied to patients from the Mayo validation group, 23 (56%) of 41 patients with MM have hyperdiploidy, 17 of these with normal cytogenetics. This percentage of hyperdiploid MM is similar to that reported with cytogenetics (Table 4), further validating this approach.
More importantly, when the index is applied to 28 patients with MGUS and SMM who have normal karyotypes, 40% were identified as hyperdiploid. Previous FISH studies have highlighted that aneuploidy and trisomies may exist in MGUS,24-26 but whether those cases are hyperdiploid is not clear, as aneuploidy and trisomies may also exist in nonhyperdiploid myeloma. As our index is highly specific for hyperdiploidy, we provide clear evidence that hyperdiploid MGUS exists. The percentage of hyperdiploid MGUS is also similar to the percentage of hyperdiploid MM reported.6,7,11,35 This suggests that hyperdiploid MM may originate early during disease evolution, and further supports its existence as a distinct entity.
Recent studies have established 2 main genetic categories of MM.6-8 Nonhyperdiploid MM is associated with primary translocations and chromosome 13 abnormalities whereas hyperdiploid MM is associated with recurrent trisomies of certain chromosomes and relative absence of IgH translocations and chromosome 13 abnormalities. The ability of the TI to identify hyperdiploid MGUS will facilitate further studies into whether a similar negative association between hyperdiploid MGUS and IgH translocation and chromosome 13 abnormalities exists.
Studies have also shown that the frequency of IgH translocations like t(4;14), t(11;14) and t(14;16), and 13q deletions in MGUS is similar to MM, suggesting these are primary oncogenic events that may not be sufficient to lead to progression into MM.27,31 We have now shown that the mechanism leading to hyperdiploidy is probably also a primary oncogenic event. The similar frequency of trisomies and the percentage of clonal plasma cells with trisomies between patients with hyperdiploid MGUS and those with SMM and MM provide strong evidence for this. Although previous studies suggest additional trisomies can be gained over time in MGUS prior to progression into MM,24,25 our data suggest that the mechanisms leading to hyperdiploidy and the selection pressure for certain trisomies (namely of chromosomes 3, 9, 11, 15, and 19) already exist at the MGUS stage. Currently available information therefore suggests that the main genetic abnormalities identified in MM, namely hyperdiploidy, primary translocations, and chromosome 13 abnormalities are primary/early oncogenic events already present in the premalignant MGUS stage. Consistent with this is that MM tends to cluster with MGUS in gene expression studies when unsupervised algorithms are used, suggesting the 2 conditions have more similarities than differences.36,37
Despite the apparent similarities in their genetic abnormalities, only a minority of cases of MGUS progress to MM.38 Defining factors leading to progression would be important for early therapeutic intervention or even chemoprevention strategies. Few genetic events are consistently associated with progression from MGUS to MM except perhaps for activating RAS mutations.39,40 Nongenetic events may therefore be important for disease progression. Epigenetic silencing of tumor suppressor genes such as p16INK4A and E-cadherin41-43 by gene promoter hypermethylation has been shown to be associated with progression from MGUS to MM. Changes in cytokines and bone marrow microenvironment favoring progression may be another mechanism.44,45 The requirement for progression is likely to be heterogeneous and may be unique to individual genetic subtypes. This is an important issue that needs to be addressed. Availability of high throughput global screens at the RNA (gene expression profiling), DNA (array-CGH), protein (protein array), and epigenetic (oligonucleotide methylation array) levels and tools like TI that allow ploidy assignment in a larger number of patients with MM and MGUS will facilitate this in the not-too-distant future.
In conclusion, we have developed a FISH-based trisomy index that is highly specific for hyperdiploid MM. This index allows the detection of hyperdiploid MGUS in a cohort of patients with MGUS with normal karyotype and establishes that the hyperdiploid and nonhyperdiploid dichotomy also exists in MGUS. This is further proof that hyperdiploid MM and nonhyperdiploid MM exist as separate entities with distinct oncogenic events.
Prepublished online as Blood First Edition Paper, May 26, 2005; DOI 10.1182/blood-2005-02-0761.
Supported by the International Waldenström Macroglobulinemia Foundation, grants R01 CA83 724-01, SPORE P50 CA100 707-01, and P01 CA62 242 from the National Cancer Institute, and the Fund to Cure Myeloma. P.R.G. is supported by ECOG grant CA21 115-25C from the National Cancer Institute. R.F. is a Clinical Investigator of the Damon Runyon Cancer Research Fund. W.J.C. is supported by an International Fellowship from the Agency for Science, Technology, and Research (A*STAR), Singapore.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal