Abstract
The AML1/EVI1 chimeric gene is created by the t(3;21)(q26;q22) chromosomal translocation seen in patients with leukemic transformation of myelodysplastic syndrome or blastic crisis of chronic myelogenous leukemia. We knocked-in the AML1/EVI1 chimeric gene into mouse Aml1 genomic locus to explore its effect in developmental hematopoiesis in vivo. AML1/EVI1/+ embryo showed defective hematopoiesis in the fetal liver and died around embryonic day 13.5 (E13.5) as a result of hemorrhage in the central nervous system. The peripheral blood had yolk-sac-derived nucleated erythroblasts but lacked erythrocytes of the definitive origin. Although E12.5 fetal liver contained progenitors for macrophage only, E13.5 fetal liver contained multilineage progenitors capable of differentiating into dysplastic myelocyte and megakaryocyte. No erythroid progenitor was detected in E12.5 or E13.5 fetal liver. Hematopoietic progenitors from E13.5 AML1/EVI1/+ fetal liver were highly capable of self-renewal compared with those from wild-type liver. Maintained expression of PU.1 gene and decreased expression of LMO2 and SCL genes may explain the aberrant hematopoiesis in AML1/EVI1/+ fetal liver. (Blood. 2005;106:2147-2155)
Introduction
The t(3;21)(q26;q22) chromosomal translocation occurs in patients with aggressive transformation of myelodysplastic syndrome (MDS) or chronic myelogenous leukemia (CML).1-4 In the joining region of t(3;21)(q26;q22), the AML1 gene on 21q22 is fused with the EVI1 (ecotropic viral integration site-1) gene on 3q26.5 The resultant AML1/EVI1 fusion gene is translated in frame to generate a chimeric transcription factor in which the N-terminal of AML1, including its DNA-binding Runt domain, is fused to almost the entire portion of EVI1. This chimeric transcription factor could be a molecular culprit for the leukemic progression of stem-cell malignancies caused by t(3;21)(q26;q22).
AML1 is involved in transcriptional regulation of a number of hematopoietic cell-specific genes. The Runt domain of AML1 binds to a specific DNA consensus sequence named polyomavirus enhancer binding protein 2 (PEBP2; ACCRCA), together with a non-DNA-binding β subunit (core-binding factor β [CBFβ]/PEBP2β) to form a heterodimeric active transcription factor complex.6-10 AML1- or CBFβ-deficient mice are embryonic lethal at embryonic day 12.5 (E12.5) and show massive hemorrhage in the central nervous system (CNS) with lack of hematopoiesis in the fetal liver.11-15 A recent study demonstrated that inactivation of AML1 in adult mice results in megakaryocyte maturation arrest, defect in T- and B-lymphocyte development, and increase in hematopoietic precursor cells.16
The EVI1 gene was initially identified as the frequent retrovirus integration site in myeloid tumors in AKXD mice.17 EVI1 is less detected in normal hematopoietic cells but highly expressed in some patients of MDS or acute myelogenous leukemia (AML).18,19 EVI1 has 2 zinc finger domains, one in the N-terminal and the other in the C-terminal region. EVI1 is reported to interfere with the transforming growth factor β (TGFβ) signaling and to antagonize its growth inhibitory effect through targeting an intracellular signal transducer Smad3.20 EVI1 is also known to enhance activator protein 1 (AP-1) activity21 or block c-Jun N-terminal kinase (JNK) activity.22 These findings suggest the versatile nature of this molecule in malignant transformation of hematopoietic cells.
The molecular characterization of AML1/EVI1 points to 2 major mechanisms of its leukemogenic effect; one is the dominant suppression of the functions of wild-type AML1, and the other is the ectopic expression of EVI1 molecule in the hematopoietic cells.23 AML1/EVI1 binds to the PEBP2 site through its Runt domain and suppresses the expression of AML1 target genes by recruiting corepressor C-terminal binding protein by the EVI1 portion.24,25 This dominant-negative effect is a common feature among AML1-related chimeric molecules, including AML1/ETO (eight-twenty-one) in t(8;21)(q22;q22) and translocation ets leukemia (TEL)/AML1 in t(12;21)(p13;q22).9,23,26-29 By suppressing the expression of canonical AML1 target genes, AML1/EVI1 distorts the normal function of AML1 and may lead to leukemia development. The second mechanism of leukemogenesis is brought by the EVI1 part; inhibition of TGFβ signaling,30 stimulation of AP-1 activity,31 or repression of JNK activity.22 Thus, EVI1 portion also contributes to the oncogenicity of AML1/EVI1.
Of the AML1-fusion proteins, AML1/EVI1 and AML1/ETO are of particular interest. They show a similar structure; N-terminal half of AML1, including the Runt domain, is fused to almost the entire of EVI1 or ETO, both of which are the zinc finger protein.23,28,29 Moreover, AML1/EVI1 and AML1/ETO mediate dominant-negative effects over wild-type AML1 by recruiting corepressors via EVI123 and ETO portions,28,29 respectively. However, despite such similarities in molecular structure and function, AML1/EVI1 and AML1/ETO are differentially associated with the disease phenotype. AML1/EVI1 aggravates MDS or CML,1-4 while AML1/ETO develops de novo AML subtype M2 of the French-American-British classification.29 A possible explanation for such different disease settings is that AML1/EVI1 and AML1/ETO perturb different signaling pathway. Thus, exploring the molecular functional differences between AML1/EVI1 and AML1/ETO in vivo would advance our understanding toward differential role of the chimeric transcription molecules in causing different forms of leukemia.
For a direct comparison of AML1/EVI1- and AML1/ETO-expressing animals, we created AML1/EVI1 knock-in mice and compared its phenotype with that of AML1/ETO knock-in mice described previously.32,33 AML1/EVI1/+ embryos died around E13.5 resulting from hemorrhage in the brain and the spinal cord as a result of impaired definitive hematopoiesis in the fetal liver. However, E13.5 fetal liver contained hematopoietic progenitors with enhanced replating efficiency. These phenotypes are shared between AML1/EVI1/+ and AML1/ETO/+ embryos. On the other hand, unique to AML1/EVI1/+ mice was that hematopoietic progenitors in the fetal liver cannot develop to erythroid cells but can develop into dysplastic myeloid and megakaryocytic cells, whereas hematopoietic progenitors in the AML1/ETO/+ fetal liver produce cells of all 3 lineages with normal morphology. Expression analysis for the crucial genes revealed that AML1/EVI1/+ fetal liver maintained normal level of the PU.1 gene expression, but lacked expression of the LMO2 and SCL genes. Normal expression of the PU.1 gene may support maintenance of the progenitor cells capable of differentiating into the myeloid and megakaryocytic lineages, and the low levels of LMO2 and SCL genes expression could lead to blockade of erythroid differentiation in AML1/EVI1/+ fetal liver. Dysplastic hematopoiesis seen in AML1/EVI1/+ embryo partly recapitulates that observed in patients with the t(3;21)(q26;q22) translocation, indicating the pathogenic role for this chimeric protein in t(3;21)(q26;q22)-related leukemia.
Materials and methods
Construction of AML1/EVI1 knock-in targeting vector
Murine Aml1 genome derived from a genomic library was amplified by polymerase chain reaction (PCR) method to produce a DNA fragment that includes intron 4 and the first 94 base pair (bp) of exon 5 with silent mutations at codons 519 and 522 of exon 5 creating a SacII cleavage site. In the same fashion, another DNA fragment including the last 16 bp of human AML1 exon 5 and 5′ part of subsequent EVI1 sequence was also amplified using human AML1/EVI1 cDNA5 as a template to create a SacII site by introducing silent mutations at identical codons. These 2 PCR products were subcloned into the human AML1/EVI1 expression plasmid, pME18S-AML1/EVI1,5 to give an in-frame fusion of the first 94 bp of murine Aml1 exon 5 to the following portion of human AML1/EVI1 cDNA (Figure 1A). The Neo-resistant gene derived from pBK-Neo32 was added downstream of the AML1/EVI1 sequence. A 4.1-kb (kilobase) XbaI-AvrII fragment from intron 4 and a 4.3-kb EcoRI fragment from intron 5 of murine Aml1 genome were used for homologous recombination. To create the final targeting vector, the DNA fragment containing the previously mentioned sequences was inserted into the NotI and ClaI sites of pBluescript-DTA (diphtheria toxin-A gene), in which the Dta gene from pMC1DTApolyA (Stratagene, La Jolla, CA) was subcloned into the SalI site of pBluescript. The construct was verified by restriction endonuclease mapping and DNA sequencing.
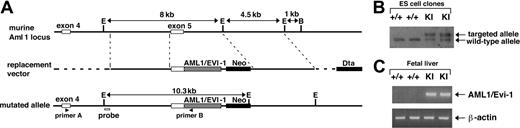
Gene targeting strategy to introduce AML1/EVI1 chimeric gene into the murine AML1 locus. (A) Schematic presentation of partial murine Aml1 genomic locus (top line), replacement vector containing a partial human AML1/EVI1 cDNA, a neomycin resistance cassette (Neo) for positive selection and a diphtheria toxin-A cassette (Dta) for negative selection (middle line), and the targeted allele (bottom line). (B) Southern analysis of wild-type (+/+) and AML1/EVI1 knock-in (KI) ES cell clones. For detecting homologous recombination, the murine Aml1-specific probe indicated in the lower line of panel A was used. (C) RT-PCR analysis on E12.5 fetal liver cells from wild-type (+/+) and KI embryos. AML1/EVI1 mRNA was amplified using primers A and B indicated in the lower line of panel A. Amplification was also performed for β-actin mRNA as a control for the presence of amplifiable RNA.
Gene targeting strategy to introduce AML1/EVI1 chimeric gene into the murine AML1 locus. (A) Schematic presentation of partial murine Aml1 genomic locus (top line), replacement vector containing a partial human AML1/EVI1 cDNA, a neomycin resistance cassette (Neo) for positive selection and a diphtheria toxin-A cassette (Dta) for negative selection (middle line), and the targeted allele (bottom line). (B) Southern analysis of wild-type (+/+) and AML1/EVI1 knock-in (KI) ES cell clones. For detecting homologous recombination, the murine Aml1-specific probe indicated in the lower line of panel A was used. (C) RT-PCR analysis on E12.5 fetal liver cells from wild-type (+/+) and KI embryos. AML1/EVI1 mRNA was amplified using primers A and B indicated in the lower line of panel A. Amplification was also performed for β-actin mRNA as a control for the presence of amplifiable RNA.
Production of chimeric mice
The linearized targeting vector (50 μg) was electroporated into TT2 embryonic stem (ES) cells.34 G418-resistant clones were analyzed for homologous recombination by Southern analysis hybridizing EcoRI-digested DNA with a probe 5′ to the 5′ homologous sequence in the targeting vector and a Neo gene probe (Figure 1B). Heterozygous AML1/EVI1/+ clones with undifferentiated morphology were injected into morulas of the ICR mouse strain. Chimeric males were bred with C57BL/6 females, and whole-body specimens of resultant embryos were analyzed for the presence of the knock-in allele by Southern analysis.
Visual inspection and histologic analysis
Embryos were removed from the uterus, dissected free from the fetal membranes, and inspected under a dissecting microscope for evidence of gross abnormalities. Embryos were fixed in formaldehyde solution and embedded in paraffin. The sections were stained with hematoxylin and eosin solution. Peripheral blood was collected in 10 mM EDTA (ethylenedia-minetetraacetic acid), and smears were stained with Wright-Giemsa solution.
Histologic images were obtained on either an Olympus AX80 microscope (Olympus, Tokyo, Japan) equipped with an Olympus DP70 digital camera, a Nikon Eclipse E600 microscope (Nikon, Tokyo, Japan) equipped with an Olympus DP12 digital camera, or an Olympus IMT-2 microscope equipped with an Olympus DP12 digital camera. Each microscope was equipped with a 10 ×/.22 ocular lens. Images were cropped in Adobe Photoshop CS (Adobe Systems, San Jose, CA) and composed in Canvas 9 (Polaroid, Waltham, MA).
Cell culture, transfection, and induction of megakaryocytic differentiation
K562 cells were maintained in RPMI-1640 supplemented with 10% fetal calf serum. To obtain bulk transfectant of mock or the AML1/EVI1 cDNA, 1 × 107 K562 cells were electroporated with 50 μg pCXN2 or pCXN2-AML1/EVI1 at 380 V and 975 μF using Gene Pulser (Bio-Rad, Hercules, CA) and selected with 0.4 mg/mL G418 (Sigma-Aldrich, St Louis, MO). Resistant cells were screened for expression of AML1/EVI1 chimeric proteins by Western analysis using anti-AML1 Ab-1 antibody (Oncogene Research Products, San Diego, CA). To induce megakaryocytic differentiation, 100 nM staurosporine (Sigma-Aldrich) was added to the culture.
DNA content analysis
Each mock or AML1/EVI1-expressing K562 bulk population treated with 100 nM staurosporine was washed, fixed, and stained with 50 mg/mL propidium iodide using Cycle TEST PLUS (BD Biosciences, San Jose, CA). DNA content of nuclei was determined by fluorescence-activated cell sorting analysis.
Ultrastructural study
E13.5 fetal livers of AML1/EVI1/+ heterozygous embryos and their wild-type littermates were examined with a JEOL 1200CX electron microscope (JEOL, Tokyo, Japan). Each mock or AML1/EVI1-expressing bulk population treated with 100 nM staurosporine for 48 hours was also examined.
In vitro culture of hematopoietic cells
Fetal livers from E12.5 or E13.5 embryos were disrupted by passing through 21-gage needles and then 26-gage needles for 3 times each. Single-cell suspensions with 1 × 104 cells were plated in triplicate in 1.2% methylcellulose in Iscoves modified Dulbecco medium containing 30% fetal calf serum, 0.1 mM β-mercaptoethanol, 2 mM glutamine, and 1% bovine serum albumin, 5 U/mL human erythropoietin, 100 ng/mL murine stem cell factor, 5 ng/mL murine interleukin 3 (IL-3; Kirin Brewery, Tokyo, Japan). Cell aggregates containing more than 50 cells were counted as colonies. Cytocentrifuge preparations of hematopoietic colonies were stained with Wright-Giemsa for morphologic examination, benzidine for the presence of erythroblasts, and choline esterase for the presence of megakaryocytes. In replating experiments, all of the cells from the preceding culture were collected, washed, and then replated at 1 × 104 cells per methylcellulose plate under the same condition as in the preceding culture. Colonies were scored as previously mentioned.
Quantitative reverse transcriptase (RT)-PCR analysis.
E12.5 and E13.5 live embryos were selected on the basis of the presence of heartbeats. Total RNA was isolated from liver cells using RNAeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Random hexamer-primed cDNA was prepared from 2 μg total RNA with mouse mammary leukemia virus reverse transcriptase (Gibco BRL, Grand Island, NY) in a total volume of 25 μL. An oligonucleotide primer set containing murine Aml1 exon 5-derived sense (5′-AAGAGCTTCACTCTGACCAT-3′) and human EVI1 noncoding region-derived antisense (5′-CCTTTCACCTACTTCGATCT-3′) sequences was used for amplification of the AML1/EVI1 fusion sequence. Amplifications of various hematopoietic regulator transcripts were performed with primer sets described previously35-39 as well as that for murine CD11b, sense (5′-CAGATCAACAATGTGACCGTATGG-3′) and antisense (5′-CATCATGTCCTTGTACTGCCGC-3′) primers, using CYBR Green PCR Master Mix and ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster, CA). Parallel reactions were carried out using a β-actin-amplifying primer set to ensure the integrity of the RNA samples.
Quantification of mRNA level was performed by measuring the cycle threshold (CT) that is defined as the fractional cycle number at which the fluorescence encounters a fixed threshold. The CT value of each gene was normalized to that of β-actin (ΔCT; CT value of target gene minus CT value of β-actin). Results were expressed as difference of ΔCT between AML1/EVI1/+ versus wild-type or AML1-/- versus wild-type animals, and fold expression.
Results
Generation of an AML1/EVI1 fusion allele
The t(3;21) breakpoint on chromosome 21 in human leukemic cells occurs in the fifth intron of the AML1 gene, which is 3′ to the exons encoding the Runt DNA-binding domain.5 To mimic in the mouse genome the mutant AML1/EVI1 chimeric gene at the human t(3;21) allele, we made a targeting construct that replaces murine Aml1 exon 5 with human AML1 exon 5 fused in frame to the human EVI1 sequence (Figure 1A). This targeting strategy renders generation of the murine/human hybrid AML1/EVI1 gene whose expression is controlled by the endogenous mouse Aml1 promoter. Using electroporation, we obtained 8 ES cell clones targeted correctly (no. 31, no. 35, no. 37, no. 42, no. 47, no. 50, no, 70, and no. 97), as determined by the presence of both AML1/EVI1 allele and endogenous murine Aml1 allele in Southern analysis (Figure 1B). These AML1/EVI1/+ ES cell clones were injected into mouse morulas, and 2 chimeric mice (no. 31 and no. 97) transmitted the mutant allele through the germ line. These chimeric mice were largely healthy up to 7 months but showed sudden death thereafter. The cause of death is unknown despite anatomic examination performed immediately after the death of the 5 chimeric mice; there were no signs of hepatosplenomegaly or lymphadenopathy, excluding at least the leukemic cause of death.
Knock-in expression of AML1/EVI1 fusion gene results in embryonic lethality with CNS hemorrhage and a lack of fetal-liver hematopoiesis
Genotyping analysis of 42 delivered F1 pups of AML1/EVI1 chimeric male and wild-type C57BL/6 female found no AML1/EVI1/+ genotypes, suggesting that AML1/EVI1/+ embryos died in the uteri. AML1+/+ agouti mice appeared normal; thus, designated major mutations did not occur in the ES cells. To examine the timing and cause of embryonic death in AML1/EVI1/+ heterozygous embryos, we killed pregnant females at various time points between E10.5 and E14.5. The AML1/EVI1 chimeric mRNA was detected in the fetal liver of the heterozygous mice by RT-PCR (Figure 1C). Between E10.5 and E11.5, all the AML1/EVI1/+ embryos were viable and showed no significant morphologic abnormalities (Table 1). However, 10% of the embryos was found dead at E12.5 and 100% by E14.5, as judged by the absence of heart beats. The size of the AML1/EVI1/+ embryos between E12.5 and E13.5 was comparable to that of the control littermates (Figure 2A). However, they had noticeable white livers, and signs of massive hemorrhage in the CNS and intersegmental regions of the presumptive spinal cord. Microscopic analysis revealed that the hemorrhage occurred as early as E12.5 in the cerebral ventricle and the dorsal root ganglia (Figure 2B-E), and such hemorrhagic sites were somewhat conserved in AML1/EVI1/+ embryos analyzed (n = 58). This hemorrhage and lethality of AML1/EVI1/+ heterozygous embryos were observed in 2 independent ES cell lines (no. 31 and no. 97). These data indicate that AML1/EVI1/+ embryos die around E13.5 from the massive hemorrhage in the CNS. We focused our further analysis on embryos derived from line no. 97 because of the efficient germ line transmission and propagation relative to those from line no. 31.
Genotype and phenotype of embryos derived from AML1/EVI1 chimeric males mated with normal females
. | . | Genotype . | . | Phenotype of KI*mice . | . | ||
|---|---|---|---|---|---|---|---|
| Stages . | No. of Pups . | +/+ . | +/KI* . | Hemorrhage . | Death . | ||
| E10.5 | 9 | 1 | 8 | 0 | 0 | ||
| E11.5 | 14 | 11 | 3 | 0 | 0 | ||
| E12.5 | 98 | 67 | 31 | 27 | 3 | ||
| E13.5 | 78 | 62 | 16 | 16 | 10 | ||
| E14.5 | 32 | 21 | 11 | 11 | 11 | ||
. | . | Genotype . | . | Phenotype of KI*mice . | . | ||
|---|---|---|---|---|---|---|---|
| Stages . | No. of Pups . | +/+ . | +/KI* . | Hemorrhage . | Death . | ||
| E10.5 | 9 | 1 | 8 | 0 | 0 | ||
| E11.5 | 14 | 11 | 3 | 0 | 0 | ||
| E12.5 | 98 | 67 | 31 | 27 | 3 | ||
| E13.5 | 78 | 62 | 16 | 16 | 10 | ||
| E14.5 | 32 | 21 | 11 | 11 | 11 | ||
Genotype analysis was performed by Southern analysis of DNA extracted from embryos. Surviving embryos were defined as those with beating hearts at the time of dissection.
KI indicates AML1/EVI1 knock-in.
Morphologic comparison of E12.5 wild-type (WT) and AML1/EVI1/+ heterozygous embryos. (A) External appearance of WT and AML1/EVI1/+ littermates. AML1/EVI1/+ embryo on the right is similar in size to WT littermate on the left but is clearly identifiable by the presence of fetal liver pallor and massive hemorrhage within the CNS and soft tissues in the back (objective lens [OL], 2 ×/0.05; original magnification [OL], × 20). (B-C) Sections of the cerebral ventricle from WT (B) and AML1/EVI1/+ (C) embryos. AML1/EVI1/+ embryo shows massive hemorrhage into the ventricle (OL, 10 ×/0.40; OM, × 100). (D-E) Sections of the dorsal root ganglia from WT (D) and AML1/EVI1/+ (E) embryos. AML1/EVI1/+ embryo shows hemorrhage in the ganglia (OL, 10 ×/0.40; OM, × 100). (F-G) Smears of the peripheral blood from WT (F) and AML1/EVI1/+ (G) embryos. AML1/EVI1/+ embryo shows the absence of definitive erythrocytes, while WT littermate shows numerous enucleated definitive erythrocytes (OL, 40 ×/0.65; OM, × 400). (H-I) Smears of the peripheral blood from WT embryo. Only WT littermate shows monocyte (H) and neutrophil (I) (OL, 100 ×/1.40; OM, × 1000). (J-K) Sections of the fetal liver from WT (J) and AML1/EVI1/+ (K) embryos. AML1/EVI1/+ embryo shows a near complete absence of hematopoietic precursors, while WT littermate shows numerous hematopoietic precursor cells (OL, 20 ×/0.70; OM, × 200).
Morphologic comparison of E12.5 wild-type (WT) and AML1/EVI1/+ heterozygous embryos. (A) External appearance of WT and AML1/EVI1/+ littermates. AML1/EVI1/+ embryo on the right is similar in size to WT littermate on the left but is clearly identifiable by the presence of fetal liver pallor and massive hemorrhage within the CNS and soft tissues in the back (objective lens [OL], 2 ×/0.05; original magnification [OL], × 20). (B-C) Sections of the cerebral ventricle from WT (B) and AML1/EVI1/+ (C) embryos. AML1/EVI1/+ embryo shows massive hemorrhage into the ventricle (OL, 10 ×/0.40; OM, × 100). (D-E) Sections of the dorsal root ganglia from WT (D) and AML1/EVI1/+ (E) embryos. AML1/EVI1/+ embryo shows hemorrhage in the ganglia (OL, 10 ×/0.40; OM, × 100). (F-G) Smears of the peripheral blood from WT (F) and AML1/EVI1/+ (G) embryos. AML1/EVI1/+ embryo shows the absence of definitive erythrocytes, while WT littermate shows numerous enucleated definitive erythrocytes (OL, 40 ×/0.65; OM, × 400). (H-I) Smears of the peripheral blood from WT embryo. Only WT littermate shows monocyte (H) and neutrophil (I) (OL, 100 ×/1.40; OM, × 1000). (J-K) Sections of the fetal liver from WT (J) and AML1/EVI1/+ (K) embryos. AML1/EVI1/+ embryo shows a near complete absence of hematopoietic precursors, while WT littermate shows numerous hematopoietic precursor cells (OL, 20 ×/0.70; OM, × 200).
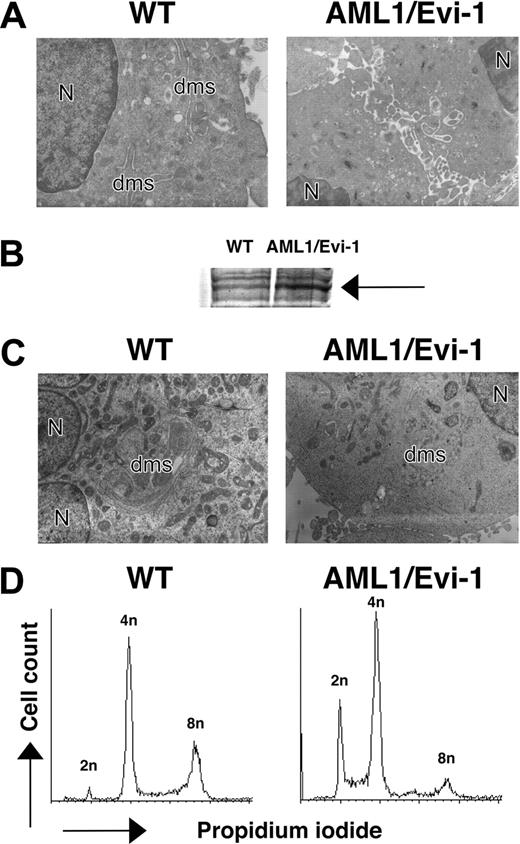
Yolk sac hematopoiesis appeared largely intact in AML1/EVI1/+ embryos, with normal number and morphology of the yolk sac-derived nucleated erythroblasts circulating in the peripheral vessel (Figure 2G). Erythroblasts in E12.5 AML1/EVI1/+ embryos looked similar to those in E11.5 wild-type littermates, suggesting that they normally developed in the yolk sac. In contrast, fetal liver hematopoiesis was severely impaired, as indicated by markedly whitish liver resulting from a lack of red cells. The microscopic examination showed near complete absence of the erythroid, myeloid, or megakaryocytic progenitors of the definitive origin (Figure 2K). Postenucleated erythrocytes were absent in the peripheral blood from E12.5 AML1/EVI1/+ embryos, whereas it was abundantly observed in the peripheral blood of E12.5 wild-type littermates (Figure 2F-G). Also, monocytes and neutrophils were absent in the smears from AML1/EVI1/+ embryos, whereas they were easily observed in the smears from wild-type littermates (Figure 2H-I). These findings indicate that AML1/EVI1/+ heterozygous mice fail to establish definitive hematopoiesis in the fetal liver but maintain primary hematopoiesis in the yolk sac, recapitulating hematopoietic defects observed in AML1-/-11,12 or AML1/ETO/+ mice.32,33 We further performed electron microscopic examination to search for hematopoietic progenitors that would have been missed by histologic microscopic examination in the E13.5 fetal liver. In addition to decreased erythroid and myeloid progenitors, we observed megakaryocytes that were defective in their demarcation membrane (Figure 3A), which resemble those in AML1 conditional knock-out mice.16 Thus, although there is massive defect in fetal liver hematopoiesis, some hematopoietic progenies do exist, although dysplastic, in the AML1/EVI1/+ fetal liver.
Morphology and DNA content of megakaryocytic cells overexpressing AML/EVI1. (A) Electron micrographs of megakaryocytes in E13.5 fetal liver. N indicates nucleus; dms, demarcation membrane (OM, × 3000). (B) Expression of AML1/EVI1 chimeric protein in transfected K562 cells. An arrow indicates overexpressed AML1/EVI1 proteins. (C) Electron micrographs of mock or AML1/EVI1-overexpressing K562 cells after staurosporine treatment for 48 hours. N indicates nucleus; dms, demarcation membrane (OM, × 3000). (D) DNA content of mock or AML1/EVI1-overexpressing K562 cells after staurosporine treatment for 48 hours.
Morphology and DNA content of megakaryocytic cells overexpressing AML/EVI1. (A) Electron micrographs of megakaryocytes in E13.5 fetal liver. N indicates nucleus; dms, demarcation membrane (OM, × 3000). (B) Expression of AML1/EVI1 chimeric protein in transfected K562 cells. An arrow indicates overexpressed AML1/EVI1 proteins. (C) Electron micrographs of mock or AML1/EVI1-overexpressing K562 cells after staurosporine treatment for 48 hours. N indicates nucleus; dms, demarcation membrane (OM, × 3000). (D) DNA content of mock or AML1/EVI1-overexpressing K562 cells after staurosporine treatment for 48 hours.
To test for the direct effect of AML1/EVI1 molecule in megakaryocyte morphology, we expressed AML1/EVI1 cDNA in human leukemic cell line K562 (Figure 3B) and differentiated one bulk population into the megakaryocytic lineage by exposing to staurosporine.40 The AML1/EVI1-expressing K562 cells treated with staurosporine showed poorly developed demarcation membrane and lower level of polyploidy as compared with control mock cells (Figure 3C), indicating that the expression of AML1/EVI1 molecule is causative for the aberrant maturation of megakaryocytes in AML1/EVI1/+ mice.
AML1/EVI1 generates dysplastic hematopoietic progenitors in the fetal liver
To analyze for the defective hematopoiesis in the AML1/EVI1/+ embryos, we performed methylcellulose colony-forming assay using fetal liver cells from E12.5 or E13.5 embryos. Because the total number of cells recovered from E12.5 or E13.5 AML1/EVI1/+ fetal livers was 20-fold less than that recovered from wild-type fetal livers, we adjusted the number of plated fetal liver cells (1 × 104 cells) to assess for the precursor frequencies. In day 10 of culture, wild-type fetal liver gave rise to mutilineage colonies, whereas E12.5 AML1/EVI1/+ fetal liver cells generated macrophage colonies only, with no detectable erythroid, myeloid, and mixed colonies (Table 2). The E13.5 AML1/EVI1/+ cell culture looked similar to that of E12.5, except for some mixed-like colonies that were 5-fold more numerous than those from wild-type cell culture in microscopic examination. These data indicate that expression of AML1/EVI1 leads to severe defects in definitive hematopoiesis, but the fetal liver contains some progenitors capable of differentiating into multilineage hematopoietic progenies.
Hematopoietic progenitors in E12.5 and E13.5 fetal liver
Stages/genotype . | No. of Pups . | BFU-E . | CFU-GM . | CFU-GEMM . | CFU-M . |
|---|---|---|---|---|---|
| E12.5 | |||||
| +/+ | 13 | 10 ± 3 | 32 ± 8 | 20 ± 9 | 8 ± 2 |
| AML1/EVI1/+ | 6 | 0 | 0 | 0 | 25 ± 11 |
| E13.5 | |||||
| +/+ | 9 | 8 ± 3 | 58 ± 14 | 25 ± 11 | 10 ± 3 |
| AML1/EVI1/+ | 5 | 0 | 0 | 154 ± 54 | 4 ± 3 |
Stages/genotype . | No. of Pups . | BFU-E . | CFU-GM . | CFU-GEMM . | CFU-M . |
|---|---|---|---|---|---|
| E12.5 | |||||
| +/+ | 13 | 10 ± 3 | 32 ± 8 | 20 ± 9 | 8 ± 2 |
| AML1/EVI1/+ | 6 | 0 | 0 | 0 | 25 ± 11 |
| E13.5 | |||||
| +/+ | 9 | 8 ± 3 | 58 ± 14 | 25 ± 11 | 10 ± 3 |
| AML1/EVI1/+ | 5 | 0 | 0 | 154 ± 54 | 4 ± 3 |
Numbers represent colonies per 1 × 104 fetal liver cells, mean ± SD.
BFU-E indicates erythroid burst-forming unit; CFU-GM, granulocyte/macrophage colony-forming unit; CFU-GEMM, granulocyte/erythrocyte/macrophage/megakaryocyte colony-forming unit; CFU-M, macrophage colony-forming unit.
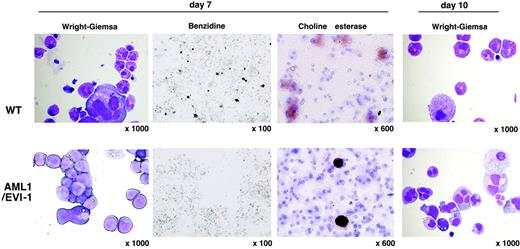
To gain more insights into the mixed-lineage-like colonies derived from E13.5 AML1/EVI1/+ fetal liver, we performed time-course microscopic analysis of the methylcellulose culture. On day 7 of culture, colonies from E13.5 AML1/EVI1/+ fetal liver consisted mainly of myelocytic blastoid cells, while those from wild-type fetal liver contained numerous erythroblasts, hypergranular myeloid cells, and megakaryocytes (Figure 4). Benzidine staining confirmed the absence of erythroblasts in AML1/EVI1/+ cell-derived colonies. However, choline esterase-positive megakaryocytes were observed in both types of the colonies. On day 10, colonies from AML1/EVI1/+ fetal liver contained a large number of dysplastic myelocytes with bilobulated or multilobulated nuclei along with scanty cytoplasmic granulation. These findings suggest that hematopoietic progenitors in the AML1/EVI1/+ fetal liver are completely blocked for differentiation into the erythroid lineage but are capable of differentiating into the myeloid and megakaryocyte lineages, although immature and dysplastic.
Morphology of dysplastic mixed-like hematopoietic colonies in the AML1/EVI1/+ fetal liver culture. On days 7 and 10 of culture, cytocentrifuge preparations of cells from mixed-like colonies were stained with Wright-Giemsa for morphologic examination (OL, 100 ×/1.40; OM, × 1000), benzidine for the presence of erythroblasts (OL, 10 ×/0.30; OM, × 100), or choline esterase for the presence of megakaryocytes (OL, 60 ×/1.40; OM, × 600). AML1/EVI1/+ mixed-like colonies in the bottom panels show retarded maturation and hypogranulation of myeloid cells as well as the absence of erythroid cells. In contrast, WT mixed colonies in the top panels show normal trilineage differentiation.
Morphology of dysplastic mixed-like hematopoietic colonies in the AML1/EVI1/+ fetal liver culture. On days 7 and 10 of culture, cytocentrifuge preparations of cells from mixed-like colonies were stained with Wright-Giemsa for morphologic examination (OL, 100 ×/1.40; OM, × 1000), benzidine for the presence of erythroblasts (OL, 10 ×/0.30; OM, × 100), or choline esterase for the presence of megakaryocytes (OL, 60 ×/1.40; OM, × 600). AML1/EVI1/+ mixed-like colonies in the bottom panels show retarded maturation and hypogranulation of myeloid cells as well as the absence of erythroid cells. In contrast, WT mixed colonies in the top panels show normal trilineage differentiation.
Hematopoietic progenitor in the AML1/EVI1/+ fetal liver is highly capable of self-renewal
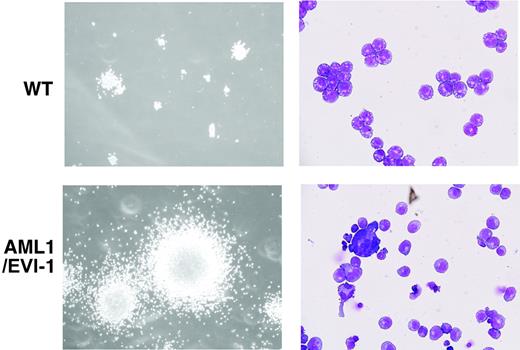
To examine self-renewal capacity of hematopoietic progenitors in the E13.5 AML1/EVI1/+ fetal liver, we performed a serial methylcellulose replating assay using bulk population of fetal liver-derived colonies under the conditions optimal for the development of multipotential hematopoietic progenitors. Cells from AML1/EVI1/+ fetal livers were able to replate beyond 7 passages, whereas cells from wild-type fetal livers lost their ability to form colonies after 5 passages (Table 3). During each replating, AML1/EVI1-expressing colonies showed increase in size and number (Figure 5, left). The cytospin samples from the fourth passage colony identified a polymorphic population of dysplastic cells, including agranular myelocytes and megakaryocytes (Figure 5, right). We conclude that expression of AML1/EVI1 in the fetal liver produces multilineage potential progenitors that are capable of more self-renewal than the wild-type progenitors and have a dysplastic maturation tendency. However, even after amplification of the cells through passage, we failed to demonstrate expression of AML1/EVI1 chimeric proteins in the colony-forming cells by Western analysis with anti-AML1 antibody, possibly because of low expression and high molecular weight (190 kDa) of the proteins.
Serial in vitro replating assays of E13.5 fetal liver hematopoietic progenitors
. | In vitro passage . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | ||||||
| +/+ | 99 ± 23 | 384 ± 86 | 164 ± 41 | 59 ± 76 | 11 ± 9 | 0 | 0 | ||||||
| AML1/EVI1/+ | 162 ± 59 | 388 ± 131 | 365 ± 150 | 403 ± 89 | 436 ± 104 | 407 ± 114 | 626 ± 3 | ||||||
. | In vitro passage . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | ||||||
| +/+ | 99 ± 23 | 384 ± 86 | 164 ± 41 | 59 ± 76 | 11 ± 9 | 0 | 0 | ||||||
| AML1/EVI1/+ | 162 ± 59 | 388 ± 131 | 365 ± 150 | 403 ± 89 | 436 ± 104 | 407 ± 114 | 626 ± 3 | ||||||
Cells from methylcellulose culture were disaggregated, washed, and replated at 1 × 104 cells per methylcellulose plate under the same condition every 7 days. Numbers represent colonies per 1 × 104 cells, mean ± SD.
Expression of PU.1 is maintained in AML1/EVI1/+ fetal liver cells but significantly decreased in AML1-/- cells
To get the molecular underpinning for the dysregulated hematopoiesis caused by AML1/EVI1, we assessed the expression of various genes implicated in definitive hematopoiesis. The total RNA was extracted from E12.5 fetal liver cells from wild-type, AML1-/-, and AML1/EVI1/+ embryos, and mRNA was quantified using semiquantitative RT-PCR analysis. The level of expression in AML1-/- and AML1/EVI1/+ fetal liver cells was compared with that in wild-type cells. Table 4 revealed the calculated fold expression of the indicated genes in AML1-/- and AML1/EVI1/+ embryos relative to that of wild-type littermates along with the cycle threshold differences. The expression of the PU.141 gene in AML1/EVI1/+ fetal liver cells was maintained to the normal, whereas its expression level was markedly decreased in AML1-/- fetal liver cells, as has been reported previously.42 Consistent with this, the expression of the CD11b gene, one of the target genes of PU.1 (an ets transcription factor family member), was maintained in AML1/EVI1/+ knock-in liver, whereas markedly decreased in AML1-/- liver. The expression of the critical transcription factor such as PU.1 in AML1/EVI1/+ fetal liver may support multilineage hematopoietic progenitors up to E13.5 and also aids the monocyte/macrophage lineage differentiation. The myeloid lineage-specific transcription factor C/EBPA43 was expressed several fold higher in AML1/EVI1/+ fetal liver cells than in wild-type cells, and this may bolster the myeloid-lineage differentiation along with PU.1. The expression levels of cytokine receptors transcripts such as G-CSF receptor and GM-CSF receptor were largely unchanged across wild-type, AML1-/- and AML1/EVI1/+ fetal liver cells. On the other hand, the expression of erythroid lineage-specific genes such as LMO244 and SCL45 was approximately 10-fold lower in AML1/EVI1/+ fetal liver cells than in wild-type cells. The decreased expression of LMO2 and SCL could impinge on successful differentiation along the erythroid lineage in AML1/EVI1/+ fetal liver cells, and the similar decrease was observed in AML1-/- fetal liver cells that lack erythroid cells as well. Other erythroid lineage-specific transcripts such as δ-aminolevulinic acid synthaseerythroid (ALASE) and β-globin were more decreased in AML1/EVI1/+ fetal liver cells than in AML1-/- cells, despite equivalent expression of their master regulator GATA binding protein 1 (GATA-1) in both cell types. The net GATA-1 activity was presumably repressed in AML1/EVI1/+ fetal liver cells because of maintained expression of PU.1 that inhibits DNA binding of GATA-1.46 The suppressed expression of ALASE and β-globin could also contribute to the complete defect of erythroid differentiation in AML1/EVI1/+ progenitor cells. These expression analyses provide a logical explanation for the distinct hematopoietic capacity intrinsic to AML1-/- and AML1/EVI1/+ fetal liver cells. Our data point that the sufficient expression of the PU.1 gene may be the first critical prerequisite for the definitive hematopoiesis to be set off in the fetal liver.
Expressional changes of various hematopoietic regulators in E12.5 AML1/EVI1/+ and AML1−/− fetal liver cells
. | ΔCTKI - ΔCTWT . | Fold (KI/WT) . | ΔCTKO - ΔCTWT . | Fold (KO/WT) . |
|---|---|---|---|---|
| C/EBPA | −1.4 ± 0.1 | 2.7 ± 0.1 | −2.9 ± 0.0 | 7.2 ± 0.1 |
| LMO2 | 3.5 ± 0.3 | 0.1 ± 0.6 | 3.3 ± 0.1 | 0.1 ± 0.3 |
| SCL | 3.9 ± 0.8 | 0.1 ± 1.7 | 2.4 ± 0.2 | 0.2 ± 0.3 |
| c-MYB | −0.5 ± 0.2 | 1.4 ± 1.5 | −2.4 ± 0.3 | 5.4 ± 0.6 |
| GATA1 | −0.4 ± 0.7 | 1.4 ± 1.3 | −1.6 ± 0.5 | 3.0 ± 1.0 |
| PU.1 | −0.4 ± 0.1 | 1.3 ± 0.1 | 8.2 ± 0.1 | 0.0 ± 0.3 |
| NFE2 | 2.9 ± 0.5 | 0.1 ± 1.0 | 0.3 ± 0.2 | 0.6 ± 0.2 |
| CD11b | 1.2 ± 0.4 | 0.4 ± 0.8 | 5.0 ± 1.8 | 0.0 ± 3.6 |
| MCSFR | 0.1 ± 0.2 | 0.9 ± 0.4 | −1.5 ± 0.9 | 2.8 ± 1.8 |
| GCSFR | −0.6 ± 0.1 | 1.5 ± 0.2 | 0.0 ± 0.0 | 1.0 ± 0.0 |
| GMCSFR | 0.1 ± 0.2 | 1.1 ± 0.4 | −0.6 ± 0.1 | 1.5 ± 0.2 |
| MPO | 10 ± 1.1 | 0.0 ± 2.2 | 7.0 ± 0.7 | 0.0 ± 1.3 |
| β-globin | 3.9 ± 0.9 | 0.1 ± 1.7 | 1.0 ± 0.4 | 0.5 ± 0.8 |
| FOG1 | 2.3 ± 0.2 | 0.2 ± 0.3 | 2.0 ± 0.2 | 0.3 ± 0.3 |
| ALASE | 5.6 ± 0.4 | 0.0 ± 0.7 | 1.9 ± 0.4 | 0.3 ± 0.8 |
. | ΔCTKI - ΔCTWT . | Fold (KI/WT) . | ΔCTKO - ΔCTWT . | Fold (KO/WT) . |
|---|---|---|---|---|
| C/EBPA | −1.4 ± 0.1 | 2.7 ± 0.1 | −2.9 ± 0.0 | 7.2 ± 0.1 |
| LMO2 | 3.5 ± 0.3 | 0.1 ± 0.6 | 3.3 ± 0.1 | 0.1 ± 0.3 |
| SCL | 3.9 ± 0.8 | 0.1 ± 1.7 | 2.4 ± 0.2 | 0.2 ± 0.3 |
| c-MYB | −0.5 ± 0.2 | 1.4 ± 1.5 | −2.4 ± 0.3 | 5.4 ± 0.6 |
| GATA1 | −0.4 ± 0.7 | 1.4 ± 1.3 | −1.6 ± 0.5 | 3.0 ± 1.0 |
| PU.1 | −0.4 ± 0.1 | 1.3 ± 0.1 | 8.2 ± 0.1 | 0.0 ± 0.3 |
| NFE2 | 2.9 ± 0.5 | 0.1 ± 1.0 | 0.3 ± 0.2 | 0.6 ± 0.2 |
| CD11b | 1.2 ± 0.4 | 0.4 ± 0.8 | 5.0 ± 1.8 | 0.0 ± 3.6 |
| MCSFR | 0.1 ± 0.2 | 0.9 ± 0.4 | −1.5 ± 0.9 | 2.8 ± 1.8 |
| GCSFR | −0.6 ± 0.1 | 1.5 ± 0.2 | 0.0 ± 0.0 | 1.0 ± 0.0 |
| GMCSFR | 0.1 ± 0.2 | 1.1 ± 0.4 | −0.6 ± 0.1 | 1.5 ± 0.2 |
| MPO | 10 ± 1.1 | 0.0 ± 2.2 | 7.0 ± 0.7 | 0.0 ± 1.3 |
| β-globin | 3.9 ± 0.9 | 0.1 ± 1.7 | 1.0 ± 0.4 | 0.5 ± 0.8 |
| FOG1 | 2.3 ± 0.2 | 0.2 ± 0.3 | 2.0 ± 0.2 | 0.3 ± 0.3 |
| ALASE | 5.6 ± 0.4 | 0.0 ± 0.7 | 1.9 ± 0.4 | 0.3 ± 0.8 |
ΔCT indicates mean CT of indicated gene - mean CT of β-actin; KI, AML1/EVI1 knock-in; KO, AML1 knock-out; fold, fold difference relative to that of WT litters calculated by 2-(ΔCTKI or KO - ΔCTWT). Data are means ± standard deviation.
Morphology of serially replated AML1/EVI1/+hematopoietic colonies. (Left) WT and typical dysplastic multilineage AML1/EVI1/+ colonies after the fourth passage. The size of AML1/EVI1/+ hematopoietic colonies is significantly larger than that of WT colonies. (Right) Wright-Giemsa-stained cytocentrifuge preparations of the WT and AML1/EVI1/+ colony. AML1/EVI1/+ hematopoietic colonies maintain the dysplastic nature. Scales bars equal 300 μm (OL, 100 ×/1.40; OM, × 1000).
Morphology of serially replated AML1/EVI1/+hematopoietic colonies. (Left) WT and typical dysplastic multilineage AML1/EVI1/+ colonies after the fourth passage. The size of AML1/EVI1/+ hematopoietic colonies is significantly larger than that of WT colonies. (Right) Wright-Giemsa-stained cytocentrifuge preparations of the WT and AML1/EVI1/+ colony. AML1/EVI1/+ hematopoietic colonies maintain the dysplastic nature. Scales bars equal 300 μm (OL, 100 ×/1.40; OM, × 1000).
Discussion
AML1-CBFβ transcription factor complex is one of the critical regulators of the hematopoietic system. A substantial amount of evidence highlights the central role for this transcription factor system in normal hematopoiesis as well as in abnormal hematopoietic disorders. Four gene-ablated animals including ours, AML1-null,11,12 CBFB-null,13-15 AML1/ETO knock-in,32,33 and AML1/EVI1 knock-in mice, show normal hematopoiesis in the yolk sac but share common defects in definitive hematopoiesis in the fetal liver. These 4 embryos also show massive hemorrhage in the CNS and spinal cord and die during midgestation. A series of biochemical analyses in AML1-CBFβ transcription factor, or AML1/ETO or AML1/EVI1 fusion molecules23,28,29 indicate that such in vivo phenotypes arise from shutting off or suppressing AML1 function in the early hematopoietic tissues. We speculate that such phenotype in AML1/EVI1 knock-in mice also comes from dominant-negative effect against wild-type-AML1 in vivo.
The microscopic analysis of the fetal liver of the 4 mutant embryos shows common lack of visible hematopoietic cells. Despite such gross similarity, close examination of the hematopoiesis reveals some intriguing differences. First, although AML1-/-11,12 or CBFB-/-13-15 fetal liver has no hematopoietic progenitors that would have produced colonies, AML1/EVI1/+ and AML1/ETO/+32,33 fetal livers have progenitors that can give rise to macrophage colonies. AML1/EVI1/+ fetal liver (E12.5) contains 3-fold more progenitors than wild-type fetal liver capable of differentiating into the monocyte/macrophage lineage, and a similar finding is reported for AML1/ETO/+ yolk sac where Yergeau et al32 observed a marked increase in the number of macrophage colonies. Therefore, the capacity to differentiate along the monocyte/macrophage lineage seems a characteristic feature of hematopoietic progenitors expressing AML1/EVI1 or AML1/ETO in the fetal hematopoietic system. Second, in contrast to AML1-/- or CBFB-/- fetal liver, AML1/EVI1/+ and AML1/ETO/+33 fetal livers (E13.5) can also give rise to colonies committed to erythrocyte/megakaryocyte and myelocyte lineages. The appearance of megakaryocytic cells in the colony assay would suggest some production of platelets in these embryos, and this might contribute to 1 day-longer survival of AML1/EVI1 and AML1/ETO knock-in embryos (lethal at E13.5) than AML1 or CBFB knock-out embryos (lethal at E12.5). Third, the serial colony-forming assay demonstrates that AML1/EVI1- or AML1/ETO33 -expressing progenitors show more self-renewal than those of the wild-type animals, although these progenitors cannot establish functional hematopoiesis in the fetal liver. These differences between AML1/EVI1- or AML1/ETO-expressing cells versus AML1 or CBFB knock-out cells may arise from either incomplete suppression of wild-type-AML1 or additional functions brought by the individual fused molecules.
Although AML1/EVI1/+ and AML1/ETO/+ fetal liver showed similar results in colony counts, there are some striking differences in their contents of colonies that are unlikely to result from minor differences in culture conditions. We used higher concentrations of erythropoietin and stem cell factor than those reported in AML1/ETO/+ fetal liver culture.32,33 Nonetheless, AML1/EVI1/+ hematopoietic progenitors fail to generate erythroid cells, whereas AML1/ETO/+ hematopoietic progenitors give rise to mixed colonies containing numerous erythroblasts.33 The observed resistance to the erythroid-lineage differentiation is highly characteristic of AML1/EVI1/+ hematopoietic progenitors. This is consistent with the reported retroviral expression of EVI1 in hematopoietic progenitor cells that blocks erythroid differentiation in vitro47 or the transgenic expression of EVI1 impairing erythropoiesis in vivo.48 Therefore, the expression of EVI1, either in canonical form or as the fusion chimeric protein, is detrimental to erythroid-lineage differentiation. Another distinct finding between the 2 cell types is the more severe dysplasticity in AML1/EVI1/+ cells. In AML1/EVI1/+ progenitors, the arising myeloid cells arrest at the myelocyte stage with no granulation, and the arising megakaryocytes show ill-developed demarcation membrane in the cytoplasm. In contrast, AML1/ETO/+ progenitors exhibited far more differentiated morphologies, although reported dysplastic in the text.33 Overexpressed EVI1 protein in G-CSF-treated 32Dcl3 cells completely blocks myeloid differentiation.49 Thus, the adverse effect of EVI1 in myeloid differentiation may contribute to the more severe defect in the myeloid maturation of the AML1/EVI1-expressing progenitors than that of the AML1/ETO-expressing cells. Finally, the abnormal expression of the AML1/EVI1 gene causes disturbance of megakaryocytic maturation, which is not observed in AML1/ETO-expressing animals. This effect seems direct as evident by AML1/EVI1-expressing K562 cells showing poor demarcation membrane and lower DNA ploidy when treated with staurosporine. In this case, however, the result seems to be caused by the dominant-negative effect over wild-type-AML1, as AML1 conditional knock-out mice show similar maturation abnormality in megakaryocytes.
AML1/EVI1 causes broad perturbation of gene expression that is key to normal hematopoietic cell development. The first key gene is PU.1 that is known to play a pivotal role in B cell/monocyte development50-54 and the maintenance of hematopoietic stem cell in the fetal liver.55 PU.1 transcripts are markedly decreased in AML1-/- fetal liver cells, as reported previously,42 whereas its expression is normal in AML1/EVI1/+ fetal liver cells, and this expression pattern is paralleled by one of its direct target gene CD11b. The maintained expression of PU.1 may afford AML1/EVI1/+ hematopoietic progenitors a capacity to produce macrophage and mixed colonies. Importantly, our data suggest that the reported complete lack of hematopoietic progenitors in AML1-/- fetal liver may result from the absence of PU.1, and the recent finding underscores the critical role for PU.1 in supporting hematopoietic stem cell maintenance in the fetal liver.55 Thus, PU.1 seems to be the key transcription factor that explains the phenotypic difference between AML1/EVI1/+ and AML1-/- animals. The second key gene CEBPA is involved in early granulocytic differentiation.56 The expression of CEBPA is increased in both AML1/EVI1/+ and AML1-/- fetal livers. In AML1/EVI1/+ liver, the increased expression of C/EBPα may enhance myeloid/monocytic differentiation of the existing hematopoietic progenitor supported by PU.1. In AML1-/- fetal liver, however, the lack of PU.1 expression dissipates the hematopoietic progenitor; thus, there is no myeloid/monocytic progeny arising from even the increased expression of C/EBPα. Third, the expression of LMO2 and SCL is severely repressed in AML1/EVI1/+ fetal liver. The LMO2 and SCL genes are essential for erythroid lineage differentiation,44,45 and low expression of these genes could account for the absence of erythroid cells in AML1/EVI1/+ fetal liver, even though the hematopoietic progenitor cells are supported by the presence of PU.1. The lack of LMO2 and SCL expression is also observed in AML1-/- fetal liver. This suggests that the defective expression of LMO2 and SCL in AML1/EVI1/+ cells is due to dominant-negative effect of AML1/EVI1 over wild-type AML1, and that LMO2 and SCL might be candidate target genes for wild-type AML1. This possibility might be worth exploring, although no PEBP2 binding sites are found at least in the proximal regulatory region of the LMO2 or SCL gene.57,58 Finally, it is worth emphasizing that GATA1 expression is kept comparable across wild-type, AML1-/-, and AML1/EVI1/+ fetal livers, and yet its target genes ALASE and β-globin are much more repressed in AML1/EVI1/+ fetal liver cells than in AML1-/- cells. These differences in expression of ALASE and β-globin genes could be due to the interplay between PU.1 and GATA-1,46 providing a case that some of the altered gene expressions are the consequences of the combinatory effect of involved transcription factors.
In summary, we have demonstrated that AML1/EVI1 knock-in mice almost completely lack effective hematopoiesis in the fetal liver, but they retain progenitors with increased self-renewal capacity and potential to differentiate into the myeloid/monocytic and megakaryocytic lineages with some dysplasticity. We provide correlative evidence that PU.1 plays a critical role in the maintenance of hematopoietic progenitors in the AML1/EVI1/+ fetal liver. Collectively, our study suggests that AML1/EVI1 is not only a dominant-negative suppressor of wild-type-AML1 but also has more a proliferative and dysplastic effect added on by the fusing EVI1 part. Clinical evidence shows that the emergence of AML1/EVI1 is associated with the progression of hematopoietic stem cell disorder, and our results explain the aggressive and dysplastic transformation of the disease course observed in patients with AML1/EVI1-expressing leukemia. Inducible expression of AML1/ETO in mouse hematopoietic tissue shows that the expression of AML1/ETO per se is not leukemogenic59,60 and requires a second hit introduced by alkylating agents to cause leukemia.60,61 It certainly needs to be tested whether the expression of AML1/EVI1 is sufficient to cause leukemia when expressed in an inducible fashion in adult mice. Our prediction would be that AML1/EVI1 expression is leukemogenic on its own, or at least more prone to leukemia when a second hit comes in. A conditional project of AML1/EVI1 knock-in expression is in progress in our laboratory.
Prepublished online as Blood First Edition Paper, May 24, 2005; DOI 10.1182/blood-2004-11-4330.
Supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology (Japan); the Ministry of Health, Labour, and Welfare (Japan); Japanese Society for the Promotion of Science; and the Japan Health Sciences Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We want to dedicate this paper to the late professor Dr Hisamaru Hirai at the University of Tokyo. He was one of the leading investigators of AML1 and made seminal contributions to the field of leukemogenesis and hematopoietic-cell regulation. TT2 ES cells are a generous gift from Dr S Aizawa (RIKEN Center for Developmental Biology, Kobe, Japan). pBK-Neo vector is also a generous gift from Dr D-E Zhang (The Scripps Research Institute, La Jolla, CA). pCXN2 is kindly provided by Dr J. Miyazaki (University of Osaka, Osaka, Japan). Recombinant human erythropoietin (EPO) and murine IL-3 and stem cell factor (SCF) are provided by KIRIN Brewery. We thank Dr Motoo Shinoda (Laboratory Animal Research Center, Dokkyo University School of Medicine, Tochigi, Japan) for technical assistance.

![Figure 2. Morphologic comparison of E12.5 wild-type (WT) and AML1/EVI1/+ heterozygous embryos. (A) External appearance of WT and AML1/EVI1/+ littermates. AML1/EVI1/+ embryo on the right is similar in size to WT littermate on the left but is clearly identifiable by the presence of fetal liver pallor and massive hemorrhage within the CNS and soft tissues in the back (objective lens [OL], 2 ×/0.05; original magnification [OL], × 20). (B-C) Sections of the cerebral ventricle from WT (B) and AML1/EVI1/+ (C) embryos. AML1/EVI1/+ embryo shows massive hemorrhage into the ventricle (OL, 10 ×/0.40; OM, × 100). (D-E) Sections of the dorsal root ganglia from WT (D) and AML1/EVI1/+ (E) embryos. AML1/EVI1/+ embryo shows hemorrhage in the ganglia (OL, 10 ×/0.40; OM, × 100). (F-G) Smears of the peripheral blood from WT (F) and AML1/EVI1/+ (G) embryos. AML1/EVI1/+ embryo shows the absence of definitive erythrocytes, while WT littermate shows numerous enucleated definitive erythrocytes (OL, 40 ×/0.65; OM, × 400). (H-I) Smears of the peripheral blood from WT embryo. Only WT littermate shows monocyte (H) and neutrophil (I) (OL, 100 ×/1.40; OM, × 1000). (J-K) Sections of the fetal liver from WT (J) and AML1/EVI1/+ (K) embryos. AML1/EVI1/+ embryo shows a near complete absence of hematopoietic precursors, while WT littermate shows numerous hematopoietic precursor cells (OL, 20 ×/0.70; OM, × 200).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/6/10.1182_blood-2004-11-4330/6/m_zh80180583930002.jpeg?Expires=1765891678&Signature=Y9RzGDit6mTSSt6UrPWU4TLC-22MW1BSmKYHpDWM9~I5IFR1XhTLO2WKvabUJOgXPMs10N7ZO6t0fpJLv7By3Edr1ZpMYC3c2KHP5FF4oRaT2-A-Jn0hQ-VQLN-CUzN4FsOgqarG7rirB6MKTUZqycyf9Z-44PuW2vT41K-Uu~GR5bzCk~LZ6zacoISch5OjIHorrEJwzeKtMqBA6E0ZSktk4eUObYShNoYXpHNGBgAChb5Y1ZwegG56ZXGKIufCiPtk7UljKXHIDdq9fMHEdZUTe4N9nDunUHW7v-ugkIrBk~TxGF~TImiE78QdRO152R9F3xzuOPtet-AvRoiIVw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal