Abstract
A proportion of patients with Hodgkin lymphoma carry Epstein-Barr virus (EBV), an oncogenic herpesvirus, in their tumor cells. Although it is generally assumed that EBV contributes to the malignant phenotype of Hodgkin lymphoma cells, direct evidence in support of this is lacking. Here we show that EBV infection of Hodgkin lymphoma cells results in the induction of autotaxin, a secreted tumor-associated factor with lysophospholipase-D activity. Up-regulation of autotaxin increased the generation of lysophosphatidic acid (LPA) and led to the enhanced growth and survival of Hodgkin lymphoma cells, whereas specific down-regulation of autotaxin decreased LPA levels and reduced cell growth and viability. In lymphoma tissues, autotaxin expression was mainly restricted to CD30+ anaplastic large-cell lymphomas and Hodgkin lymphoma; in the latter, high levels of autotaxin were strongly associated with EBV positivity (P = .006). Our results identify the induction of autotaxin and the subsequent generation of LPA as key molecular events that mediate the EBV-induced growth and survival of Hodgkin lymphoma cells and suggest that this pathway may provide opportunities for novel therapeutic intervention. (Blood. 2005;106:2138-2146)
Introduction
Epstein-Barr virus (EBV) is associated with B-lymphoid (eg, Burkitt lymphoma, posttransplantation lymphoma) and nonlymphoid malignancies (eg, nasopharyngeal carcinoma). EBV is also present in the malignant Hodgkin and Reed-Sternberg (HRS) cells of some Hodgkin lymphomas (HLs)1,2 in which the viral genome is monoclonal, implying that EBV infection is an early pathogenic event.3 EBV also persists throughout the course of HL, suggesting that it is important to the maintenance of the transformed phenotype.4 However, the contribution of EBV to the pathogenesis of HL has yet to be established.
In contrast to EBV-transformed B-lymphoblastoid cell lines and most B-cell posttransplantation lymphomas in which all the known latent virus genes are expressed, other EBV-associated B-cell malignancies show a more restricted pattern of virus gene expression. For example, in most primary Burkitt lymphomas, EBV expresses only the EBV-encoded RNAs (EBERs), the Epstein-Barr nuclear antigen-1 (EBNA1), and the BamH1A rightward transcripts5 (BARTs). Primary EBV-positive HRS cells express not only these genes but also latent membrane protein 1 (LMP1) and LMP2.6-8
We show that EBV infection of HL cells leads to the induction of autotaxin, an autocrine motility factor originally isolated from A2058 human melanoma cells9 that has been shown to augment many cellular characteristics associated with tumor aggressiveness,10 including cell proliferation,11 cell survival, invasion,10 and angiogenesis.12 Autotaxin is a phospholipase D (PLD) that can generate lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) from lysophosphatidylcholine (LPC) and sphingosylphosphorylcholine (SPC), respectively.11,13 A role for LPA in human oncogenesis was suggested by the observation that LPA is present at elevated levels in ascites of patients with ovarian cancer.14 Subsequently, it has been shown that LPA is a potent tumor-promoting small molecule in a range of biologic systems and that it binds to specific cell surface heterotrimeric G-protein-coupled receptors (the endothelial cell differentiation gene [EDG] receptors) stimulating the proliferation, migration, and survival of cancer cells.15,16 Therefore, mechanisms that lead to aberrant LPA production are likely to be associated with the initiation or progression of cancer.16
We demonstrate that the induction of autotaxin by EBV leads to the increased generation of LPA and to the proliferation and survival of HL cells. This is the first report that autotaxin and the tumor-promoting lipid LPA are the molecular targets of a transforming virus.
Materials and methods
Generation and phenotype of cell lines
EBV-negative KM-H2 cells17 were infected with Akata-derived recombinant EBV18 and cultured in 1 mg/mL G418. KM-H2 cells have been shown to be representative of HL cells.19,20 Control KM-H2 cells were generated by electroporation with vector only (pzipLNSNeo). EBV-positive L591 cells21 were serially diluted for up to 6 weeks to generate EBV-negative clones. Clone L591-SD3 was used in later experiments. Immunohistochemistry for EBNA1,22 reverse transcription-polymerase chain reaction (RT-PCR) and in situ hybridization for EBERs,23 and quantitative PCR for the detection of viral DNA24 were used to demonstrate the presence of EBV. Immunoglobulin heavy (IgH) gene sequence analysis of cell lines was performed as previously described.25 Viability of cell lines was determined using trypan blue reagent (Sigma-Aldrich, Poole, United Kingdom) and proliferation by WST-1 reagent (Roche Diagnostics, Lewes, United Kingdom). The WST-1 reagent measures the activity of mitochondrial dehydrogenases in the sample. The tetrazolium salts in the reaction are cleaved to formazan, and the augmentation in enzyme activity leads to an increase in the amount of formazan dye formed, which directly correlates with the number of metabolically active cells in the culture.
RT-PCR
RNA was extracted using the NucleoSpin RNA II kit (AB Gene, Epsom, United Kingdom). RT-PCR analysis for EBV gene expression was as previously described.26 Expression was detected using the following primers: autotaxin forward, 5′-CACCAGAGGCTAAATATGATGC-3′; autotaxin reverse, 5′-GAAGGAGGACACAGAGAGAG-3′; EDG1 forward, 5′-ATTCAGCCGCAGCAAATC-3′; EDG1 reverse, 5′-TAACTCTACCCACCAACACC-3′; EDG2 forward, 5′-AATTCAACTCTGCCATGAACCC-3′; EDG2 reverse 5′-ACTTTTCTCCTCTCTCACACCC-3′. Probes were 5′-CACCAGCTGTCTGGATTTCA-3′ (autotaxin), 5′-AAATCTCTGGGCTTCGACTG-3′ (EDG1), and 5′-CATCTTGGCTGGAGTTCACA-3′ (EDG2).
Gene expression analysis
Affymetrix HU133A arrays27 were used for all experiments. Total RNA from mycoplasma-free cell lines was used to prepare biotinylated RNA.28 A complete description of procedures is available at http://bioinf.picr.man.ac.uk/mbcf/downloads/GeneChip_Target_Prep_Protocol-CR-UK_v2.pdf. Ratios for GAPDH and β-actin (3′/5′) were within acceptable limits (glyceraldehyde phosphate dehydrogenase [GAPDH], 0.80-1.18; β-actin, 0.83-1.89), and BioB spike controls were present on 5 of 8 chips, with BioC, BioD, and CreX also present in increasing intensity. When scaled to a target intensity of 100 (Affymetrix MAS 5.0), scaling factors for all arrays were within acceptable limits (HU133A chips, 0.591-1.134), as were background, Q values, and mean intensities.
Images of GeneChips were analyzed using Affymetrix Microarray Suite 5.0. Probe level quantile normalization29 and robust multiarray analysis30 on the raw .CEL files were performed using the Affymetrix package of the Bioconductor (http://www.bioconductor.org) project. Differentially expressed probe sets were identified using significance analysis of microarrays (SAM)31,32 ; only those with positive or negative changes of 2.5-fold or more and a false discovery rate of 5% or less were included. Hierarchic clustering was performed using dChip (http://www.dchip.org).
Generation of autotaxin-specific monoclonal antibodies
A polypeptide (amino acids 58-182 of human autotaxin) was expressed in Escherichia coli as a glutathione-S-transferase (GST) fusion protein using pGEX-4T vector (Amersham-Pharmacia, Little Chalfont, United Kingdom). Rats (WKY/Izm strain) were immunized with purified fusion protein, and medial iliac lymph nodes were used for fusion with PAI mouse myeloma cells. Hybridomas were selected by screening with enzyme-linked immunosorbent assay (ELISA), immunofluorescence, and Western blotting. Two antibody-secreting hybridoma cell lines were generated (2A12 and 4F1).
Protein analysis
Cells were lysed in buffer (50 mM Tris, pH 7.5, 9 M urea, 0.15 M β-mercaptoethanol) and protein quantified by Bio-Rad DC Protein Assay Kit (Bio-Rad, Hemel Hempstead, United Kingdom). Gel sample buffer was added to samples before sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transfer to BioTrace NT membrane (VWR International Ltd, Poole, United Kingdom), and incubation with monoclonal antibody (mAb) 2A12 (1:100) and then horseradish peroxidase (HRP)-conjugated rabbit anti-rat IgG (DAKO, Ely, United Kingdom) (1:1000). Detection was with enhanced chemiluminescence (ECL; Amersham-Pharmacia Biotech, Little Chalfont, United Kingdom). Immunohistochemistry was performed on 4-μm sections, as previously described.33 HL was classified as autotaxinhi if autotaxin-staining intensity in HRS cells was equivalent to or higher than that observed in endothelial cells in the same section or as autotaxinneg/autotaxinlo if autotaxin-staining intensity in HRS cells was lower than that in endothelial cells.
Conditioned media experiments and addition of exogenous LPA and S1P
Control KM-H2 cells (KM-H2-neo) were grown in their own conditioned media or in media from EBV-infected KM-H2 cells (KM-H2-Akata). KM-H2-Akata cells were grown in conditioned media from KM-H2-neo cells or their own media. Cell proliferation was assessed by WST-1 assay. For assessment of the effects of exogenous LPA or S1P, 5 × 104 cells were washed in serum-free media, resuspended in 100 μL RPMI containing 1% B-cell serum (BCS) alone (control), RPMI containing 1% BCS and 5 μM LPA, or RPMI containing 1% BCS and 1 μM S1P. Then WST-1 assays were performed.
Analysis of autotaxin activity
Cells (7.5 × 106) were cultured in 15 mL serum-free and phenol red-free OptiMEM (Invitrogen, Renfrew, United Kingdom) for 16 hours. After the addition of phosphatase inhibitors, conditioned media were incubated with 20 μM LPC or 20 μM SPC. Five hundred nanograms each of 17:0-LPA, D31-16:0-LPC, and C17 S1P were added as internal standards. Standard nomenclature was adopted in describing the lipids (ie, number of carbon atoms/number of double bonds). Samples were extracted with water-saturated butanol and dried. Contaminants were removed with chloroform/methanol/water. Samples were dissolved in chloroform/methanol/water (5:5:1) and subjected to liquid chromatography/mass spectrometry (LC-MS) separation. Detection was by selected ion monitoring (SIM) in -ve and +ve electrospray ionization (ESI) modes using a probe voltage of ± 4 kV, a nebulizer nitrogen flow of 4 L/min, and a desolvation line temperature of 300°C.
LPA-stimulated PLD activity
Cells were incubated in medium containing 4 μCi/mL (0.148 MBq/mL) [3H]-palmitic acid before resuspension in RPMI containing 10 mM HEPES (N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid) pH 7.4 and 0.1% bovine serum albumin (BSA). Cells were stimulated for 20 minutes in 30 mM butan-1-ol with 5 μM LPA at 37°C, and PLD activity was determined by analyzing the generation of [3H]phosphatidylbutanol.34
Knockdown of autotaxin expression
KM-H2-Akata cells in serum-free OptiMEM were incubated in the presence of 10 nM autotaxin-specific siRNA (5′-GAAAGGCAGCAAAGUCAUG-3′) in RiboJuice according to the protocol for 6-well plates (Novagen, Nottingham, United Kingdom). Cells were incubated at 37°C for 4 hours, after which an equivalent volume of OptiMEM media containing Pen/Strep and 20% BCS was added, followed by further incubation at 37°C. RT-PCR, immunoblotting for autotaxin and proliferation, and viability assays were performed as described earlier. Ribojuice only, OptiMEM only, and irrelevant siRNAs (scrambled LMP1 siRNA 5′-GGGUAGAUAGACUCUCGCU-3′) acted as negative controls.
Statistical analysis
All quantifiable data were subjected to statistical analysis in Microsoft Excel using a 2-tailed Student t test assuming the 2 samples displayed unequal variance. The generated P values were taken to indicate a significant difference between the data sets when P was below .05. Any data that satisfied these criteria are indicated by an asterisk above the relevant column (Figures 1C, 1D, 4A, 4B, 4D-F, 5C, and 5D). On the charts themselves, the error bars represent the standard error of the means for a particular data set.
Results
HL cells expressing a limited repertoire of virus genes display enhanced growth and survival
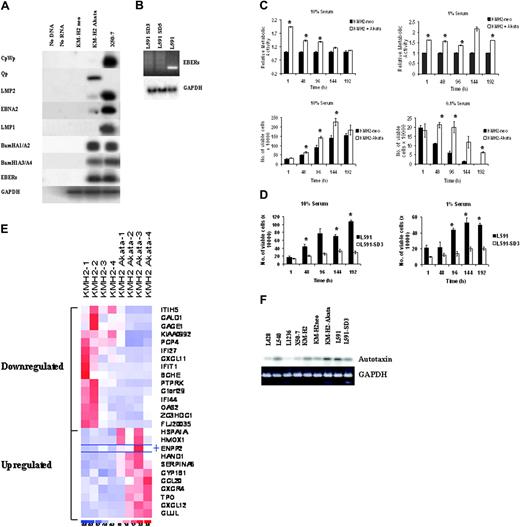
We initially infected EBV-negative KM-H2 cells with Akata-derived recombinant EBV and confirmed its presence by RT-PCR or in situ hybridization for EBER and immunohistochemistry for EBNA1 (data not shown). During latency, EBV uses one of several promoters to transcribe EBNA1—the Qp promoter, which initiates transcription of EBNA1 alone, or the Cp and Wp promoters, which generate a long transcript that is multiply spliced to form EBNA1 and the other EBNA proteins.35 RT-PCR analysis of EBV-infected KM-H2 cells revealed that most EBNA1 transcripts originated from Qp with very low levels of transcription from Cp/Wp and associated low levels of EBNA2. LMP2 was expressed (Figure 1A), but neither LMP1 RNA (Figure 1A) nor LMP1 protein (data not shown) were detectable. There was no induction of the replicative cycle in infected cells, as demonstrated by the absence of BZLF1 expression (data not shown). Thus, EBV-infected KM-H2 cells show a pattern of virus gene expression different from that observed in primary HRS cells, including the notable absence of LMP1 expression. Despite this restricted pattern of virus gene expression, EBV-infected KM-H2 cells displayed significantly increased proliferation (P < .001, P < .01, P < .01, P < .01) at 1, 48, 96, and 192 hours in 1% serum and enhanced viability (P = .01, P < .05, P < .01) at 48, 96, and 192 hours, respectively, in 0.1% serum compared with EBV-negative cells (Figure 1C). To confirm these effects of EBV infection, EBV-negative L591 cells were isolated from parental EBV-positive cells (Figure 1B): loss of the EBV genome from L591 cells resulted in a dramatic decrease in cell proliferation (data not shown) and viability (Figure 1D) (P = .001, P < .05, P < .001) at 96, 144, and 192 hours in 1% serum. Sequence analysis of the IgH gene from EBV-negative L591 cells and EBV-positive KM-H2 cells confirmed their origin from the respective parental lines (data not shown).
Autotaxin is a transcriptional target of EBV in HL
Gene expression analysis revealed 11 up-regulated and 15 down-regulated probe sets after EBV infection of KM-H2 cells (Figure 1E; Table 1). The most highly up-regulated probe was autotaxin. The induction of autotaxin mRNA in EBV-infected KM-H2 was confirmed by RT-PCR analysis (Figure 1F). Autotaxin mRNA levels were decreased after the loss of EBV from L591 cells and were low in other EBV-negative HL cell lines (Figure 1F).
Differentially expressed genes in EBV-infected KM-H2 compared with control cells
Gene . | Accession no. . | Mean fold change . | Ontology . |
|---|---|---|---|
| ENPP2 | L35594 | 4.16 | Cell motility, GPCR protein signaling pathway |
| CXCL12 | U19495 | 4.12 | Immune response, cell adhesion, chemotaxis, signal transduction |
| GLUL | AL161952 | 3.33 | Glutamine biosynthesis |
| SERPINA6 | NM_001756 | 3.10 | Transport |
| HMOX1 | NM_002133 | .2.87 | Positive regulation of lkB kinase/NF-kB cascade |
| CXCR4 | AJ224869 | 2.80 | Immune response, chemotaxis, GPCR protein signaling pathway |
| CCL20 | NM_004591 | 2.77 | Immune response, chemotaxis, cell-cell signaling |
| TPO | M17755 | 2.77 | Thyroid hormone generation |
| HSPA1A | NM_005345 | 2.70 | Ubiquitin-proteasome pathway, protein folding, signal transduction |
| CYP1B1 | NM_000104 | 2.55 | Eye morphogenesis |
| HAND1 | NM_004821 | 2.55 | Heart development, transcription from Pol II promoter |
| GAGE1 | NM_001472 | −2.54 | Cellular defense response |
| OAS2 | NM_016817 | −2.60 | Immune response |
| FLJ20035 | NM_017631 | −2.62 | Protein biosynthesis |
| IFI44 | NM_006417 | −2.68 | Viral response |
| ZC3HDC1 | NM_022750 | −2.72 | Nucleic acid binding |
| ITIH5 | NM_030569 | −2.95 | Regulation of localization, synthesis, and degradation of hyaluronan |
| PTPRK | NM_002844 | −2.99 | Transmembrane receptor protein tyrosine phosphatase activity |
| CALD1 | AL583520 | −3.10 | Muscle contraction and development |
| PCP4 | NM_006198 | −3.41 | Central nervous system development |
| BCHE | NM_000055 | −3.50 | Cocaine metabolism |
| CXCL11 | AF002985 | −3.60 | Immune response, chemotaxis, cell-cell signaling |
| IFIT1 | NM_001548 | −3.75 | Immune response |
| KIAA0992 | NM_016081 | −3.90 | Cell shape, adhesion, and contraction |
| Clorf29 | NM_006820 | −6.01 | Unknown |
| IFI27 | NM_005532 | −19.38 | Immune response |
Gene . | Accession no. . | Mean fold change . | Ontology . |
|---|---|---|---|
| ENPP2 | L35594 | 4.16 | Cell motility, GPCR protein signaling pathway |
| CXCL12 | U19495 | 4.12 | Immune response, cell adhesion, chemotaxis, signal transduction |
| GLUL | AL161952 | 3.33 | Glutamine biosynthesis |
| SERPINA6 | NM_001756 | 3.10 | Transport |
| HMOX1 | NM_002133 | .2.87 | Positive regulation of lkB kinase/NF-kB cascade |
| CXCR4 | AJ224869 | 2.80 | Immune response, chemotaxis, GPCR protein signaling pathway |
| CCL20 | NM_004591 | 2.77 | Immune response, chemotaxis, cell-cell signaling |
| TPO | M17755 | 2.77 | Thyroid hormone generation |
| HSPA1A | NM_005345 | 2.70 | Ubiquitin-proteasome pathway, protein folding, signal transduction |
| CYP1B1 | NM_000104 | 2.55 | Eye morphogenesis |
| HAND1 | NM_004821 | 2.55 | Heart development, transcription from Pol II promoter |
| GAGE1 | NM_001472 | −2.54 | Cellular defense response |
| OAS2 | NM_016817 | −2.60 | Immune response |
| FLJ20035 | NM_017631 | −2.62 | Protein biosynthesis |
| IFI44 | NM_006417 | −2.68 | Viral response |
| ZC3HDC1 | NM_022750 | −2.72 | Nucleic acid binding |
| ITIH5 | NM_030569 | −2.95 | Regulation of localization, synthesis, and degradation of hyaluronan |
| PTPRK | NM_002844 | −2.99 | Transmembrane receptor protein tyrosine phosphatase activity |
| CALD1 | AL583520 | −3.10 | Muscle contraction and development |
| PCP4 | NM_006198 | −3.41 | Central nervous system development |
| BCHE | NM_000055 | −3.50 | Cocaine metabolism |
| CXCL11 | AF002985 | −3.60 | Immune response, chemotaxis, cell-cell signaling |
| IFIT1 | NM_001548 | −3.75 | Immune response |
| KIAA0992 | NM_016081 | −3.90 | Cell shape, adhesion, and contraction |
| Clorf29 | NM_006820 | −6.01 | Unknown |
| IFI27 | NM_005532 | −19.38 | Immune response |
Only genes that had a positive or negative change of 2.5-fold or greater and a false discovery rate of 5% or less were included. Autotaxin is represented by its gene symbol, ENPP2.
GPCR indicates G-protein-coupled receptor.
Generation of autotaxin-specific monoclonal antibodies and up-regulation of autotaxin protein in EBV-infected HL cells
A purified polypeptide from human autotaxin was used to derive 2 antibody-secreting hybridoma cell lines, 2A12 and 4F1. Immunofluorescence (data not shown) and immunoblotting (Figure 2A) were used to confirm the specificity of each monoclonal antibody for autotaxin. Immunoblotting confirmed the up-regulation of autotaxin protein in EBV-infected KM-H2 and L591 cells (Figure 2B). Analysis of other EBV-negative HL cell lines, such as L428, L1236, and HDLM2, revealed lower levels of autotaxin protein than was found in EBV-positive cell lines (Figure 2B). Autotaxin was absent from EBV-negative BL cells (Ramos, DG75, BJAB) and EBV-positive BL cells, including those displaying type 1 latency (Akata, Rael, Daudi, Elijah) or type 3 latency (Raji, Namalwa) (Figure 2C). Of the EBV-transformed lymphoblastoid cell lines (LCLs) studied, only B95.8 cells expressed detectable levels of autotaxin protein. Nasopharyngeal carcinoma cells, including EBV-negative HONE-1 cells and EBV-positive C666-1 cells, lacked autotaxin protein (Figure 2D). HONE-1 cells infected with the same Akata-derived recombinant EBV used to generate EBV-positive KM-H2 cells failed to show induction of autotaxin after infection (Figure 2D).
Expression of autotaxin in primary lymphoma tissues
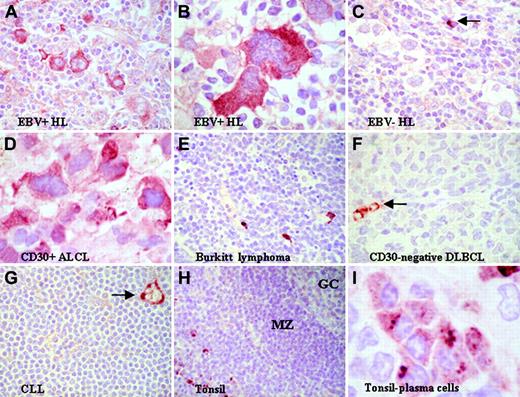
When immunohistochemistry of pelleted cell lines demonstrated that both autotaxin-specific antibodies were effective on paraffin-embedded samples (data not shown), these were used to examine the expression of autotaxin in a series of classic HL tumors with known EBV status. HL samples that contained HRS cells with high-level expression of autotaxin (autotaxinhi) were significantly more frequent in EBV-positive than in EBV-negative samples (Figure 3A-C; Table 2) (Fisher exact test, P = .006). Autotaxin was absent from most EBV-positive and EBV-negative Burkitt lymphomas (Figure 3E) and primary EBV-positive undifferentiated nasopharyngeal carcinoma (NPC) (n = 9; data not shown), confirming the results of in vitro studies suggesting that the induction of autotaxin by EBV was restricted to HL cells. With respect to NPC, the absence of autotaxin expression might be considered surprising because autotaxin expression has previously been reported in other tumors of epithelial origin. Analysis of other B-cell non-Hodgkin lymphomas (B-NHLs) (Table 3) revealed that autotaxin was frequently expressed in the CD30+ and anaplastic variants of diffuse large B-cell lymphoma (DLBCL; Figure 3D) but was absent from most high-grade (including CD30- DLBCLs) and low-grade B-NHLs. (Figure 3F-G). In normal lymphoid tissues, autotaxin was not detectable in most B-cell compartments, including B cells of germinal centers and mantle zones (Figure 3H), but it was expressed by a subset of postgerminal center CD138+ B cells (Figure 3I). These data suggest that the restricted expression of autotaxin in tumors is in part a reflection of the stage of B-cell differentiation from which each is derived.
Autotaxin expression in EBV-positive and EBV-negative HL
Autotaxin status of HRS cells . | EBV-positive . | EBV-negative . |
|---|---|---|
| Autotaxinhi | 10 | 6 |
| Autotaxinneg/autotaxinlo | 1 | 11 |
| Autotaxinhi, % | 91 (10 of 11) | 35 (6 of 17) |
Autotaxin status of HRS cells . | EBV-positive . | EBV-negative . |
|---|---|---|
| Autotaxinhi | 10 | 6 |
| Autotaxinneg/autotaxinlo | 1 | 11 |
| Autotaxinhi, % | 91 (10 of 11) | 35 (6 of 17) |
HL cells were classified as autotaxinhi if autotaxin expression in HRS cells was equivalent to or higher than that observed in endothelial cells in the same section or as autotaxinneg/autotaxinlo if autotaxin expression in HRS cells was lower than in endothelial cells. Fisher exact test results show there was a significantly higher proportion of autotaxinhi tumors in the EBV-positive group (P = .006).
Autotaxin expression in NHL
Lymphoma subtype . | No. positive . | Total . | % positive . |
|---|---|---|---|
| Low-grade B-cell NHL | |||
| Follicular lymphoma | 2 | 25 | 8 |
| Chronic lymphocytic lymphoma | 0 | 6 | 0 |
| Mantle cell lymphoma | 1 | 5 | 20 |
| MALT lymphoma | 1 | 1 | 100 |
| High-grade B-cell NHL | |||
| Burkitt lymphoma | 2 | 14 | 14 |
| DLBCL | |||
| Centroblastic and immunoblastic* | 7 | 26 | 27 |
| CD30+/anaplastic* | 12 | 12 | 100 |
| Primary cutaneous | 2 | 5 | 40 |
| Posttransplantation lymphoproliferative disorder | 1 | 1 | 100 |
| T-cell neoplasms | |||
| Peripheral T-cell NHL unspecified | 2 | 4 | 50 |
| Mycosis fungoides | 0 | 1 | 0 |
| NK/T -cell lymphoma (angiocentric) | 0 | 1 | 0 |
| ALCL (null/T-cell type) | 2 | 5 | 40 |
Lymphoma subtype . | No. positive . | Total . | % positive . |
|---|---|---|---|
| Low-grade B-cell NHL | |||
| Follicular lymphoma | 2 | 25 | 8 |
| Chronic lymphocytic lymphoma | 0 | 6 | 0 |
| Mantle cell lymphoma | 1 | 5 | 20 |
| MALT lymphoma | 1 | 1 | 100 |
| High-grade B-cell NHL | |||
| Burkitt lymphoma | 2 | 14 | 14 |
| DLBCL | |||
| Centroblastic and immunoblastic* | 7 | 26 | 27 |
| CD30+/anaplastic* | 12 | 12 | 100 |
| Primary cutaneous | 2 | 5 | 40 |
| Posttransplantation lymphoproliferative disorder | 1 | 1 | 100 |
| T-cell neoplasms | |||
| Peripheral T-cell NHL unspecified | 2 | 4 | 50 |
| Mycosis fungoides | 0 | 1 | 0 |
| NK/T -cell lymphoma (angiocentric) | 0 | 1 | 0 |
| ALCL (null/T-cell type) | 2 | 5 | 40 |
DLBCL indicates diffuse large B-cell lymphoma; ALCL, anaplastic large-cell lymphoma.
Morphologic variants of DLBCL.
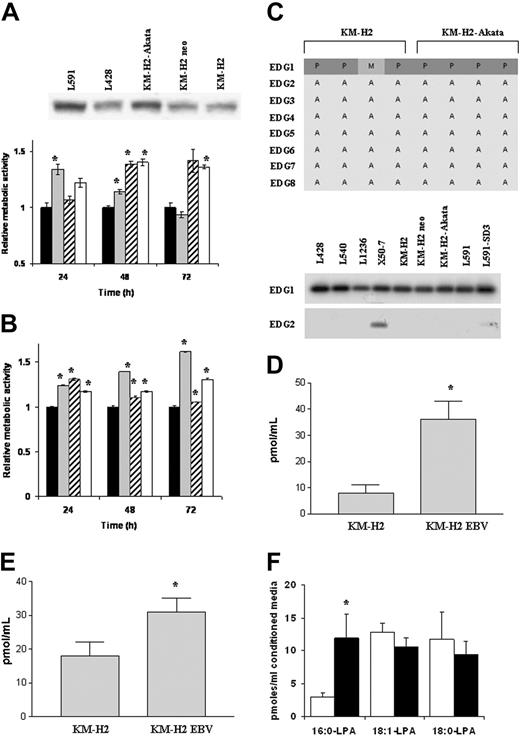
Increased production of soluble autotaxin in the supernatant of EBV-infected KM-H2 cells is associated with enhanced cell growth
Because autotaxin can be cleaved from the cell membrane, it was important to establish whether the induction of autotaxin also resulted in increased levels of soluble autotaxin. Although all HL cell lines generated soluble autotaxin, levels were higher in EBV-infected cells (Figure 4A). HL cell lines were grown in conditioned growth medium from either EBV-positive or EBV-negative KM-H2 cells. EBV-negative KM-H2 cells grown in their own conditioned medium provided a baseline, and EBV-positive KM-H2 cells provided a positive control. EBV-negative KMH2 cells grown in conditioned medium from EBV-infected KM-H2 cells showed an increase in cell proliferation after the first 24 hours, P equaling .028 (Figure 4A). This was followed by a reduction in cell growth, presumably the result of gradual depletion of growth factors in the conditioned medium. In contrast, EBV-infected KM-H2 cells grown in conditioned medium from EBV-negative cells showed no change in proliferation after 24 hours, but thereafter cell growth increased.
Increased growth of EBV-negative KM-H2 cells after treatment with LPA or S1P
To determine whether HL cells were responsive to LPA or S1P stimulation, KM-H2 cells were cultured in the presence of either LPA or S1P. Both LPA (Figure 4B) and S1P (data not shown) significantly increased the growth of EBV-negative KM-H2 cells (P < .001) at 24, 48, and 72 hours. The same effect, but to a lesser degree, was seen for the growth of EBV-positive cells above their baseline on the addition of LPA or S1P. These data suggest that KM-H2 cells are responsive to the biologic mediators generated by autotaxin but that there is maximal activation by these mediators in EBV-infected cells. Although EBV-positive and EBV-negative KM-H2 cells expressed the S1PR1 receptor (EDG1), they did not express the major LPA receptor, LPAR1 (EDG2), or the other known LPA or S1P receptors (Figure 4C). These findings were confirmed by RT-PCR analysis (Figure 4C). The expression of an unknown LPA receptor in KM-H2 cells was supported by the detection of a small but significant stimulation of PLD activity by 5 μM LPA (control, 0.126 ± 0.011, + LPA 0.155 ± 0.009; results are the mean ± SD dpm in phosphatidylbutanol as a percentage of dpm in total phospholipids).
Increased growth and survival of EBV-positive HL cells expressing a limited repertoire of EBV genes is associated with the transcriptional up-regulation of autotaxin. (A) RT-PCR analysis demonstrates a pattern of virus gene expression in EBV-infected KM-H2 (KM-H2-Akata) cells predominated by expression of the EBERs, Qp-driven EBNA1, and the BamH1A transcripts. There was also very low-level transcription from Cp/Wp and associated low levels of EBNA2 and LMP2 transcripts in these cells. Neither LMP1 RNA nor protein (data not shown) could be detected in these cells. (B) Loss of EBER expression in 2 clones (SD3 and SD5) of EBV-negative L591 cells derived from the EBV-positive parental line by serial dilution. Quantitative PCR was also used to confirm the loss of the EBV genome from these cells (data not shown). Clone SD3 was used in later experiments. (C) (upper panels) WST1 assays showing relative metabolic activity (cell growth) of control cells (KMH2-neo) adjusted to a value of 1 compared with EBV-infected KM-H2 cells (KM-H2 + Akata) in 10% and 1% sera. A clear and significant difference in cell growth between control and EBV-infected cells is observed. (lower panels) Cell viability assays at 10% and 0.1% serum, demonstrating consistent and significant increases in the viability of EBV-infected KM-H2 cells compared with controls. ▪ indicates KM-H2-neo; and □, KM-H2 + Akata. (D) L591 SD3 cells showed dramatically reduced viability compared with EBV-positive parental L591 cells in 10% and 1% sera. Cell proliferation was similarly affected by EBV loss (data not shown). ▪ indicates L591; and □, L591-SD3. (E) Heat map showing gene expression differences across 4 replicates of EBV-negative KM-H2 cells and EBV-infected KM-H2 cells (KM-H2 Akata); 26 probe sets met the criteria of a positive or negative change of 2.5-fold or greater and a false discovery rate of 5% or less. Eleven probe sets were up-regulated and 15 were down-regulated after EBV infection. The most highly up-regulated probe set (mean fold increase, 4.16) was autotaxin (ENPP2). (F) Confirmation of increased autotaxin mRNA in EBV-infected KM-H2 (KM-H2 Akata) and L591 cells by RT-PCR analysis compared with EBV-negative parental KM-H2, KM-H2 neo control, and L591-SD3 cells. Other EBV-negative HL cell lines (L428, L540, L1236) and the EBV-transformed lymphoblastoid cell line X50-7 expressed lower levels of autotaxin mRNA.
Increased growth and survival of EBV-positive HL cells expressing a limited repertoire of EBV genes is associated with the transcriptional up-regulation of autotaxin. (A) RT-PCR analysis demonstrates a pattern of virus gene expression in EBV-infected KM-H2 (KM-H2-Akata) cells predominated by expression of the EBERs, Qp-driven EBNA1, and the BamH1A transcripts. There was also very low-level transcription from Cp/Wp and associated low levels of EBNA2 and LMP2 transcripts in these cells. Neither LMP1 RNA nor protein (data not shown) could be detected in these cells. (B) Loss of EBER expression in 2 clones (SD3 and SD5) of EBV-negative L591 cells derived from the EBV-positive parental line by serial dilution. Quantitative PCR was also used to confirm the loss of the EBV genome from these cells (data not shown). Clone SD3 was used in later experiments. (C) (upper panels) WST1 assays showing relative metabolic activity (cell growth) of control cells (KMH2-neo) adjusted to a value of 1 compared with EBV-infected KM-H2 cells (KM-H2 + Akata) in 10% and 1% sera. A clear and significant difference in cell growth between control and EBV-infected cells is observed. (lower panels) Cell viability assays at 10% and 0.1% serum, demonstrating consistent and significant increases in the viability of EBV-infected KM-H2 cells compared with controls. ▪ indicates KM-H2-neo; and □, KM-H2 + Akata. (D) L591 SD3 cells showed dramatically reduced viability compared with EBV-positive parental L591 cells in 10% and 1% sera. Cell proliferation was similarly affected by EBV loss (data not shown). ▪ indicates L591; and □, L591-SD3. (E) Heat map showing gene expression differences across 4 replicates of EBV-negative KM-H2 cells and EBV-infected KM-H2 cells (KM-H2 Akata); 26 probe sets met the criteria of a positive or negative change of 2.5-fold or greater and a false discovery rate of 5% or less. Eleven probe sets were up-regulated and 15 were down-regulated after EBV infection. The most highly up-regulated probe set (mean fold increase, 4.16) was autotaxin (ENPP2). (F) Confirmation of increased autotaxin mRNA in EBV-infected KM-H2 (KM-H2 Akata) and L591 cells by RT-PCR analysis compared with EBV-negative parental KM-H2, KM-H2 neo control, and L591-SD3 cells. Other EBV-negative HL cell lines (L428, L540, L1236) and the EBV-transformed lymphoblastoid cell line X50-7 expressed lower levels of autotaxin mRNA.
Up-regulation of autotaxin in EBV-infected HL cells is associated with increased use of LPC and generation of LPA
Although purified autotaxin can hydrolyze LPC and SPC, producing LPA and S1P, respectively, it has not yet been demonstrated that medium conditioned with cell-derived autotaxin can generate these signaling lipids. Analysis of medium conditioned by either EBV-positive or EBV-negative KM-H2 cells and incubated with LPC and SPC showed that SPC was not hydrolyzed and that no S1P was generated; in contrast, hydrolysis of LPC was significant, with 4-fold more LPC hydrolyzed by the EBV-positive KM-H2 cell medium than the EBV-negative medium (Figure 4D; P < .001). In parallel, more LPA content was detected in the EBV-positive than in the EBV-negative medium (Figure 4E; P < .05). The LPA content of the conditioned medium was low and was not equivalent to the loss of LPC; this was attributed to the short half-life of LPA in conditioned medium, with almost half the exogenously added LPA degraded within 10 minutes (data not shown). Despite the relatively small amplification in total LPA generation by the EBV-positive KM-H2 cell medium, there was a selective increase in 16:0-LPA generation (P < .01) (Figure 4F). This acyl structure was highly represented in the egg LPC added to the medium as the substrate and was not found in the unsupplemented culture medium; thus, the increase was exclusively attributed to LPC-PLD activity, which can be concluded to have been autotaxin.
Generation of autotaxin-specific monoclonal antibodies and the specific up-regulation of autotaxin protein by EBV-infection of HL cells. (A) Immunoblotting of sf9 cells infected with autotaxin (ATX)-baculovirus using 2A12 antibody demonstrates strong reactivity, whereas sf9 cells infected with wild-type (wt) baculovirus show no reactivity. Also shown here is reactivity against autotaxin in human serum, human plasma, culture supernatant and cells of breast cancer cell line MBA-MD-231, and supernatant and cells of glioma cell line SF539. (B) Western blotting using 2A12 antibody demonstrates increased autotaxin protein in EBV-infected KM-H2 cells (KM-H2 Akata) compared with uninfected KM-H2 cells (left panel). Most other EBV-negative HL cell lines (L428, L540, L1236, HD-MyZ, HDLM2) expressed lower levels of autotaxin protein. (right panel) Down-regulation of autotaxin protein in L591 cells after the loss of the EBV genome. (C) Autotaxin could not be detected in EBV-negative BL cells (Ramos, DG75, BJAB) or in a number of EBV-positive BL cells, including those displaying a type 1 form of EBV latency (Akata, Rael, Elijah), a type 3 form of EBV latency (Raji, Namalwa), or a Wp-restricted pattern of EBV latency (Daudi) that expresses EBNA1 and the EBNA3 family. BL lines are negative for autotaxin, though there is an unidentified lower molecular weight band in the DG75, BJAB, Akata, Elijah, and Rael BL lines and in the IB4 LCL line. Furthermore, of several lymphoblastoid cells lines (X50-7, B95.8 LCL, IB4), only B95.8 cells expressed detectable levels of autotaxin. (D) NPC cells, including EBV-positive C666-1 and EBV-negative HONE-1 cells, lacked autotaxin protein. HONE-1 cells infected with the same Akata-derived recombinant EBV used to generate EBV-positive KM-H2 cells failed to up-regulate autotaxin expression after infection.
Generation of autotaxin-specific monoclonal antibodies and the specific up-regulation of autotaxin protein by EBV-infection of HL cells. (A) Immunoblotting of sf9 cells infected with autotaxin (ATX)-baculovirus using 2A12 antibody demonstrates strong reactivity, whereas sf9 cells infected with wild-type (wt) baculovirus show no reactivity. Also shown here is reactivity against autotaxin in human serum, human plasma, culture supernatant and cells of breast cancer cell line MBA-MD-231, and supernatant and cells of glioma cell line SF539. (B) Western blotting using 2A12 antibody demonstrates increased autotaxin protein in EBV-infected KM-H2 cells (KM-H2 Akata) compared with uninfected KM-H2 cells (left panel). Most other EBV-negative HL cell lines (L428, L540, L1236, HD-MyZ, HDLM2) expressed lower levels of autotaxin protein. (right panel) Down-regulation of autotaxin protein in L591 cells after the loss of the EBV genome. (C) Autotaxin could not be detected in EBV-negative BL cells (Ramos, DG75, BJAB) or in a number of EBV-positive BL cells, including those displaying a type 1 form of EBV latency (Akata, Rael, Elijah), a type 3 form of EBV latency (Raji, Namalwa), or a Wp-restricted pattern of EBV latency (Daudi) that expresses EBNA1 and the EBNA3 family. BL lines are negative for autotaxin, though there is an unidentified lower molecular weight band in the DG75, BJAB, Akata, Elijah, and Rael BL lines and in the IB4 LCL line. Furthermore, of several lymphoblastoid cells lines (X50-7, B95.8 LCL, IB4), only B95.8 cells expressed detectable levels of autotaxin. (D) NPC cells, including EBV-positive C666-1 and EBV-negative HONE-1 cells, lacked autotaxin protein. HONE-1 cells infected with the same Akata-derived recombinant EBV used to generate EBV-positive KM-H2 cells failed to up-regulate autotaxin expression after infection.
Expression of autotaxin in lymphoid tissues. (A-B) Strong reactivity for autotaxin in HRS cells from EBV-positive primary HL tumors. (C) Lower levels of autotaxin expression in EBV-negative HRS cells despite strong labeling of nontumor cells (arrow). The difference in expression between EBV-negative and EBV-positive tumors was highly significant (see “Expression of autotaxin in primary lymphoma tissues”). In B-NHL, autotaxin expression was largely restricted to CD30+ ALCL (D), whereas most high-grade B-NHLs, such as BL (E) and CD30- DLBCL (F), and low-grade B-NHLs, such as chronic lymphocytic lymphoma (CLL) (G), were negative. (F-G, arrows) Strong labeling of endothelial cells. This restricted pattern may be partly attributed to the finding that among normal B cells, autotaxin expression was absent from germinal center (GC) and mantle zone (MZ) (H) but was expressed by CD138+ postgerminal center B cells (I). Images were acquired using Nikon TE2000 with 60 ×/1.4 NA oil immersion lens (Nikon, Kingston-upon-Thames, United Kingdom). Cells were stained with immunoperoxidase. Images were captured with a Nikon Coolpix 2100, and Paint Shop Pro 8.0 (Jase Software, Maidenhead, United Kingdom).
Expression of autotaxin in lymphoid tissues. (A-B) Strong reactivity for autotaxin in HRS cells from EBV-positive primary HL tumors. (C) Lower levels of autotaxin expression in EBV-negative HRS cells despite strong labeling of nontumor cells (arrow). The difference in expression between EBV-negative and EBV-positive tumors was highly significant (see “Expression of autotaxin in primary lymphoma tissues”). In B-NHL, autotaxin expression was largely restricted to CD30+ ALCL (D), whereas most high-grade B-NHLs, such as BL (E) and CD30- DLBCL (F), and low-grade B-NHLs, such as chronic lymphocytic lymphoma (CLL) (G), were negative. (F-G, arrows) Strong labeling of endothelial cells. This restricted pattern may be partly attributed to the finding that among normal B cells, autotaxin expression was absent from germinal center (GC) and mantle zone (MZ) (H) but was expressed by CD138+ postgerminal center B cells (I). Images were acquired using Nikon TE2000 with 60 ×/1.4 NA oil immersion lens (Nikon, Kingston-upon-Thames, United Kingdom). Cells were stained with immunoperoxidase. Images were captured with a Nikon Coolpix 2100, and Paint Shop Pro 8.0 (Jase Software, Maidenhead, United Kingdom).
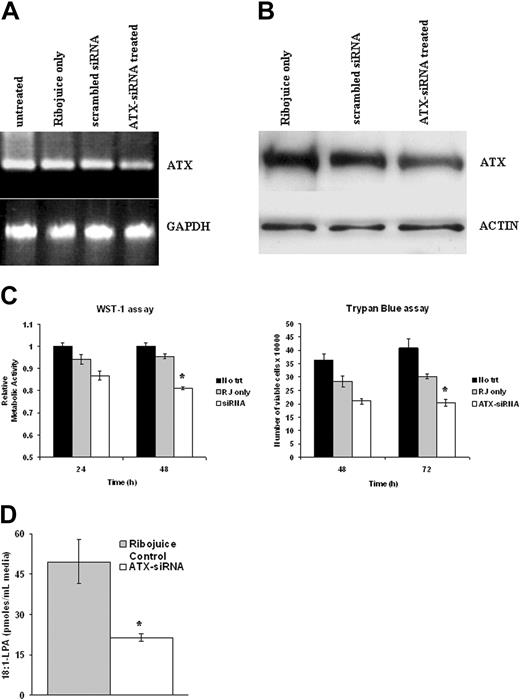
Autotaxin promotes the growth and survival of EBV-infected HL cells
To determine the contribution of autotaxin to the increased growth and survival of EBV-infected HL cells, we used siRNAs. Autotaxin mRNA levels in EBV-positive KM-H2 cells were reduced after 48 hours of treatment with autotaxin-specific siRNAs compared with control cells (cells treated with transfection reagent alone or with scrambled siRNA sequence); GAPDH transcription was unaffected by these treatments (Figure 5A). Knockdown of autotaxin protein in EBV-infected KM-H2 cells was also demonstrated after exposure to autotaxin-specific siRNAs (Figure 5B). Autotaxin knockdown resulted in significantly reduced cell growth (48 hours; P = .001) and viability (48 hours; P < .01) (Figure 5C) of HL cells compared with Ribojuice-only controls. Importantly, knockdown of autotaxin expression also resulted in a substantial decrease in the levels of LPA generated by conditioned medium derived from EBV-infected KM-H2 cells (Figure 5D) (P < .001).
Discussion
Despite the frequent detection of EBV in the tumor cells of Hodgkin lymphoma, the mechanisms underlying the contribution of virus infection to the development and maintenance of the transformed phenotype in this disease remains to be established. Here we studied the impact of EBV infection on the growth and survival of the HL-derived cell line, KM-H2. Although this cell line has not been definitively shown (ie, by examination of immunoglobulin gene rearrangements) to arise from the primary tumor, it is closely related cytogenetically19 and by gene expression20 to other established HL-derived cell lines. EBV-infected KM-H2 cells, displaying a pattern of virus gene expression largely restricted to Qp-driven EBNA1, LMP2, EBERs, and the BamH1A transcripts, show significantly increased cell growth and survival compared with uninfected controls. In vivo, EBV-infected HRS cells also show expression of this restricted subset of virus genes but, in addition, show very high levels of the latent membrane protein LMP1.6-8 Although it is generally supposed that the major contribution of EBV to the growth and survival of HRS cells is mediated through LMP1, our data demonstrate an important contribution from any one or a combination of the other genes. This scenario is similar to that of Burkitt lymphoma, in which the expression of a restricted subset of latent genes is associated with apoptosis resistance and increased tumorigenicity.36 Recently, Kis et al37 also failed to detect LMP1 expression in KMH2 cells infected with recombinant EBV. Interestingly, in their study, LMP1 expression could be induced in infected KMH2 cells after exposure to CD40 ligand and interleukin-4 (IL-4), suggesting that the latency II phenotype observed in vivo may be a consequence of the tumor microenvironment.
Increased soluble autotaxin production by EBV-infected HL cells is associated with increased use of LPC and generation of LPA. (A) (upper panel) Immunoblotting demonstrates increase in autotaxin protein levels in the supernatant of EBV-infected KM-H2 cells compared with controls. Other EBV-negative HL cells produced lower but detectable levels of soluble autotaxin compared with EBV-positive variants. (lower panel) EBV-negative KM-H2 cells cultured in supernatant from EBV-infected KM-H2 cells (neo cells, EBV media) showed a transient increase in growth compared with control (EBV-negative KM-H2 cells grown in their own supernatant; neo cells, neo media). EBV-positive KM-H2 cells grown in medium from EBV-negative KM-H2 cells (EBV cells, neo media) showed reduced growth compared with control (EBV-positive KM-H2 cells grown in their own supernatant EBV cells, EBV media). ▪ indicates Neo cells, Neo media; ▦, Neo cells, EBV media; ▨, EBV cells, Neo media; and □, EBV cells, EBV media. (B) KM-H2 cells treated with exogenously supplied LPAfollowed by cell growth assay (WST-1). LPA significantly increased the growth of EBV-negative KM-H2 cells (KM-H2-neo) but not of EBV-positive cells (KM-H2-EBV). ▪ indicates KM-H2-neo; ▦, KM-H2-neo + LPA; ▨, KM-H2-EBV; and □, KM-H2-EBV + LPA. (C) (top panel) Microarray analysis of LPA/SIP receptor (EDG receptor) expression in EBV-negative KMH-2 cells and EBV-positive KM-H2 cells (KM-H2 Akata) represented as Affymetrix “calls.” P indicates mRNA present; A, mRNA absent; M, mRNA called marginal. Although EBV-negative and EBV-positive KM-H2 cells expressed the S1PR1 receptor (EDG1), they lacked expression of the major LPA receptor, LPAR1 (EDG2). The other known LPA or S1P receptors (EDG3-8) were lacking in these cells. (bottom panel) These findings were confirmed by RT-PCR analysis of a range of EBV-positive and EBV-negative HL cells (data for EDG1 and EDG2 shown). (D) Hydrolysis of 20 μM LPC by serum-free-conditioned medium, collected after 16 hours of culture over a 5-hour incubation. (E) Generation of LPA by incubation of 20 μM LPC for 3 hours with KM-H2 or KM-H2-EBV cell-conditioned medium. (F) Acyl species analysis of LPA generated by incubation of egg LPC with conditioned media. The 18:1 and 18:0 LPAs were generated by the cells without added LPC. □ indicates KM-H2 neo; and ▪, KMH2-EBV. See the “Statistical analysis” section under “Materials and methods” for explanation of error bars and asterisks.
Increased soluble autotaxin production by EBV-infected HL cells is associated with increased use of LPC and generation of LPA. (A) (upper panel) Immunoblotting demonstrates increase in autotaxin protein levels in the supernatant of EBV-infected KM-H2 cells compared with controls. Other EBV-negative HL cells produced lower but detectable levels of soluble autotaxin compared with EBV-positive variants. (lower panel) EBV-negative KM-H2 cells cultured in supernatant from EBV-infected KM-H2 cells (neo cells, EBV media) showed a transient increase in growth compared with control (EBV-negative KM-H2 cells grown in their own supernatant; neo cells, neo media). EBV-positive KM-H2 cells grown in medium from EBV-negative KM-H2 cells (EBV cells, neo media) showed reduced growth compared with control (EBV-positive KM-H2 cells grown in their own supernatant EBV cells, EBV media). ▪ indicates Neo cells, Neo media; ▦, Neo cells, EBV media; ▨, EBV cells, Neo media; and □, EBV cells, EBV media. (B) KM-H2 cells treated with exogenously supplied LPAfollowed by cell growth assay (WST-1). LPA significantly increased the growth of EBV-negative KM-H2 cells (KM-H2-neo) but not of EBV-positive cells (KM-H2-EBV). ▪ indicates KM-H2-neo; ▦, KM-H2-neo + LPA; ▨, KM-H2-EBV; and □, KM-H2-EBV + LPA. (C) (top panel) Microarray analysis of LPA/SIP receptor (EDG receptor) expression in EBV-negative KMH-2 cells and EBV-positive KM-H2 cells (KM-H2 Akata) represented as Affymetrix “calls.” P indicates mRNA present; A, mRNA absent; M, mRNA called marginal. Although EBV-negative and EBV-positive KM-H2 cells expressed the S1PR1 receptor (EDG1), they lacked expression of the major LPA receptor, LPAR1 (EDG2). The other known LPA or S1P receptors (EDG3-8) were lacking in these cells. (bottom panel) These findings were confirmed by RT-PCR analysis of a range of EBV-positive and EBV-negative HL cells (data for EDG1 and EDG2 shown). (D) Hydrolysis of 20 μM LPC by serum-free-conditioned medium, collected after 16 hours of culture over a 5-hour incubation. (E) Generation of LPA by incubation of 20 μM LPC for 3 hours with KM-H2 or KM-H2-EBV cell-conditioned medium. (F) Acyl species analysis of LPA generated by incubation of egg LPC with conditioned media. The 18:1 and 18:0 LPAs were generated by the cells without added LPC. □ indicates KM-H2 neo; and ▪, KMH2-EBV. See the “Statistical analysis” section under “Materials and methods” for explanation of error bars and asterisks.
Autotaxin promotes the growth and survival of EBV-infected HL cells. (A) (top left) Semiquantitative RT-PCR analysis of equivalent starting amounts of cDNA (shown here are results of 30 cycles of amplification) from untreated EBV-positive KM-H2 cells or these cells treated for 48 hours with transfection reagent alone (Ribojuice only), Ribojuice plus scrambled siRNA, or autotaxin (ATX)-specific siRNAs. A clear difference in autotaxin mRNA levels between siRNA-transfected and control cells was detectable. (bottom panel) GAPDH mRNA levels were unaffected by these treatments. (B) Immunoblotting demonstrates knockdown of autotaxin protein in EBV-positive KM-H2 cells treated with autotaxin-specific siRNAs compared with controls. (C) Treatment of EBV-infected KM-H2 cells with autotaxin-specific siRNAs resulted in significant reduction in proliferation after 48 hours (top panel) and cell viability after 72 hours (bottom panel) compared with cells treated with transfection reagent alone or with scrambled siRNA (data not shown). RJ indicates ribojuice transfection reagent. (D) Down-regulation of autotaxin expression in EBV-infected KM-H2 cells resulted in reduced generation of LPA from LPC.
Autotaxin promotes the growth and survival of EBV-infected HL cells. (A) (top left) Semiquantitative RT-PCR analysis of equivalent starting amounts of cDNA (shown here are results of 30 cycles of amplification) from untreated EBV-positive KM-H2 cells or these cells treated for 48 hours with transfection reagent alone (Ribojuice only), Ribojuice plus scrambled siRNA, or autotaxin (ATX)-specific siRNAs. A clear difference in autotaxin mRNA levels between siRNA-transfected and control cells was detectable. (bottom panel) GAPDH mRNA levels were unaffected by these treatments. (B) Immunoblotting demonstrates knockdown of autotaxin protein in EBV-positive KM-H2 cells treated with autotaxin-specific siRNAs compared with controls. (C) Treatment of EBV-infected KM-H2 cells with autotaxin-specific siRNAs resulted in significant reduction in proliferation after 48 hours (top panel) and cell viability after 72 hours (bottom panel) compared with cells treated with transfection reagent alone or with scrambled siRNA (data not shown). RJ indicates ribojuice transfection reagent. (D) Down-regulation of autotaxin expression in EBV-infected KM-H2 cells resulted in reduced generation of LPA from LPC.
In the present study, the loss of EBV from L591 cells also resulted in dramatically impaired cell growth and survival, probably the result of the combined loss of all latent virus gene expression given that EBV-positive L591 cells displayed a latency 3 form of virus gene expression in which all known latent genes, including LMP1 and LMP2, are expressed. Although the provenance of L591 remains uncertain, it is possible it originated from a type 2 latency expressing primary tumor that subsequently drifted to latency 3 phenotype, analogous to a similar drift that occurs in some type 1 latency BL cells after serial passage. In this respect it is interesting that, unlike LCLs, latency 3-expressing L591 cells maintained the expression of autotaxin.
We used microarray analysis to determine whether the growth- and survival-promoting effects of EBV we observed were a consequence of cellular transcriptional changes. Of the genes differentially expressed in EBV-infected HL cells, autotaxin is not only the most highly up-regulated probe set, it is also a critical mediator of many of the biologic processes associated with malignancy. In particular, autotaxin-expressing NIH3T3 cells display a more invasive phenotype, and autotaxin expression in ras-transformed NIH3T3 cells enhances tumorigenesis and metastatic potential compared with ras-transformed controls.10 Autotaxin expression results in increased motility and invasiveness of breast cancer cells38 and is reported to be up-regulated in metastatic compared with nonmetastatic variants of the MDA-MB-435 breast cancer cell line.39 Furthermore, autotaxin has been shown to be an angiogenic factor that stimulates human endothelial cells grown on Matrigel to form tubules similar to those induced by vascular endothelial growth factor.12 Many of the biologic effects of autotaxin are the result of its PLD activity, which can generate LPA11 and S1P13 from LPC and SPC, respectively. LPA and S1P bind to specific G-protein-coupled receptors (formerly the EDG receptors) to exert their biologic effects. A potential role for LPA in human oncogenesis was first suggested by the observation that LPA is present at elevated levels in ascites of patients with ovarian cancer.14 Subsequently, it has been shown that LPA is a potent tumor-promoting small molecule in a range of biologic systems, stimulating the proliferation, migration, and survival of cancer cells.15
Induction of autotaxin by EBV infection of KM-H2 cells was initially confirmed by RT-PCR analysis and subsequently by immunoblotting using the monoclonal antibodies we had developed. This effect was not unique to the KM-H2 HL cell line because the down-regulation of autotaxin expression also accompanied the loss of the EBV genome from L591 cells. Furthermore, most EBV-negative HL cell lines displayed low levels of autotaxin mRNA and protein. It should be noted that some of these lines may not be representative of most HL cells given that L540 is derived from T cells (it harbors TCR gene rearrangements) and HD-My-Z is probably of monocytic origin. It should be noted that despite the existence of a subset of HL tumors in which there is tumor expression of T-cell antigens, only a small minority of these are genuinely of T-cell origin, as defined by the presence of TCR rearrangements. Importantly, the highly significant association of autotaxin expression with the presence of EBV in primary HRS cells strongly suggests that EBV also induces autotaxin in primary HL tumors. Unfortunately, it has been impossible to date to study cultured primary HRS cells from tissue biopsies, so we were unable to determine whether the induction of autotaxin in primary HRS cells was also associated with phenotypic effects similar to those observed in the cell lines. Absent or low-level expression of autotaxin expression in EBV-transformed lymphoblastoid cells, and in a range of Burkitt lymphoma cells lines irrespective of EBV status, demonstrates that EBV-mediated induction of autotaxin is likely to be restricted to HL cells. In fact, we were also unable to observe the induction of autotaxin by EBV in nasopharyngeal carcinoma cells, even when we infected such cells with the same virus used to derive the EBV-positive KM-H2 cells. In addition to these observations, we were able to show that autotaxin, though frequently expressed in primary HL, was only infrequently observed in most NHLs with the exception of CD30+ anaplastic large-cell lymphomas (ALCLs)—tumors phenotypically closely related to HLs. Recent microarray analysis of primary effusion lymphomas that compared their gene expression with that of other lymphoid cells lines identified autotaxin expression only in the PEL- and HL-derived cell lines, confirming the apparently restricted expression of autotaxin to tumors derived from certain specific B-cell lineages.40
The mechanisms that underlie the cell-type specificity of autotaxin induction by EBV infection of HL cells are intriguing. Recently, constitutively activated AP-1 with c-Jun and JunB overexpression has been described in the tumor cells of HL and ALCL but not in other lymphoma types.41 Furthermore, the transformation of chick embryo fibroblasts by v-Jun has been shown to result in an approximately 100-fold induction of autotaxin mRNA,42 suggesting that AP-1 activity might be responsible for the restricted expression of autotaxin. It is also unclear which of the EBV genes expressed in the EBV-positive HRS cells is responsible for the induction of autotaxin. This is under investigation, as is whether autotaxin is induced through a pathway involving the activation of AP-1.
We investigated whether the induction of autotaxin we observed in HL cells was of biologic significance. The demonstration of increased levels of soluble autotaxin in the supernatant of EBV-infected HL cells, together with the finding that conditioned media from EBV-infected HL cells were able to increase the growth of EBV-negative cells, strongly suggested this was the case. Furthermore, elevated autotaxin levels were associated with the increased use of LPC and the increased generation of LPA. Secreted autotaxin can increase LPA production. Our finding that EBV-infected HL cells degraded greater than 50% of generated LPA within 10 minutes emphasized the significance of this. Thus, the sustained release of autotaxin is necessary for the continued LPA generation necessary for increased proliferation and survival. We were unable to detect the formation of S1P from SPC by the autotaxin-conditioned medium, demonstrating that though purified autotaxin can hydrolyze SPC, its physiologically relevant substrate is likely to be LPC. It is probable that S1P generation is through a sphingosine kinase-catalyzed route.
Although these data strongly suggested that the induction of autotaxin and the subsequent generation of LPA were responsible for the growth- and survival-promoting effects of EBV in HL cells, definitive proof of this required a more direct approach. Therefore, we used siRNAs to down-regulate autotaxin expression at the mRNA and the protein levels. Reduction in autotaxin expression was associated with significant decreases in cell growth and viability of EBV-infected KM-H2 cells. Furthermore, the increased generation of LPA we had observed after the induction of autotaxin in EBV-positive cells was partially reversed by treatment with autotaxin-specific siRNAs.
Taken together, our data demonstrate an important and specific consequence of EBV infection of the malignant cells of HL: namely, the induction of autotaxin. In vitro, this induction leads to the generation of LPA and to the increased growth and survival of EBV-infected HRS cells. This is the first demonstration that virus infection leads directly to the synthesis of the growth-promoting lipid LPA and that it could represent a more general pathway used by herpesviruses during their normal life cycle or during the initiation and maintenance of virus-associated tumors. Targeting this pathway could represent a novel approach to the treatment of EBV-associated HL. The development of specific inhibitors of autotaxin production or activity should lead to a decrease in LPA levels within the tumor microenvironment. This, in turn, would lead to a reduction in the levels of LPA receptor activation and a subsequent decrease in LPA-mediated downstream intracellular signaling. Within the context of EBV-associated primary HL, the increased levels of autotaxin and LPA may function in an autocrine or a paracrine manner, directly affecting the malignant HRS cells themselves, the reactive lymphocytic infiltrate, or both.
Prepublished online as Blood First Edition Paper, June 2, 2005; DOI 10.1182/blood-2005-02-0471.
Supported by the Leukaemia Research Fund.
K.R.N.B. and J.R.F. contributed equally to this study.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal