Abstract
Mutations in the kinase domain (KD) of BCR-ABL are the leading cause of acquired imatinib resistance. In some cases, identical mutations were detected at relapse and in pretherapeutic specimens, consistent with selection of resistant clones in the presence of drug. However, the incidence of KD mutations in imatinibnaive patients, irrespective of response to therapy, is unknown. We studied mutation frequency in 66 patients with chronic myelogenous leukemia (CML), using cDNA sequencing and allele-specific oligonucleotide-polymerase chain reaction (ASO-PCR) assays for 8 common mutations. Thirteen patients were positive by ASO-PCR only, 1 by ASO-PCR and sequencing, and 1 by sequencing only (overall frequency, 22.7%). T315I was most frequent (12% of patients). Eleven of the 14 patients with positive ASO-PCR had follow-up samples available for sequencing. Wild-type sequence was detected in 6 of 11, 2 different mutations in 1 of 11, and identical mutations in 4 of 11 patients, 2 of whom had achieved major cytogenetic response. In multivariate analysis mutation detection was associated with clonal cytogenetic evolution, exposure to 6-Thioguanine, and a low platelet count, but not with response to imatinib, event-free survival, and overall survival. KD mutants present at low levels do not invariably lead to relapse, and additional factors are required to induce a fully drug-resistant phenotype. (Blood. 2005; 106:2128-2137)
Introduction
Imatinib is a highly effective therapy in all stages of chronic myelogenous leukemia (CML), but relapse after an initial response is common in patients with advanced disease.1-3 In 50% to 90% of patients with acquired resistance to imatinib, mutations in the kinase domain (KD) of BCR-ABL have been detected. These mutations result in impaired drug binding and are thought to cause clinical resistance.4-8 Although mutations have been identified in more than 20 different amino acids, the 8 most common mutations account for greater than 70% of cases.9 In some patients who relapsed with KD mutations, analysis of archived material stored prior to therapy revealed the identical nucleotide exchanges.5,10-12 In most of these cases the proportion of mutant allele was small, and detection required sensitive techniques, such as polymerase chain reaction with allele-specific oligonucleotides (ASO-PCR), but isolated cases with up to 40% of mutant allele were also reported.5 Overall, these data are consistent with selection of resistant clones in the presence of imatinib. The fact that some KD domain mutant clones expand to the level of detection even before initiation of therapy suggests that some mutations may confer a growth advantage even in the absence of imatinib. Such mutants would be gain-of-function over wild type and could thus contribute to disease progression. The available data from imatinib-naive samples are limited to patients in whom mutations were detected at the time of relapse, and the incidence of mutations prior to therapy in unselected patients has not been established. Furthermore, it is not known whether KD mutant clones that are detectable prior to therapy are regularly selected in the presence of imatinib, and whether they may predict for an adverse outcome on therapy. To address these questions, we analyzed the frequency of KD mutations in a cohort of imatinib-naive patients with CML in chronic phase, accelerated phase, or blast crisis. We used a panel of newly developed fluorescence-based allele-specific PCR assays for high-sensitivity detection of the most common mutations as well as conventional sequencing of PCR products amplified from cDNA. The detection of KD mutants was then correlated with baseline variables and outcome.
Patients, materials, and methods
Patients
Sixty-six patients with CML in first chronic phase (n = 20), accelerated phase (n = 27), and myeloid blast crisis (n = 19) were treated with imatinib within Novartis-sponsored multi-institutional trials approved by the Institutional Review Board of the University of Leipzig, Leipzig, Germany. Prior to the study, informed consent was obtained according to the Declaration of Helsinki. Patient characteristics are given in Table 1. Accelerated phase was defined as any of the following: basophils in the peripheral blood (PB) of at least 20%, blasts in PB or bone marrow (BM) greater than 15% but less than 30%, blasts plus promyelocytes in the PB greater than 30%, platelet count less than 100 × 109/L (unless related to therapy), or clonal cytogenetic abnormalities in addition to the Philadelphia chromosome. Blastic phase was diagnosed when the blast count in the BM or PB exceeded 30%. Patients with chronic phase received an initial dose of 400 mg imatinib daily, and patients with accelerated phase or blast crisis received 600 mg daily. Complete hematologic response (CHR), partial hematological response (PHR), and major cytogenetic response (MCR) were defined according to published response criteria.1,2,13 The patients were routinely followed by peripheral blood counts and clinical examinations at 2- to 4-week intervals. Cytogenetics and quantitative reverse transcription (RT)-PCR14 were done every 3 to 6 months.
Patient characteristics
Characteristic . | Data . |
|---|---|
| Sex, no. (%) | |
| Female | 38 (57.6) |
| Male | 28 (42.4) |
| Age, y, median (range) | 57 (31-80) |
| Disease duration, mo, median (range) | 30 (0-190) |
| Disease phase, no. (%) | |
| Chronic | 20 (30.3) |
| Accelerated | 27 (40.1) |
| Clonal cytogenetic evolution only | 10 (15.2) |
| Blastic | 19 (28.8) |
| Clonal cytogenetic evolution, no. (%)† | 26 (39.3) |
| Prior therapy, no. (%) | |
| Hydroxyurea | 51 (77.3) |
| Interferon-α | 37 (56.1) |
| Cytarabine | 18 (27.3) |
| Topoisomerase II inhibitors | 11 (16.7) |
| 6-Mercaptopurine | 6 (9.1) |
| 6-Thioguanine | 6 (9.1) |
| Busulfan | 5 (7.8) |
| Melphalan | 3 (4.5) |
| Other* | 4 (6.1) |
| None | 4 (6.1) |
Characteristic . | Data . |
|---|---|
| Sex, no. (%) | |
| Female | 38 (57.6) |
| Male | 28 (42.4) |
| Age, y, median (range) | 57 (31-80) |
| Disease duration, mo, median (range) | 30 (0-190) |
| Disease phase, no. (%) | |
| Chronic | 20 (30.3) |
| Accelerated | 27 (40.1) |
| Clonal cytogenetic evolution only | 10 (15.2) |
| Blastic | 19 (28.8) |
| Clonal cytogenetic evolution, no. (%)† | 26 (39.3) |
| Prior therapy, no. (%) | |
| Hydroxyurea | 51 (77.3) |
| Interferon-α | 37 (56.1) |
| Cytarabine | 18 (27.3) |
| Topoisomerase II inhibitors | 11 (16.7) |
| 6-Mercaptopurine | 6 (9.1) |
| 6-Thioguanine | 6 (9.1) |
| Busulfan | 5 (7.8) |
| Melphalan | 3 (4.5) |
| Other* | 4 (6.1) |
| None | 4 (6.1) |
Includes vincristine (1 patient), fludarabine (1), splenic radiotherapy (1), and total-body irradiation before allografting (1).
Data available for 62 of 66 patients.
Cell lines
BaF3 cells expressing the Q252H, Y253F, Y253H, E255K, E255V, T315I, M351T, and F359V mutants of p210BCR-ABL have been described or were generated accordingly.7,15,16 The CML cell line K562 was used as a positive control for wild-type BCR-ABL, while HL60 cells were used as negative control. All cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and penicillin and streptomycin.
Allele-specific real-time PCR
RNA was extracted with affinity columns (RNAeasy kit, Qiagen, Hilden, Germany) from 5 to 10 × 106 nucleated bone marrow cells isolated by red cell lysis. cDNA was synthesized with random hexamer primers in 40-μL reactions, as described.17 ABL was amplified as a quality control for the cDNA preparation.17
Conditions for allele-specific PCR amplification of the 8 most common BCR-ABL mutants were optimized on an MJ Research Opticon 2 System (MJ Research, Reno, NV). PCR amplification was performed in a single step using 1 of 2 universal 5′ primers located on BCR exon 13 and individual allele-specific 3′ primers (Table 1). Fluorescence was detected by SybrGreen (Applied Biosystems, Foster City, CA). For optimization of conditions, serial dilutions of mutant plasmid DNA in distilled water were used as well as cDNA prepared from serial dilutions of RNA from BaF3 cells expressing mutant BCR-ABL into RNA from BaF3 cells expressing wild-type BCR-ABL. Preliminary experiments showed that the sensitivity of the assay could be increased by 1:10 dilution of the cDNA. The precise reason for this phenomenon is unknown, although similar observations were made for ultrasensitive BCR-ABL amplification for detection of BCR-ABL mRNA in healthy individuals.18 Assays were deemed acceptable if they (1) detected mutant BCR-ABL with a sensitivity of at least 1:103 in cell lines (RNA from BaF3 cells expressing mutant BCR-ABL diluted in RNA from BaF3 cells expressing wild-type BCR-ABL), (2) consistently showed no amplification of wild-type only and BCR-ABL-negative controls after 45 (Q252H, Y253F, Y253H, E255K) or 50 cycles (E255V, T315I, M351T, F359V) of PCR, and (3) generated PCR products with a consistent melting curve in all positive control reactions.12
Patient samples were tested twice in triplicate 20-μL reactions for all 8 mutations. Each reaction contained 1 × SybrGreen Master Mix (Applied Biosystems), 20 pmol each primer, 0.5 × PCR Enhancer (Invitrogen, Carlsbad, CA), and 2 μL diluted cDNA (equivalent to 2.5-5 × 104 cells, respectively). cDNA equivalent to at least 1.5 × 105 cells was screened for each of the 8 mutations. In each individual experiment, the sensitivity was monitored by amplification in duplicate of a serial dilution of mutant plasmid DNA. Strict precautions were taken to avoid false-positive results. The RNA extraction and cDNA synthesis were performed in a diagnostic laboratory (Leipzig, Germany) that had never handled recombinant mutant BCR-ABL. All reagents, including primers and water, pipette tips, tubes, and pipettors were new and exclusively used for this study. The reaction mixes were prepared, and patient samples were added in a “BCR-ABL-naive” laboratory located in a separate building at Oregon Health and Science University. The plates were sealed over the patient reactions and transported to the investigators' laboratory, where the controls were added to an unsealed portion of the plate. Negative controls (all in duplicate) included HL60 and K562 cells as well as no template tubes. Samples were scored positive, when all of the following criteria were met: (1) at least 1 of 3 wells positive in at least 1 of 2 assays, (2) amplification detected at least 3 cycles before the maximum cycle number of the assay (ie, at no more than 42 cycles for Q252H, Y253F, Y253H, E255K and at no more than 47 cycles for E255V, T315I, M351T, F359V), (3) shape of the melting curve identical to control (to avoid that fluorescence from nonspecific amplification products was scored as a true positive), (d) no specific amplification in any of the BCR-ABL wild-type or BCR-ABL-negative controls.
Mutation analysis by sequencing of PCR products
Patient cDNA was PCR amplified using B2A forward (TTCAGAAGCTTCTCCCTGACAT) and Abl 4065 reverse (CCTTCTCTAGCAGCTCATACACCTG). One microliter of this reaction was used as template in the nested reaction using Bcr F4 forward (ACAGCATTCCGCTGACCATCAATA) and U396 reverse (GCCATAGGTAGCAATTTCCC). All PCRs were performed with Accuprime polymerase (Invitrogen). PCR products were sequenced with both forward primer 3306F (TGGTTCATCATCATTCAACGG) and reverse primer 4000R (GGACATGCCATAGGTAGCA). Sequences were aligned and analyzed with Sequencher sequence analysis software (Gene Codes, Ann Arbor, MI).
Statistical analysis
We analyzed a number of pretreatment characteristics for association with the detection of KD mutations, hematologic response, or MCR, including sex, age (≤ 60 years, > 60 years), disease duration (≤ 12 months, > 12 months), therapy prior to imatinib, disease phase, clonal cytogenetic evolution (CE), splenomegaly, hemoglobin level (≤ 100 g/L, > 100 g/L), white blood cell count (≤ 10 × 109/L, > 10 × 109/L), platelet count (≤ 100 × 109/L, >100 × 109/L but ≤ 450 × 109/L, > 450 × 109/L), blasts in PB (≤ 7%, > 7%), basophils in the PB (≤ 7%, > 7%), blasts in BM (≤ 5%, > 5%). Data were analyzed by univariate logistic regression analysis with all pretreatment characteristics treated as independent variables. For the multivariate logistic regression, the stepwise procedure was used to identify pretreatment characteristics for KD mutation, PHR, or MCR among pretreatment characteristics with a P value less than .1 from univariate logistic regression. Only patients with complete data sets were included in the multivariate model. Event-free survival was defined as the interval from starting imatinib therapy to loss of PHR, CHR, or MCR, whichever occurred first, to initiation of alternative treatment or to death. Patients who underwent allogeneic stem cell transplantation without fulfilling any of these criteria were censored at the time of the allograft. Overall survival was defined as the interval between starting imatinib and death, regardless of the time of discontinuation of imatinib. Patients who underwent allografting were censored at the time of transplantation. Patients who discontinued imatinib because of toxicity or unrelated causes before progression had occurred were excluded from the analysis of event-free survival and overall survival.
Results
Establishment of allele-specific PCR assays for detection of KD mutations.
Because the purpose of our study was to screen for KD mutations with high sensitivity, we chose ASO-PCR as the screening strategy. ASO-PCR has been reported to achieve a sensitivity of up to 1 copy of mutant allele in 104 copies of wild-type allele.19 However, given that a separate assay, performed in multiple replicates, is required to test for each of the mutations, the number of different mutations included in the screening had to be limited. At least 28 different kinase domain mutations have been reported in patients with resistance to imatinib.4-8,15,16,20-22 From these, we selected mutations based on their prevalence in published series. Initial experiments were carried out to optimize the sensitivity and specificity of ASO-PCR for each of the selected mutations. Evaluation of primers designed to amplify ABL versus a combination of primers on BCR with the mutant ABL reverse primer showed the latter to be more specific. Attempts to use nested PCR to increase sensitivity led to loss of specificity, with detection of signals in wild-type BCR-ABL specimens (not shown). Eventually, optimal results were achieved with a 1-step approach using 1 of 2 universal 5′ primers on BCR exon 13 in conjunction with allele-specific primers complementary to the respective mutations on ABL. In several instances, the primer design included additional mismatches in the ABL primer to suppress amplification of wild-type allele. The melting curves of the amplification products were routinely compared with the melting curves of the simultaneously amplified standards, and only reactions with identical melting curves were scored positive (Figure 1). Overall, we established assays with a sensitivity of at least 1:103 for 8 common mutations, representing approximately 72% of clinical isolates.9 For 7 of these mutations, the sensitivity was reproducibly more than 105, while it was lower for Y253H (≥ 103) (Table 2).
Primer sequences, PCR conditions, and sensitivity of the ASO-PCR assays used in this study
Mutant . | Base exchange . | Frequency, %* . | Primer names . | Sequence (5′ → 3′) . | Annealing temperature, °C . | No. cycles . | Product length, bp . | Extension time, s . | Detection limit, mutant/wild type . |
|---|---|---|---|---|---|---|---|---|---|
| Q252H | G-C | 5.1 | 252H C3R | TCGTACACCTCCCCGCAG | 60.0 | 45 | 789 | 60 | ≤ 1/100 000 |
| Bcr 2687F | CGCTGACCATCAATAAGG | ||||||||
| Y253F | A-T | 3.4 | 253F R | CTCGTACACCTCCCCGA | 60.0 | 45 | 790 | 60 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG | ||||||||
| Y253H | T-C | 6.8 | 253H 16R | CGTACACCTCCCCGTG | 60.4 | 45 | 805 | 60 | ≤ 1/1000 |
| Bcr 2671F | ACTCCAGACTGTCCACAGCAT | ||||||||
| E255K | G-A | 19.2 | 255K T3R | CACGCCCTCGTACACTTT | 60.0 | 45 | 798 | 60 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG | ||||||||
| E255V | A-T | 2.3 | 255V T3R | ACACGCCCTCGTACATCA | 60.0 | 45 | 797 | 60 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG | ||||||||
| T315I | C-T | 20.9 | 315 R | CGTAGGTCATGAACTCAA | 60.0 | 50 | 994 | 80 | ≤ 1/100 000 |
| Bcr2671F | ACTCCAGACTGTCCACAGCAT | ||||||||
| M351T | T-C | 15.3 | 351 G3R | TCTTCTCCAGGTACTGCG | 60.0 | 50 | 1085 | 90 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG | ||||||||
| F359V | T-G | 4.0 | 359 R | AAGATCTCTGTGGATGAC | 60.0 | 50 | 1107 | 90 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG |
Mutant . | Base exchange . | Frequency, %* . | Primer names . | Sequence (5′ → 3′) . | Annealing temperature, °C . | No. cycles . | Product length, bp . | Extension time, s . | Detection limit, mutant/wild type . |
|---|---|---|---|---|---|---|---|---|---|
| Q252H | G-C | 5.1 | 252H C3R | TCGTACACCTCCCCGCAG | 60.0 | 45 | 789 | 60 | ≤ 1/100 000 |
| Bcr 2687F | CGCTGACCATCAATAAGG | ||||||||
| Y253F | A-T | 3.4 | 253F R | CTCGTACACCTCCCCGA | 60.0 | 45 | 790 | 60 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG | ||||||||
| Y253H | T-C | 6.8 | 253H 16R | CGTACACCTCCCCGTG | 60.4 | 45 | 805 | 60 | ≤ 1/1000 |
| Bcr 2671F | ACTCCAGACTGTCCACAGCAT | ||||||||
| E255K | G-A | 19.2 | 255K T3R | CACGCCCTCGTACACTTT | 60.0 | 45 | 798 | 60 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG | ||||||||
| E255V | A-T | 2.3 | 255V T3R | ACACGCCCTCGTACATCA | 60.0 | 45 | 797 | 60 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG | ||||||||
| T315I | C-T | 20.9 | 315 R | CGTAGGTCATGAACTCAA | 60.0 | 50 | 994 | 80 | ≤ 1/100 000 |
| Bcr2671F | ACTCCAGACTGTCCACAGCAT | ||||||||
| M351T | T-C | 15.3 | 351 G3R | TCTTCTCCAGGTACTGCG | 60.0 | 50 | 1085 | 90 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG | ||||||||
| F359V | T-G | 4.0 | 359 R | AAGATCTCTGTGGATGAC | 60.0 | 50 | 1107 | 90 | ≤ 1/100 000 |
| Bcr2687F | CGCTGACCATCAATAAGG |
The last nucleotide of the reverse primer matches the mutated base. Additional mismatches versus wild type are in bold and underlined.
N = 177.9
KD mutations are detectable in patients with CML in accelerated phase or blast crisis but not in chronic phase
Pretherapeutic samples from 66 patients were analyzed for all 8 mutations. These 66 patients consisted of 20 patients in first chronic phase, 27 in accelerated phase, and 19 in myeloid blast crisis, all previously untreated with imatinib. Mutations were detected by ASO-PCR in 14 (21.2%) of 66 patients. T315I was detected in 8 patients and was thus the most frequently detected mutation (12.1% of all samples). The Y253H mutant was detected in 2 patients. Q252H, as well as F359V, was seen in 1 patient each. In contrast, the E255V, E255K, and M351T mutations were not detected (Table 3). In most cases, the mutations were detected only at high cycle numbers, and only a fraction of the replicate reactions was positive (Table 4). This is consistent with low levels of mutant allele.
Frequency and types of mutations detected according to disease phase
. | Accelerated phase, no. (%) . | Myeloid blast crisis, no. (%) . | Total, no. (%) . |
|---|---|---|---|
| No. patients | 27 | 19 | NA |
| Q252H | 1 | 0 | 1 (1.5) |
| Y253F | 1 | 1 | 2 (3.0) |
| Y253H | 1 | 1 | 2 (3.0) |
| E255K | 0 | 0 | 0 (0) |
| E255V | 0 | 0 | 0 (0) |
| T315I | 7 | 1 | 8 (12.1) |
| M351T | 0 | 0 | 0 (0) |
| F359V | 0 | 1 | 1 (1.5) |
| V289I* | 0 | 1 | 1 (1.5) |
| Total (% of patients) | 10 (37.0) | 5 (23.8) | NA |
. | Accelerated phase, no. (%) . | Myeloid blast crisis, no. (%) . | Total, no. (%) . |
|---|---|---|---|
| No. patients | 27 | 19 | NA |
| Q252H | 1 | 0 | 1 (1.5) |
| Y253F | 1 | 1 | 2 (3.0) |
| Y253H | 1 | 1 | 2 (3.0) |
| E255K | 0 | 0 | 0 (0) |
| E255V | 0 | 0 | 0 (0) |
| T315I | 7 | 1 | 8 (12.1) |
| M351T | 0 | 0 | 0 (0) |
| F359V | 0 | 1 | 1 (1.5) |
| V289I* | 0 | 1 | 1 (1.5) |
| Total (% of patients) | 10 (37.0) | 5 (23.8) | NA |
No mutations were detected in the 20 patients with chronic-phase disease.
NA indicates not applicable
This mutation was not included in the ASO-PCR assay and was detected by sequencing.
Details on patients with detection of kinase domain mutations
Patient no. . | Disease phase . | Therapy prior to imatinib . | Karyotype . | Mutation by ASO-PCR . | Ct mut . | Positive reactions, no./total . | Sequencing . | Best response . | Follow-up sequencing/disease status . | Last follow-up/disease status . |
|---|---|---|---|---|---|---|---|---|---|---|
| 7 | AP | IFN | 46,XY,t(9,22)[18]; 47,XY,t(9;22), +der(22)[6];47, XY,+8[1] | T3151 | 45.8 | 1/6 | WT | CHR | WT day +42/PHR; WT day +107/PHR;WT day +785/CHR | Alive in CHR day +1591 |
| 13 | AP | HU | 46,XY,t(9;22)[24]/47,XY,t(9;22), +der(22)[1] | Q252H | 35.6-40.4 | 3/6 | WT | MCR (20% Ph+) | Q252H day +176/MCR; Q252H+F359V day +325/MCR | Alive in MCR day +1087 |
| 17 | AP | IFN, HU | 46,XY,t(9;22)(q34; q11)[25] | T3151 | 46.9 | 1/6 | WT | PHR | WT day + 80/PHR; WT day+270/PHR; WT day +444/hematologic relapse | Hematologic relapse day + 531; lost to follow-up |
| 25 | AP | HU | 46,XX,add(6)(p25), t(9;22)(q34;q11) [20] | T3151 | 46.7 | 1/6 | WT | CHR | E255K+M351T day +365/CHR | Allogeneic transplant day +512 for lack of MCR |
| 29 | AP | HU, IFN | 45X-Y,t(9;22) (q34;q11) | Y253H | 40.2 | 1/6 | WT | mCR (52% Ph+) | WT day +85/CHR; WT day +267/CHR | Hematologic relapse day +452; lost to follow-up |
| 36 | AP | HU, IFN, 6-TG | 47,XY,t(9;22) (q34;q11),+der(22) [23];46,XY,t(9;22) (q34;q11)[1] | T3151 | 46.6-47.9 | 2/6 | K247R | CHR | K247R day +107/CHR | Hematologic relapse day +212; alive day +1011 |
| 41 | AP | none | 46,XX,t(9;22) (q34;q11),−der(22), +idic der (22)[12] | T3151 | 43.7 | 1/6 | WT | CCR | T3151 day +93/CCR; T3151 day +182/CCR | Alive in molecular remission day + 1009 |
| 42 | AP | IDA, ARA-C, IFN | 48,XY,+8,t(9; 22)(q34;q11),+der (22)t(9;22)(q34; q11)[8] | T3151 | 44.1-45.7 | 2/6 | WT | CCR | WT day+193/CCR | Alive in CCR day +661 |
| 45 | AP | HU, 6-TG | 44,XX,t(9;22)(q34; q11),der(14q+), t(17;18)(p11;q12?), −18,−20[23] | Y253F | 38.2 | 1/6 | WT | NA | ND | Died day +9 from sepsis |
| 46 | AP | HU, 6-TG | 46,XX,t(9;22)[10] | T3151 | 37.92-41.68 | 2/6 | WT | CHR | T3151 day +158/hematologic relapse | Hematologic relapse + 158; died day + 1350 from progressive disease |
| 48 | BC | HU, IFN | 47,XX,+8,t(9; 22)(q34;11) [16],46,XX,t(9; 22)[4] | T3151 | 45.6 | 1/6 | WT | NR | T3151 day +19/continuous blast crisis | Allogeneic transplant day + 140 for progressive disease; died from TRM |
| 55 | BC | HU | ND | F359V | 39.4-44.8 | 5/6 | F359V | NR | ND | Died day + 18 from progressive disease |
| 59 | BC | IFN, HU, 6-TG, MEL | 57,XY,+1,+4, +6,+8,t(9;22) (q34;q11),+10,+11, +16,+18,+19,+21, +der2 2[5] | Y253F | 41.2 | 1/6 | WT | NR | ND | Died day +35 from progressive disease |
| 62 | BC | IFN, HU | 45,XY,t(9;22)(q34; q11),del(10)(p13?), −13[26] | Y253H | 36.2 | 1/6 | WT | NR | WT day +92/continuous blast crisis | Died day +208 from progressive disease |
| 56 | BC | IFN, ARA-C | 46,XY,t(9;22) (q34;q11)[15] | ND | NA | NA | V2891 | NR | ND | Died day + 105 from progressive disease |
Patient no. . | Disease phase . | Therapy prior to imatinib . | Karyotype . | Mutation by ASO-PCR . | Ct mut . | Positive reactions, no./total . | Sequencing . | Best response . | Follow-up sequencing/disease status . | Last follow-up/disease status . |
|---|---|---|---|---|---|---|---|---|---|---|
| 7 | AP | IFN | 46,XY,t(9,22)[18]; 47,XY,t(9;22), +der(22)[6];47, XY,+8[1] | T3151 | 45.8 | 1/6 | WT | CHR | WT day +42/PHR; WT day +107/PHR;WT day +785/CHR | Alive in CHR day +1591 |
| 13 | AP | HU | 46,XY,t(9;22)[24]/47,XY,t(9;22), +der(22)[1] | Q252H | 35.6-40.4 | 3/6 | WT | MCR (20% Ph+) | Q252H day +176/MCR; Q252H+F359V day +325/MCR | Alive in MCR day +1087 |
| 17 | AP | IFN, HU | 46,XY,t(9;22)(q34; q11)[25] | T3151 | 46.9 | 1/6 | WT | PHR | WT day + 80/PHR; WT day+270/PHR; WT day +444/hematologic relapse | Hematologic relapse day + 531; lost to follow-up |
| 25 | AP | HU | 46,XX,add(6)(p25), t(9;22)(q34;q11) [20] | T3151 | 46.7 | 1/6 | WT | CHR | E255K+M351T day +365/CHR | Allogeneic transplant day +512 for lack of MCR |
| 29 | AP | HU, IFN | 45X-Y,t(9;22) (q34;q11) | Y253H | 40.2 | 1/6 | WT | mCR (52% Ph+) | WT day +85/CHR; WT day +267/CHR | Hematologic relapse day +452; lost to follow-up |
| 36 | AP | HU, IFN, 6-TG | 47,XY,t(9;22) (q34;q11),+der(22) [23];46,XY,t(9;22) (q34;q11)[1] | T3151 | 46.6-47.9 | 2/6 | K247R | CHR | K247R day +107/CHR | Hematologic relapse day +212; alive day +1011 |
| 41 | AP | none | 46,XX,t(9;22) (q34;q11),−der(22), +idic der (22)[12] | T3151 | 43.7 | 1/6 | WT | CCR | T3151 day +93/CCR; T3151 day +182/CCR | Alive in molecular remission day + 1009 |
| 42 | AP | IDA, ARA-C, IFN | 48,XY,+8,t(9; 22)(q34;q11),+der (22)t(9;22)(q34; q11)[8] | T3151 | 44.1-45.7 | 2/6 | WT | CCR | WT day+193/CCR | Alive in CCR day +661 |
| 45 | AP | HU, 6-TG | 44,XX,t(9;22)(q34; q11),der(14q+), t(17;18)(p11;q12?), −18,−20[23] | Y253F | 38.2 | 1/6 | WT | NA | ND | Died day +9 from sepsis |
| 46 | AP | HU, 6-TG | 46,XX,t(9;22)[10] | T3151 | 37.92-41.68 | 2/6 | WT | CHR | T3151 day +158/hematologic relapse | Hematologic relapse + 158; died day + 1350 from progressive disease |
| 48 | BC | HU, IFN | 47,XX,+8,t(9; 22)(q34;11) [16],46,XX,t(9; 22)[4] | T3151 | 45.6 | 1/6 | WT | NR | T3151 day +19/continuous blast crisis | Allogeneic transplant day + 140 for progressive disease; died from TRM |
| 55 | BC | HU | ND | F359V | 39.4-44.8 | 5/6 | F359V | NR | ND | Died day + 18 from progressive disease |
| 59 | BC | IFN, HU, 6-TG, MEL | 57,XY,+1,+4, +6,+8,t(9;22) (q34;q11),+10,+11, +16,+18,+19,+21, +der2 2[5] | Y253F | 41.2 | 1/6 | WT | NR | ND | Died day +35 from progressive disease |
| 62 | BC | IFN, HU | 45,XY,t(9;22)(q34; q11),del(10)(p13?), −13[26] | Y253H | 36.2 | 1/6 | WT | NR | WT day +92/continuous blast crisis | Died day +208 from progressive disease |
| 56 | BC | IFN, ARA-C | 46,XY,t(9;22) (q34;q11)[15] | ND | NA | NA | V2891 | NR | ND | Died day + 105 from progressive disease |
Note that K247R is a previously unreported polymorphism.
Ct mut indicates PCR cycle at which mutant signal was detected; AP, accelerated phase; IFN, interferon-α; WT, wild type; CHR, complete hematologic response; PHR, partial hematologic response; HU, hydroxyurea; MCR, major cytogenetic response; mCR, minor cytogenetic response; 6-TG, 6-thioguanine; CCR, complete cytogenetic response; IDA, idarubicin; ARA-C, cytarabine; NA, not applicable; ND, not done; BC, blast crisis; NR, no response; TRM, transplant-related mortality; MEL, melphalan.
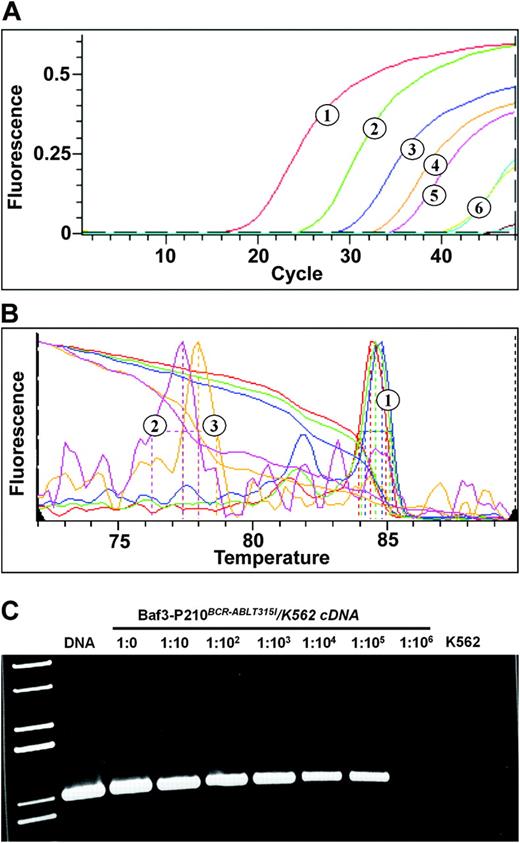
Fluorescent ASO-specific PCR assay. RNA was extracted from a dilution of BaF3 cells expressing the T315I mutant of BCR-ABL in K562 cells, reversely transcribed into cDNA, and subjected to fluorescent ASO-specific PCR with primers optimized for detection of the T315I mutant. Plasmid DNA was included as a positive control. (A) Detection of amplification products by SybrGreen. 1 indicates plasmid DNA (control); 2 to 5, dilutions from 1:0 to 1:103; and 6, dilutions of 1:104 and 1:105 (curves overlap). (B) Melting curve analysis. Only selected melting curves are shown, and the colors do not correspond to panel A. 1 indicates curves corresponding to dilutions of 1:10, 1:103, and 1:105; 2, dilution of 1:106; and 3, K562 cells. (C) The PCR products of the fluorescent assay were run on a 1% agarose gel. Lane 1 indicates molecular weight marker; DNA, diluted plasmid DNA. Note the absence of PCR product in the 1:106 dilution and the K562-only specimen. The quality of cDNA was routinely checked by amplification of ABL as a control gene (not shown).
Fluorescent ASO-specific PCR assay. RNA was extracted from a dilution of BaF3 cells expressing the T315I mutant of BCR-ABL in K562 cells, reversely transcribed into cDNA, and subjected to fluorescent ASO-specific PCR with primers optimized for detection of the T315I mutant. Plasmid DNA was included as a positive control. (A) Detection of amplification products by SybrGreen. 1 indicates plasmid DNA (control); 2 to 5, dilutions from 1:0 to 1:103; and 6, dilutions of 1:104 and 1:105 (curves overlap). (B) Melting curve analysis. Only selected melting curves are shown, and the colors do not correspond to panel A. 1 indicates curves corresponding to dilutions of 1:10, 1:103, and 1:105; 2, dilution of 1:106; and 3, K562 cells. (C) The PCR products of the fluorescent assay were run on a 1% agarose gel. Lane 1 indicates molecular weight marker; DNA, diluted plasmid DNA. Note the absence of PCR product in the 1:106 dilution and the K562-only specimen. The quality of cDNA was routinely checked by amplification of ABL as a control gene (not shown).
We also determined the rate of mutations detectable by conventional sequencing. In our hands, sequencing reliably detects mutations if at least 20% to 30% of the amplicon is mutant.23 Sixty-five samples (98.5%) were successfully amplified and gave high-quality readings. One patient (no. 55) was found to have a F359V mutation, confirming the results of the ASO-PCR assay, and 1 patient (no. 56) was found to have V289I, which is not included in the ASO-PCR assays. To our knowledge, the V289I mutation has not yet been described in patients, but V289S has been found in an in vitro mutagenesis screen for imatinib-resistant mutants, where it conferred a moderate level of drug resistance. In 2 additional patients (no. 8, no. 36) K247R was detected, and wild-type allele was completely absent. The K247R amino acid exchange has not previously been reported and appears to represent a rare polymorphism (L.C., unpublished observations, 2005). All other samples were wild type.
Overall (ie, ASO-PCR and conventional sequencing combined) mutations were detected in 10 (37.0%) of 27 patients with accelerated phase and 5 (26.3%) of 19 patients with blast crisis. None of the patients in chronic phase tested positive (Table 2). The Fisher exact test was used to compare mutation detection between patients with chronic phase, accelerated phase, and blast crisis. Mutations were detected at higher frequency in blast crisis phase versus chronic phase (P = .020) and accelerated phase versus chronic phase (P = .002). There was no difference between accelerated phase and blast crisis (P = .532).
Response to imatinib in patients with mutations
Fifteen patients had evidence for KD mutations, 2 of whom tested positive by sequencing, indicating a high proportion of mutant allele. One patient (no. 45) died from urosepsis on day +9 and was thus not evaluable for response.
Patients with imatinib-refractory disease
Five patients were refractory to imatinib, all were in myeloid blast crisis. Both patients (no. 55 with F359V and no. 56 with V289I) with mutations detected by sequencing died of progressive disease on day +35 and day +105, respectively. No follow-up samples were available for sequencing. In patient no. 48 (positive for T315I by ASO-PCR) T315I was detected by sequencing on day +19. She received salvage chemotherapy and eventually died of complications of allogeneic stem cell transplantation. Patients no. 59 (positive for Y253F by ASO-PCR) and no. 62 (positive for Y253H by ASO-PCR) died of progressive disease on day +35 and day +208, respectively. In patient no. 59, no follow-up specimens were available for sequencing, whereas only wild-type sequence was detected in patient no. 62 (day +92).
Patients with imatinib-responsive disease
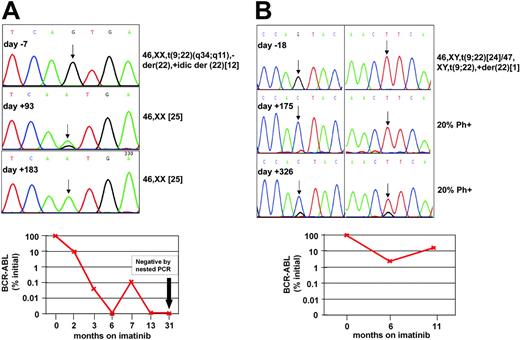
Best responses to imatinib in the remaining 9 patients were PHR in 1, CHR in 5, and MCR in 3. In 5 patients sequencing of follow-up samples failed to detect mutations, including 4 patients with CHR or PHR (nos. 7, 17, 29, 36) and 1 with MCR (no. 42). Two of these 5 patients subsequently relapsed on days +212 (no. 36) and +444 (no. 17), and no mutations were detected at the time of disease recurrence. In 1 patient (no. 46) the T315I mutations detected by ASO-PCR prior to the treatment was detected by sequencing at the time of relapse. Patient no. 25 (positive for T315I by ASO-PCR) was positive by sequencing for E255K and M351T on day +365, while in CHR. She subsequently underwent allogeneic stem cell transplantation. In patients nos. 13 and 41, the mutation identified by ASO-PCR prior to therapy was detected by sequencing at the time of MCR (Figure 2). Patient no. 41 achieved CCR at 3 months and was in complete molecular response at last follow-up on day +1003. Sequencing revealed a mix of wild type and T315I at 3 months and 100% T315I at 6 months. Patient no. 13 achieved a partial cytogenetic response (20% Philadelphia chromosome [Ph]-positive metaphases) and an almost 2-log reduction of BCR-ABL transcripts (2.3% of pretherapeutic levels) at 6 months, while sequencing showed 100% Q252H. At 11 months, his BCR-ABL levels rose to 16.5% of pretherapeutic levels, while he continued to be in MCR (20% Ph-positive). Sequencing showed an F359V mutation in addition to Q252H. This evolution is most consistent with emergence of F359V in a separate cell clone that may have caused the rise in BCR-ABL mRNA.
Follow-up of patients no. 13 and no. 41. (A) Patient no. 41. (Top) Sequence analysis of PCR products on days -7, +93, and +183 of imatinib therapy. ASO-PCR detected T315I on day -7, while conventional sequencing was negative. A mix of T315I and wild type (arrow) was detected on day +93, the time of first documented complete cytogenetic response. On day +183, only T315I was detected, while the patient remained in complete cytogenetic remission. (Bottom) Follow-up with quantitative and qualitative RT-PCR showed a gradual although not uninterrupted decrease of BCR-ABL transcripts and eventually complete molecular remission. (B) Patient no. 13. (Top) Sequence analysis of PCR products on days -18, +175, and +316 of imatinib therapy. ASO-PCR detected Q252H in the specimen from day -18. One hundred percent Q252H was detected on day +175 (left chromatogram, arrow). On day +326, a mix of Q252H and F359V was detected (arrows). Note that a small wild-type signal has reemerged in position 252. (Bottom) Follow-up with quantitative RT-PCR for BCR-ABL.
Follow-up of patients no. 13 and no. 41. (A) Patient no. 41. (Top) Sequence analysis of PCR products on days -7, +93, and +183 of imatinib therapy. ASO-PCR detected T315I on day -7, while conventional sequencing was negative. A mix of T315I and wild type (arrow) was detected on day +93, the time of first documented complete cytogenetic response. On day +183, only T315I was detected, while the patient remained in complete cytogenetic remission. (Bottom) Follow-up with quantitative and qualitative RT-PCR showed a gradual although not uninterrupted decrease of BCR-ABL transcripts and eventually complete molecular remission. (B) Patient no. 13. (Top) Sequence analysis of PCR products on days -18, +175, and +316 of imatinib therapy. ASO-PCR detected Q252H in the specimen from day -18. One hundred percent Q252H was detected on day +175 (left chromatogram, arrow). On day +326, a mix of Q252H and F359V was detected (arrows). Note that a small wild-type signal has reemerged in position 252. (Bottom) Follow-up with quantitative RT-PCR for BCR-ABL.
Detection of KD mutations is associated with clonal cytogenetic evolution, low platelet count, and prior therapy with 6-Thioguanine but not with event-free or overall survival
A number of baseline disease characteristics were tested for their association with detection of mutations prior to therapy (Table 5). In univariate analysis, clonal cytogenetic evolution, disease phase, a low platelet count (compared with patients with normal platelet count), age older than 60 years, and previous therapy with 6-thioguanine (but not other cytotoxic drugs) were associated with mutation detection at P less than .1. These factors were then analyzed in a multivariate stepwise logistic regression model. Here, clonal cytogenetic evolution (estimated odds ratio [OR], 14.163), a low platelet count (estimated OR, 19.7), and prior treatment with 6-thioguanine (estimated OR, 43.3) retained statistical significance. It should be noted that all 3 associations have wide confidence intervals, particularly the association with 6-thioguanine (Table 5). No unusual pattern of clonal evolution was noted in patients with KD mutations, an additional Ph (Philadelphia chromosome) being the most frequent finding.
Analysis of factors for association with mutation detection
Factor . | No. subjects . | Mutation, no. . | Univariate P . | Multivariate P (n = 60) . | Estimated odds ratio . | 95% Wald confidence interval . |
|---|---|---|---|---|---|---|
| Sex | ||||||
| Male | 38 | 10 | 420 | NA | NA | NA |
| Female | 28 | 5 | ||||
| Age* | ||||||
| 60 y or younger | 39 | 6 | 093 | NS | NA | NA |
| Older than 60 y | 27 | 9 | ||||
| Disease duration | ||||||
| 12 mo or fewer | 17 | 2 | 211 | NA | NA | NA |
| Longer than 12 mo | 48 | 13 | ||||
| Prior therapy | ||||||
| None, HU, IFN | 35 | 9 | 593 | NA | NA | NA |
| Others | 31 | 6 | ||||
| Prior treatment with 6-thioguanine*† | ||||||
| Yes | 6 | 4 | 018 | 006 | 43.295 | 2.916-642.928 |
| No | 60 | 11 | ||||
| Phase* | ||||||
| Chronic | 20 | 0 | 003 | NS | NA | NA |
| Accelerated + blastic | 46 | 15 | ||||
| Clonal evolution* | ||||||
| Yes | 26 | 11 | 003 | 006 | 14.163 | 2.083-96.316 |
| No | 36 | 3 | ||||
| Splenomegaly | ||||||
| Yes | 46 | 14 | 127 | NA | NA | NA |
| No | 13 | 1 | ||||
| Hemoglobin level | ||||||
| More than 100 g/L | 26 | 7 | 548 | NA | NA | NA |
| Less than 100 g/L | 39 | 8 | ||||
| White blood cell count | ||||||
| More than 10 × 109/L | 19 | 2 | 148 | NA | NA | NA |
| Fewer than 10 × 109/L | 47 | 13 | ||||
| Platelets ()*‡ | ||||||
| Fewer than 100 × 109/L | 17 | 8 | 013 | 007 | 19.682 | 2.160-179.361 |
| At least 100 × 109/L but fewer than 450 × 109/L | 30 | 4 | ||||
| At least 450 × 109/L | 17 | 3 | ||||
| Blasts in PB | ||||||
| 5% or less | 43 | 11 | 563 | NA | NA | NA |
| More than 5% | 21 | 4 | ||||
| Basophils in PB | ||||||
| 7% or less | 43 | 12 | 122 | NA | NA | NA |
| More than 7% | 20 | 2 | ||||
| Blasts in BM | ||||||
| 5% or less | 30 | 6 | 401 | NA | NA | NA |
| More than 5% | 27 | 8 |
Factor . | No. subjects . | Mutation, no. . | Univariate P . | Multivariate P (n = 60) . | Estimated odds ratio . | 95% Wald confidence interval . |
|---|---|---|---|---|---|---|
| Sex | ||||||
| Male | 38 | 10 | 420 | NA | NA | NA |
| Female | 28 | 5 | ||||
| Age* | ||||||
| 60 y or younger | 39 | 6 | 093 | NS | NA | NA |
| Older than 60 y | 27 | 9 | ||||
| Disease duration | ||||||
| 12 mo or fewer | 17 | 2 | 211 | NA | NA | NA |
| Longer than 12 mo | 48 | 13 | ||||
| Prior therapy | ||||||
| None, HU, IFN | 35 | 9 | 593 | NA | NA | NA |
| Others | 31 | 6 | ||||
| Prior treatment with 6-thioguanine*† | ||||||
| Yes | 6 | 4 | 018 | 006 | 43.295 | 2.916-642.928 |
| No | 60 | 11 | ||||
| Phase* | ||||||
| Chronic | 20 | 0 | 003 | NS | NA | NA |
| Accelerated + blastic | 46 | 15 | ||||
| Clonal evolution* | ||||||
| Yes | 26 | 11 | 003 | 006 | 14.163 | 2.083-96.316 |
| No | 36 | 3 | ||||
| Splenomegaly | ||||||
| Yes | 46 | 14 | 127 | NA | NA | NA |
| No | 13 | 1 | ||||
| Hemoglobin level | ||||||
| More than 100 g/L | 26 | 7 | 548 | NA | NA | NA |
| Less than 100 g/L | 39 | 8 | ||||
| White blood cell count | ||||||
| More than 10 × 109/L | 19 | 2 | 148 | NA | NA | NA |
| Fewer than 10 × 109/L | 47 | 13 | ||||
| Platelets ()*‡ | ||||||
| Fewer than 100 × 109/L | 17 | 8 | 013 | 007 | 19.682 | 2.160-179.361 |
| At least 100 × 109/L but fewer than 450 × 109/L | 30 | 4 | ||||
| At least 450 × 109/L | 17 | 3 | ||||
| Blasts in PB | ||||||
| 5% or less | 43 | 11 | 563 | NA | NA | NA |
| More than 5% | 21 | 4 | ||||
| Basophils in PB | ||||||
| 7% or less | 43 | 12 | 122 | NA | NA | NA |
| More than 7% | 20 | 2 | ||||
| Blasts in BM | ||||||
| 5% or less | 30 | 6 | 401 | NA | NA | NA |
| More than 5% | 27 | 8 |
NS indicates not significant; PB, peripheral blood; BM, bone marrow; and NA, not applicable.
Factors with P < .1 in univariate analysis were included in the multivariate model.
Other antileukemic therapies were also analyzed individually for their effect on mutation frequency, but no significant associations were detected.
Significant only for comparison between platelets fewer than 100 and platelets of at least 100 but fewer than 450 (normal range).
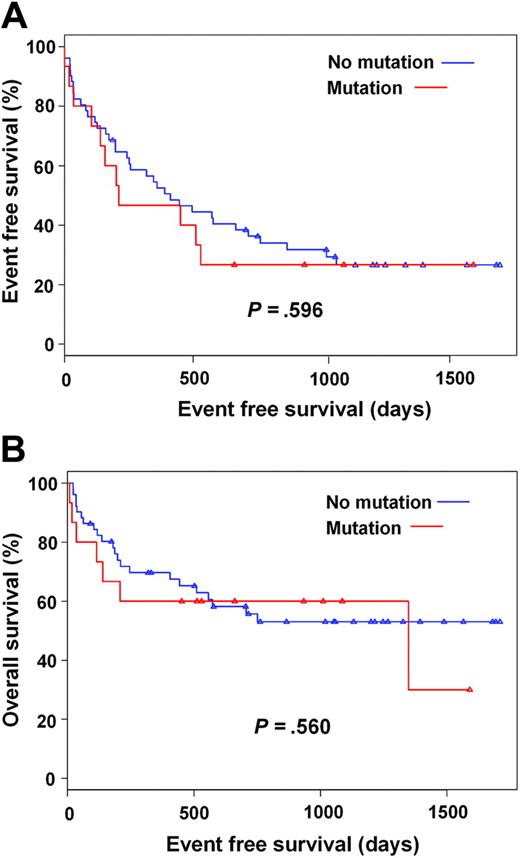
We hypothesized that mutant clones, even if present only at low levels, may be rapidly selected in the presence of imatinib. Thus, patients with detectable mutations may relapse more rapidly than patients without mutations and may have a shorter survival. Three patients were excluded from this analysis, because of early death (neutropenic sepsis and urosepsis, respectively) or early discontinuation of imatinib for skin toxicity. In the remaining 63 cases, no association was found between mutation detection and hematologic response or major cytogenetic response, while the adverse effect of established prognostic factors such as advanced disease phase was confirmed (Tables 6 and 7). Consistent with the lack of a correlation between mutation detection and these endpoints, we did not find a correlation between mutation detection and event-free (P = .596) and overall (P = .560) survival in a univariate cox regression model (Figure 3A-B).
Analysis of baseline variables for their impact on hematologic response
Factors . | No. subjects . | No. subjects with hematologic response . | Univariate P . | Multivariate P(n = 56) . |
|---|---|---|---|---|
| Sex | ||||
| Male | 38 | 28 | .179 | NA |
| Female | 25 | 22 | ||
| Age* | ||||
| Younger than 60 y | 38 | 33 | .078 | NA |
| Older than 60 y | 25 | 17 | ||
| Disease duration | ||||
| Fewer than 12 mo | 16 | 14 | .426 | NA |
| Longer than 12 mo | 46 | 36 | ||
| Prior therapy | ||||
| None, HU, IFN | 35 | 30 | .170 | NA |
| Others | 28 | 20 | ||
| Prior 6- Thioguanine | ||||
| Yes | 58 | 46 | .971 | NA |
| No | 5 | 4 | ||
| Phase* | ||||
| Chronic | 20 | 20 | .005 | NS |
| Accelerated + blastic | 43 | 30 | ||
| Clonal evolution*† | ||||
| Yes | 25 | 16 | .016 | .010 |
| No | 34 | 31 | ||
| Splenomegaly | ||||
| Yes | 43 | 35 | .772 | NA |
| No | 13 | 10 | ||
| Hemoglobin level* | ||||
| At least 100 g/L | 24 | 15 | .016 | NS |
| Less than 100 g/L | 38 | 34 | ||
| White blood cell count | ||||
| At least 10 × 109/L | 19 | 16 | .543 | NA |
| Fewer than 10 × 109/L | 44 | 34 | ||
| Platelet count* | ||||
| No more than 100 × 109/L | 16 | 9 | .095 | NS |
| More than 100 × 109/L but less than 450 × 109/L | 28 | 24 | ||
| More than 450 | 17 | 14 | ||
| Blasts in PB*‡ | ||||
| 5% or less | 41 | 38 | <.001 | .001 |
| Greater than 5% | 20 | 10 | ||
| Basophils in PB | ||||
| 7% or less | 42 | 32 | .540 | NA |
| Greater than 7% | 18 | 15 | ||
| Blasts in BM | ||||
| 5% or less | 29 | 29 | .942 | NA |
| Greater than 5% | 25 | 15 | ||
| Mutation | ||||
| Yes | 14 | 9 | .123 | NA |
| No | 49 | 41 |
Factors . | No. subjects . | No. subjects with hematologic response . | Univariate P . | Multivariate P(n = 56) . |
|---|---|---|---|---|
| Sex | ||||
| Male | 38 | 28 | .179 | NA |
| Female | 25 | 22 | ||
| Age* | ||||
| Younger than 60 y | 38 | 33 | .078 | NA |
| Older than 60 y | 25 | 17 | ||
| Disease duration | ||||
| Fewer than 12 mo | 16 | 14 | .426 | NA |
| Longer than 12 mo | 46 | 36 | ||
| Prior therapy | ||||
| None, HU, IFN | 35 | 30 | .170 | NA |
| Others | 28 | 20 | ||
| Prior 6- Thioguanine | ||||
| Yes | 58 | 46 | .971 | NA |
| No | 5 | 4 | ||
| Phase* | ||||
| Chronic | 20 | 20 | .005 | NS |
| Accelerated + blastic | 43 | 30 | ||
| Clonal evolution*† | ||||
| Yes | 25 | 16 | .016 | .010 |
| No | 34 | 31 | ||
| Splenomegaly | ||||
| Yes | 43 | 35 | .772 | NA |
| No | 13 | 10 | ||
| Hemoglobin level* | ||||
| At least 100 g/L | 24 | 15 | .016 | NS |
| Less than 100 g/L | 38 | 34 | ||
| White blood cell count | ||||
| At least 10 × 109/L | 19 | 16 | .543 | NA |
| Fewer than 10 × 109/L | 44 | 34 | ||
| Platelet count* | ||||
| No more than 100 × 109/L | 16 | 9 | .095 | NS |
| More than 100 × 109/L but less than 450 × 109/L | 28 | 24 | ||
| More than 450 | 17 | 14 | ||
| Blasts in PB*‡ | ||||
| 5% or less | 41 | 38 | <.001 | .001 |
| Greater than 5% | 20 | 10 | ||
| Basophils in PB | ||||
| 7% or less | 42 | 32 | .540 | NA |
| Greater than 7% | 18 | 15 | ||
| Blasts in BM | ||||
| 5% or less | 29 | 29 | .942 | NA |
| Greater than 5% | 25 | 15 | ||
| Mutation | ||||
| Yes | 14 | 9 | .123 | NA |
| No | 49 | 41 |
NS indicates not significant; NA, not available.
Factors with P < .1 in univariate analysis were included in the multivariate model.
Estimated odds ratio = .086; 95% Wald confidence interval = 0.013-0.568.
Estimated odds ratio = .050; 95% Wald confidence interval = 0.008-0.322.
Analysis of baseline variables for their effect on major cytogenetic response
Factors . | No. subjects . | No. subjects with major cytogenetic response . | Univariate P . | Multivariate P(n = 48) . |
|---|---|---|---|---|
| Sex | ||||
| Male | 37 | 9 | .107 | NA |
| Female | 25 | 11 | ||
| Age | ||||
| Younger than 60 y | 37 | 14 | .256 | NA |
| Older than 60 y | 25 | 6 | ||
| Disease*duration | ||||
| Fewer than 12 mo | 16 | 9 | .024 | NS |
| Longer than 12 mo | 45 | 11 | ||
| Prior therapy*† | ||||
| None, HU, IFN | 35 | 17 | .003 | .037 |
| Others | 27 | 3 | ||
| Prior treatment with 6-thioguanine | ||||
| No | 57 | 20 | .971 | NA |
| Yes | 5 | 0 | ||
| Phase* | ||||
| Chronic | 20 | 13 | <.001 | NS |
| Accelerated + blastic | 42 | 7 | ||
| Clonal evolution* | ||||
| Yes | 24 | 4 | .082 | NA |
| No | 34 | 13 | ||
| Splenomegaly | ||||
| Yes | 43 | 16 | .435 | NA |
| No | 12 | 3 | ||
| Hemoglobin level* | ||||
| More than 100 g/L | 24 | 2 | .005 | NS |
| Less than 100 g/L | 37 | 17 | ||
| White blood cell count* | ||||
| More than 10 × 109/L | 19 | 9 | .095 | NS |
| Fewer than 10 × 109/L | 43 | 11 | ||
| Platelet count | ||||
| 100 × 109/L or fewer | 16 | 5 | >.99 | NA |
| More than 100 × 109/L but less than 450 × 109/L | 28 | 10 | ||
| More than 450 × 109/L | 16 | 5 | ||
| Blasts (PB) | ||||
| 5% or less | 40 | 20 | .940 | NA |
| More than 5% | 20 | 0 | ||
| Basophils (PB) | ||||
| 7% or less | 41 | 14 | .951 | NA |
| More than 7% | 18 | 6 | ||
| Blasts (BM)*‡ | ||||
| 5% or less | 28 | 16 | .001 | .0146 |
| More than 5% | 25 | 3 | ||
| Mutation | ||||
| Yes | 14 | 3 | .330 | NA |
| No | 48 | 17 |
Factors . | No. subjects . | No. subjects with major cytogenetic response . | Univariate P . | Multivariate P(n = 48) . |
|---|---|---|---|---|
| Sex | ||||
| Male | 37 | 9 | .107 | NA |
| Female | 25 | 11 | ||
| Age | ||||
| Younger than 60 y | 37 | 14 | .256 | NA |
| Older than 60 y | 25 | 6 | ||
| Disease*duration | ||||
| Fewer than 12 mo | 16 | 9 | .024 | NS |
| Longer than 12 mo | 45 | 11 | ||
| Prior therapy*† | ||||
| None, HU, IFN | 35 | 17 | .003 | .037 |
| Others | 27 | 3 | ||
| Prior treatment with 6-thioguanine | ||||
| No | 57 | 20 | .971 | NA |
| Yes | 5 | 0 | ||
| Phase* | ||||
| Chronic | 20 | 13 | <.001 | NS |
| Accelerated + blastic | 42 | 7 | ||
| Clonal evolution* | ||||
| Yes | 24 | 4 | .082 | NA |
| No | 34 | 13 | ||
| Splenomegaly | ||||
| Yes | 43 | 16 | .435 | NA |
| No | 12 | 3 | ||
| Hemoglobin level* | ||||
| More than 100 g/L | 24 | 2 | .005 | NS |
| Less than 100 g/L | 37 | 17 | ||
| White blood cell count* | ||||
| More than 10 × 109/L | 19 | 9 | .095 | NS |
| Fewer than 10 × 109/L | 43 | 11 | ||
| Platelet count | ||||
| 100 × 109/L or fewer | 16 | 5 | >.99 | NA |
| More than 100 × 109/L but less than 450 × 109/L | 28 | 10 | ||
| More than 450 × 109/L | 16 | 5 | ||
| Blasts (PB) | ||||
| 5% or less | 40 | 20 | .940 | NA |
| More than 5% | 20 | 0 | ||
| Basophils (PB) | ||||
| 7% or less | 41 | 14 | .951 | NA |
| More than 7% | 18 | 6 | ||
| Blasts (BM)*‡ | ||||
| 5% or less | 28 | 16 | .001 | .0146 |
| More than 5% | 25 | 3 | ||
| Mutation | ||||
| Yes | 14 | 3 | .330 | NA |
| No | 48 | 17 |
NS indicates not significant; NA, not available.
Factors with P < .1 in univariate analysis were included in the multivariate model.
Estimated odds ratio = 0.195; 95% Wald confidence interval = 0.042-0.912.
Estimated odds ratio = 0.150; 95% Wald confidence interval = 0.033-0.688.
Discussion
Using a highly sensitive and specific assay, we show that KD mutations are detectable in a substantial fraction of patients with imatinib-naive CML with advanced phase disease. As mutations were detected by conventional sequencing in only 2 cases (including one mutation that is not represented in the panel of ASO-PCR assays), we interpret this to mean that the proportion of mutant allele is usually low. In fact, detection of the signals by ASO-PCR usually required more than 40 PCR cycles, corresponding to less than 0.1% mutant cells as estimated by limiting dilution experiments using BaF3 cells expressing mutant BCR-ABL diluted in K562 cells. As repeated experiments demonstrated that at low concentrations of mutant allele these assays are not quantitative (Figure 1A; data not shown), it is impossible to estimate the proportion of KD mutant cells with greater precision.
In multivariate analysis, low platelet counts, clonal cytogenetic evolution, and prior exposure to 6-thioguanine were associated with mutation detection. It should be noted that clonal evolution was one of the defining criteria for accelerated phase, and that a relatively large proportion (37%) of the patients in accelerated phase had clonal evolution as the only diagnostic feature establishing a diagnosis of accelerated phase. The correlation between clonal evolution and mutation detection suggests that a common underlying mechanism may lead to an increased risk of additional cytogenetic abnormalities as well as point mutations. Bcr-Abl-expressing cells exhibit increased levels of reactive oxygen species that are thought to induce DNA damage.24 Bcr-Abl kinase activity appears to promote various mechanisms of DNA repair, However, the repair is frequently unfaithful, which is thought to be the result of defects in S phase25 and G2/M checkpoints,26 in combination with the antiapoptotic effects of Bcr-Abl.27 In this scenario, DNA repair occurs in situations that would normally induce an apoptosis response, leading to the accumulation of mutations.28 Notably, we also found a significant association between mutation detection and exposure to 6-Thioguanine, a drug that is mutagenic because of mispairing with thymine after incorporation into DNA,29 while patients with exposure to other cytotoxic drugs, including cytarabine and topoisomerase II inhibitors, were not at increased risk. Because the number of patients exposed to 6-Thioguanine was small, this finding should be interpreted with caution. Nonetheless it is plausible that patients with CML exposed to drugs with the potential to induce point mutations may be at higher risk of acquiring KD mutations, a factor that needs to be taken into account in the design of drug combination studies.
Several case reports and small studies described the detection of KD mutations in material stored prior to therapy.10-12,30 As these studies looked specifically for mutations that had been detected at the time of drug resistance, these findings are consistent with selection of resistant clones in the presence of imatinib. Two of our patients (no. 46 and no. 48) fit in this category. In both cases, the mutation detected prior to imatinib therapy became dominant at the time of relapse. However, in 2 other patients (no. 13 and no. 41) the mutations detected prior to therapy by ASO-PCR were found by sequencing at the time of partial and complete cytogenetic response. Patient no. 41 has maintained CCR for several years and was in complete molecular response at last follow-up on day +1009. In patient no. 13, Q252H was detected at 6 months, when he had achieved MCR (20% Ph-positive). At 11 months, MCR was maintained but a rise in BCR-ABL transcripts was apparent. At this time sequencing revealed a F359V mutation in addition to the preexisting Q252H, strongly suggesting the F359V had emerged in a different leukemic cell clone. It is conceivable that in both cases the mutant clone became detectable by the less-sensitive direct-sequencing technique because wild-type Bcr-Abl had been mostly eliminated by imatinib therapy. However, the mutant clones clearly failed to expand. This unexpected observation may have several reasons. First, it is possible that mutations occur in cell clones that have limited self-renewal capacity. Such clones, although at a proliferative advantage over wild type in the presence of imatinib, would not be able to sustain leukemic hematopoiesis long term. This would be in analogy to the observation that BCR-ABL transcripts are detectable in a substantial fraction of healthy individuals, most of whom will never develop CML.18 Second, the presence of a mutation alone may not be sufficient to confer a fully resistant phenotype in the absence of additional mechanisms. For example, the expression level of Bcr-Abl protein may not be sufficient to afford at least partial independence from exogenous cytokines, a phenomenon that has been described in Bcr-Abl-positive cell lines.31 Such cell clones, although Bcr-Abl positive, may not be able to compete against normal hematopoiesis. The same phenomena may account for the fact that in several patients sequencing of follow-up samples detected only wild-type sequence or (in patient no. 25) mutations that had not been detected by ASO-PCR and may explain why mutation detection was neither correlated with hematologic or cytogenetic response nor with overall or event-free survival. Thus, an important conclusion from our experiments is that high-sensitivity screening of imatinib-naive patients for KD mutations cannot be recommended, because it is impossible to predict whether the mutant clone will expand and cause relapse. In contrast, both patients (nos. 55 and 56) in whom the mutant clone was detected by sequencing in pretherapeutic samples did not respond to imatinib and died rapidly of progressive disease. The presence of mutant clones at high levels (detectable by sequencing) indicates that they were able to maintain leukemic hematopoiesis. This suggests that mutant clones detectable by sequencing in either imatinib-naive patients or patients with a rising disease burden on imatinib therapy have demonstrated the ability to sustain hematopoiesis and are almost invariably associated with subsequent relapse.32,33
Outcomes of patients on imatinib therapy. (A) Event free survival and (B) overall survival according to detection of kinase domain mutations prior to imatinib therapy. Censored patients indicated by triangles.
Outcomes of patients on imatinib therapy. (A) Event free survival and (B) overall survival according to detection of kinase domain mutations prior to imatinib therapy. Censored patients indicated by triangles.
We are fully aware of the fact that a negative result, even with an extremely sensitive assay, does not exclude the presence of mutant clones below the level of detection. In addition, the ASO-PCR assays do not cover approximately one fourth of clinically isolated mutations.9 Not unexpectedly, a mutation not included in the ASO-PCR assays was detected in one patient. Therefore, it is possible that the frequency of positive calls would increase with more sensitive assays covering more types of mutations. Unfortunately, attempts to improve sensitivity by a nested PCR strategy were not successful, because of an unacceptable loss of specificity. The other concern is false-positive results. Although, in lieu of an alternative assay with comparable sensitivity, one cannot exclude false-positive results with certainty, it should be noted that extraordinary precautions were taken to avoid contamination by mutant BCR-ABL, and stringent criteria were applied to the interpretation of results. In addition, the strong correlation between mutation detection and biologic features such as clonal evolution and disease phase argues against false-positives results.
It is unclear why we detected certain mutations much more frequently than others. This is particularly obvious for the E255K and M351T mutations that account for 15% and 17% of clinical isolates.9 In both cases, limiting dilution experiments showed that the sensitivity of the assays was comparable to T315I, the most frequently detected mutation in our study. Given the relatively small numbers, these differences may be a chance finding. On the other hand, it is also possible that some mutations alter the biology of Bcr-Abl irrespective of their sensitivity to imatinib. In this scenario, mutants that are gain-of-function compared with wild type would be more likely to expand to the level of detection in the absence of imatinib than mutants that have a loss-of-function phenotype. For example, the M351T mutation appears to reduce Bcr-Abl kinase activity and transforming capacity compared with wild-type Bcr-Abl,34 which would explain why we failed to detect this mutation in any of our patients. Conversely, mutations that increase the transformation potency of BCR-ABL may represent a novel mechanism of disease progression. It must be stressed though that the numbers in this study are small; thus, results should not be overinterpreted.
In summary, we show that mutations of the BCR-ABL kinase domain are detectable in a fraction of imatinib-naive patients with advanced but not with chronic phase CML, and that their presence is correlated with clonal cytogenetic evolution, a low platelet count, and exposure to 6-Thioguanine. The fact that imatinib-resistant mutant clones present at low levels prior to therapy may disappear during treatment indicates that screening for mutations with highly sensitive assays may not be useful, at least in imatinib-naive patients. To adequately interpret the results of KD mutation analysis, the size of the mutant clone and the clinical context must be taken into account.
Prepublished online as Blood First Edition Paper, May 24, 2005; DOI 10.1182/blood-2005-03-1036.
Supported by grants from the Doris Duke Charitable Foundation (B.J.D.) and the Leukemia and Lymphoma Society (B.J.D.).
S.G.W. developed and performed the ASO-PCR assays, did part of the sequencing, and helped draft the manuscript; T.L. organized the generation of cDNA from imatinib-naive patients; S.D. helped with the construction of the KD mutant constructs; S.O. collected and organized the patient information; L.C. helped with sequencing; D.N. helped with collection of clinical patient information; E.P.S. helped with sequence analysis and manuscript preparation; S.McW. and B.P. did the statistical analysis; I.K. did the RNA extraction and cDNA synthesis; B.J.D. contributed to the design of this study; and M.W.D. developed the concept of the study, supervised the establishment of the ASO-PCR assay, and finalized the manuscript.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
M.W.D. is a Junior Faculty Scholar of the American Society of Hematology.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal