Abstract
X-linked lymphoproliferative disease (XLP) is characterized by abnormal immune responses to Epstein-Barr virus attributed to inactivating mutations of the SAP gene. Previous studies showed immunoglobulin E (IgE) deficiency and low serum IgG levels in Sap-deficient mice before and after viral infections, which are associated with impaired CD4+ T-helper function. In the present work, we find that signaling lymphocytic activation molecule (SLAM)-associated protein (SAP) is expressed in B cells and this expression is down-regulated after stimulation with lipopolysaccharide (LPS) and interleukin 4 (IL-4). We demonstrate that B cells from Sap-deficient mice exhibit reduced IgG and IgA production in vitro. This impairment correlates with decreased circular transcript levels of Iα, Iγ2a, Iγ2b, and Iγ3 after stimulation, which indicate a defective Ig switch recombination in Sap-deficient B cells. While XLP is believed to cause defects in T, natural killer T (NKT), and natural killer (NK) cells, our results indicate that B cells are also affected. (Blood. 2005;106:2069-2075)
Introduction
X-linked lymphoproliferative disease (XLP) is an inherited immunodeficiency with variable phenotypes often associated with Epstein-Barr virus (EBV) infection. The most frequent clinical manifestations of XLP are fatal or severe infectious mononucleosis, hypo- or agammaglobulinemia, and non-Hodgkin lymphoma of Burkitt type.1,2 It has been demonstrated that inactivation or dysfunction of SAP (also called SH2D1A or DSHP) is responsible for this disease.3-5
The signaling lymphocytic activation molecule (SLAM)-associated protein (SAP), a small cytoplasmic protein that contains a single Src homologyy 2 (SH2) domain, is expressed at high levels in T and natural killer (NK) cells. SAP interacts through its SH2 domain with tyrosine residues located in the cytoplasmic domains of several molecules of the SLAM/CD2 receptor family, including SLAM,5 2B4,6 NTB-A,7 CD84 and Ly9,8,9 and CRACC.10 Earlier studies showed that SAP regulates the signal transduction of the SLAM/CD2 family receptors by interfering with the recruitment of the tyrosine phosphatase SHP-2.5,11 Recently, compelling evidence was provided that SAP behaves as a signaling adapter permitting the recruitment of the protein tyrosine kinase FynT11-14 and the 5′ inositol phosphatase SHIP.12,15 In patients with XLP, the SAP protein is either absent or structurally altered due to germ line mutations of the SAP gene, and may thus fail to regulate the SLAM/CD2 receptors signaling events.3-5,16-19
Sap-deficient mice have been generated to study the cellular basis of XLP. In 2 Sap-deficient mouse models, infection with lymphocytic choriomeningitis virus (LCMV) or Toxoplasma gondii led to hyperproliferation of interferon gamma (IFN-γ)-producing T cells and decreased antibody responses.20-22 We have reported the generation and characterization of a third Sap-deficient mouse model that displays hyperactivation of the CD8+ cells and decreased level of total serum immunoglobulin E (IgE) and IgG both before and after murine gammaherpesvirus 68 (MHV-68) infection.23 Moreover, a defect in natural killer T (NKT) cell development has been observed in the above-mentioned mouse models and in humans.24-26 Previous investigations revealed an altered in vivo Th2 cytokine profile in Sap-deficient mice, indicating that impaired Th2 differentiation accounts for the uncontrolled expansion of Th1 cells,20-23 as well as the impaired Ig production by B cells20,21,27 and the lack of long-lived plasma cells/memory B cells in Sap-deficient mice.22 Only recently, the widely accepted hypothesis that XLP is exclusively a T and NK defect was challenged by Morra et al, who demonstrated defective B-cell responses in the absence of SAP by an in vivo approach.28 In parallel, we have addressed the question of whether SAP has a direct role in B cells by an in vitro assay. In the present study, we report the impaired Ig switch recombination in Sap-deficient B cells, indicating an intrinsic B-cell defect.
Materials and methods
Mice
The generation of the Sap-deficient mice in 129/Sv×C57BL/6 mixed background was previously reported.23 The mutant mice were back-crossed to a C57BL/6 background for more than 10 generations. Mice were bred and housed under specific pathogen-free conditions at the International Agency for Research on Cancer (IARC). Age-matched mutant and wild-type male mice, aged 2 to 6 months, were used.
Flow cytometry
Single-cell suspensions from indicated organs were depleted of erythrocytes by lysis in hemolytic gey solution. Cells (1.0 × 106) were preincubated with Fc block (rat IgG2b anti-CD16/32, clone 2.4G2; BD/PharMingen, San Diego, CA) for 5 minutes at 4°C prior to staining with antibodies conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), tricolor (TC), allophycocyanin (APC), or biotin. Biotin conjugates were visualized with fluochrome-conjugated Strepavidin (BD/PharMingen). The following antimouse monoclonal antibodies (mAbs) were purchased from BD/PharMingen, Caltag (Burlingame, CA), or SouthernBiotech (Birmingham, AL): anti-B220 (RA3-6B2), anti-CD43 (S7), anti-BP-1 (6C3), anti-CD24 (M1/69), anti-IgM (R6-60.2), anti-IgD (11-26c.2a), anti-CD5 (53-7.3), anti-CD21/CD35 (7G6), anti-CD23 (B4B4), anti-CD69 (H1.2F3), anti-CD3e (500-A2), anti-CD4 (CT-CD4), anti-CD8a (CT-CD8a), anti-NK1.1 (PK136), and anti-2B4. Cells (10 000) were acquired on a FACSCalibur flow cytometer (BD Labware, Franklin Lakes, NJ) and analyzed with WinMDI.
Positive selection of B cells and amplification of the Sap transcript
Splenocytes were stained with anti-CD3-FITC (500-A2) and anti-CD19-Tricolor (6D5; Caltag), and sorted into CD19+ and CD3+ populations using FACSVantage SE (BD Labware). Total RNA was prepared from the sorted cells by RNeasy kit (Qiagen, Hilden, Germany) and was reverse transcribed by GeneAmp RT-PCR Core kit (Applied Biosystems/Roche, Branchburg, NJ). The SAP,23 β-actin,29 and CD330 transcripts were amplified as described previously.
Negative selection of B cells and Western blot hybridization
Splenocytes were stained with PE-conjugated anti-mouse CD3e (500-A2), CD4 (GK1.5), CD8a (53-6.7), NK1.1 (PK136), and 2B4 (BD/PharMingen). The cells positive for these markers were depleted by the MACS anti-PE MicroBeads and MACS separation columns (Milteni Biotec, Bergisch Gladbach, Germany). After the depletion, plastic-adhering cells were removed by incubation at 37°C for 1.5 hours. SAP protein in cell extracts after immunoprecipitation was visualized by immunoblotting using polyclonal rabbit anti-SAP antibodies as described previously.12
Purification and culture of the small resting B cells
B cells were purified in 2 stages. First, they were negatively selected from splenocytes by immunomagnetic depletion with mouse Pan T, CD4, and CD8 beads from Dynal Biotech (Oslo, Norway). Second, small resting B cells were fractionated by centrifugation through a discontinuous Percoll gradient.31 Cells (0.5 × 106 cells/mL) were stimulated in 96-well plates, 24-well plates, or T75 cell culture flasks with 10 μg/mL lipopolysaccharide (LPS; Sigma, St Louis, MO) or 1 μg/mL anti-mouse CD40 (BD/PharMingen), in combination with the following cytokines from R&D Systems (Menneapolis, MN): 10 ng/mL interleukin 4 (IL-4), 10 ng/mL IFN-γ, or 0.5 ng/mL transforming growth factor-β1 (TGF-β1). To obtain an optimal cell growth, stimulation by TGF-β1 is carried out in the presence of 8 ng/mL IL-4 and 0.25 ng/mL IL-5, as described by Snapper et al.32 The cells were cultured in RPMI 1640 medium or Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum, 2 mM glutamine, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptolethanol, 1 mM sodium pyruvate, 100 U/mL penicillin, 100 μg/mL streptomycin (all from Invitrogen/GIBCO, Groningen, The Netherlands). The proliferative response of B cells under the above conditions was measured by assessing [3H] thymidine incorporation.
Measurement of immunoglobulin secretion and circular transcripts
Ig titers from the supernatant of cultured B cells (1.0 × 106 cells/24-well at the start of stimulation) were determined by enzyme-linked immunosorbent assay (ELISA) as previously described,23 except for IgG2a, whose counter part is IgG2c in C57BL/6 mouse background.33 For measuring IgG2c, goat anti-mouse IgG (H+L) (Jackson Immunoresearch Laboratories, West Grove, PA) was used as capture antibody, and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG2c (SouthernBiotech) as detecting antibody. Cells (20 × 106) were harvested at different time points after stimulation. Total RNA was extracted from the stimulated cells by Trizol Reagent (Invitrogen), and was reverse transcribed as described under “Positive selection of B cells and amplification of the Sap transcript.” The circular transcripts of Iα and all subtypes of Iγ were amplified following the protocols reported by Kinoshita et al.34 The β-actin transcript was amplified by polymerase chain reaction (PCR) conditions described previously.29
Results
Normal B-cell development in Sap-deficient mice
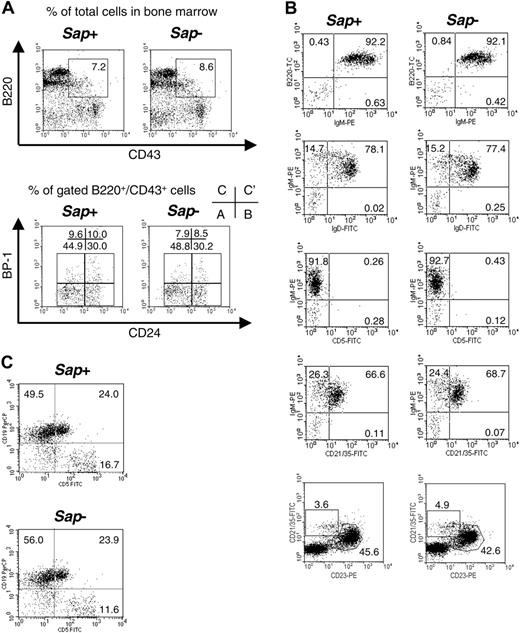
The striking defect in the 3 Sap-deficient mouse models is the dysregulation in Ig production.20-23 We previously showed comparable cell numbers and percentages of CD19+ as well as CD3+, CD4+, and CD8+ populations in Sap-deficient and wild-type littermate mice.23 In order to study whether impaired Ig production is due to a lack of specific B-cell subtypes, in addition to defective CD4+ function, we analyzed the B-cell subsets in the bone marrow, spleen, and peritoneal cavity. As shown in Figure 1, the proportions of early B cells in the bone marrow (according to Hardy fraction classification35 (B220/CD43/BP-1/CD24) (Figure 1A), mature B cells in the spleen (B220+IgM+, IgM+IgD+, IgM+CD5+, IgM+CD21+, and CD21+CD23+ cells) (Figure 1B), and peritoneal B1 population (B220+CD5+) (Figure 1C) were comparable in wild-type and mutant mice, indicating that the major B-cell subtypes develop normally in Sap-deficient mice.
Expression of SAP in B cells
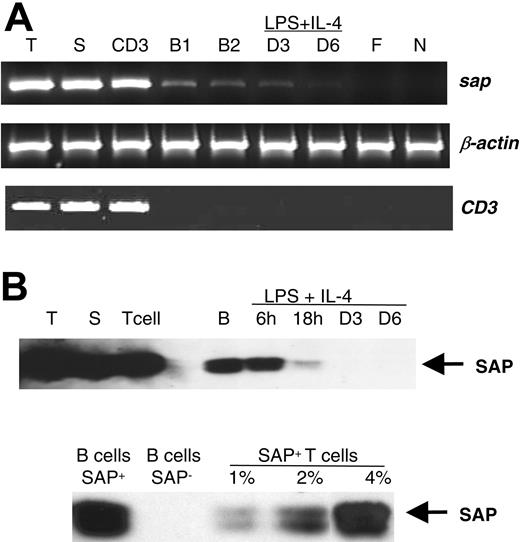
To address whether SAP plays a role in B-cell function, we assessed SAP expression in wild-type B cells. We purified B lymphocytes and performed reverse transcriptase (RT)-PCR and immunoprecipitation/immunoblot to examine Sap mRNA and SAP protein expressions. Using positive selection by fluorescence activated cell sorting (FACS), we obtained a CD19+ population with less than 0.12% of the CD3+, CD4+, CD8+, NK1.1+, and 2B4+ cells (data not shown). Using negative selection, we obtained B-cell preparations with less than 1% of the T or NK contaminants (data not shown). Semiquantitative RT-PCR analysis of RNA prepared from the sorted CD19+ cells and from the negatively enriched B cells revealed the presence of the Sap mRNA, albeit at a lower level than in thymocytes, splenocytes, or sorted CD3+ cells (Figure 2A). Interestingly, Sap expression was down-regulated after B-cell stimulation with LPS plus IL-4 (Figure 2A). Due to the limited number of cells available from FACS, the SAP protein was studied in negatively selected B cells. Consistent with the aforementioned expression of the Sap mRNA (Figure 2A), the SAP protein was present at low levels in B-cell extracts, and decreased gradually following LPS plus IL-4 stimulation (Figure 2B). To test the purity of the enriched B cells, CD3+ transcript was measured in the RNA samples by RT-PCR. Although no amplification band was found in the positively selected B cells or in the B cells after stimulation, a very faint amplification band was detected in the negatively enriched B-cell preparation (Figure 2A). Subsequently, we performed a mixing experiment to rule out the possibility that the Sap-transcript or the SAP protein observed in the RT-PCR and Western assays originate from trace T-cell contamination. Purified B cells from Sap-deficient mice that are devoid of SAP were mixed with T cells from wild-type mice to mimic various extents of T-cell contamination and then examined for SAP protein expression. As shown in Figure 2B, 1% and 2% of T-cell contamination generate faint signals in comparison to SAP expression in wild-type B-cell preparations. Only when T-cell contamination reaches 4% does the T-cell-derived SAP signal become comparable with that from the wild-type B cells. Since, as shown by FACS analysis, T cells represent fewer than 1% of the enriched B-cell population used in our experiments, it could be excluded as the source of the positive SAP signal in purified B cells.
B-cell subpopulations in the bone marrow, spleen, and peritoneal cavity. (A) Bone marrow. Percentages of gated cells or percentages of each fraction by Hardy classification35 (A, B, C, and C') are indicated. Spleen (B) and peritoneal cavity (C). Percentages of different B-cell subpopulations are indicated in the quadrants.
B-cell subpopulations in the bone marrow, spleen, and peritoneal cavity. (A) Bone marrow. Percentages of gated cells or percentages of each fraction by Hardy classification35 (A, B, C, and C') are indicated. Spleen (B) and peritoneal cavity (C). Percentages of different B-cell subpopulations are indicated in the quadrants.
Decreased IgG and IgA production in Sap-deficient B cells
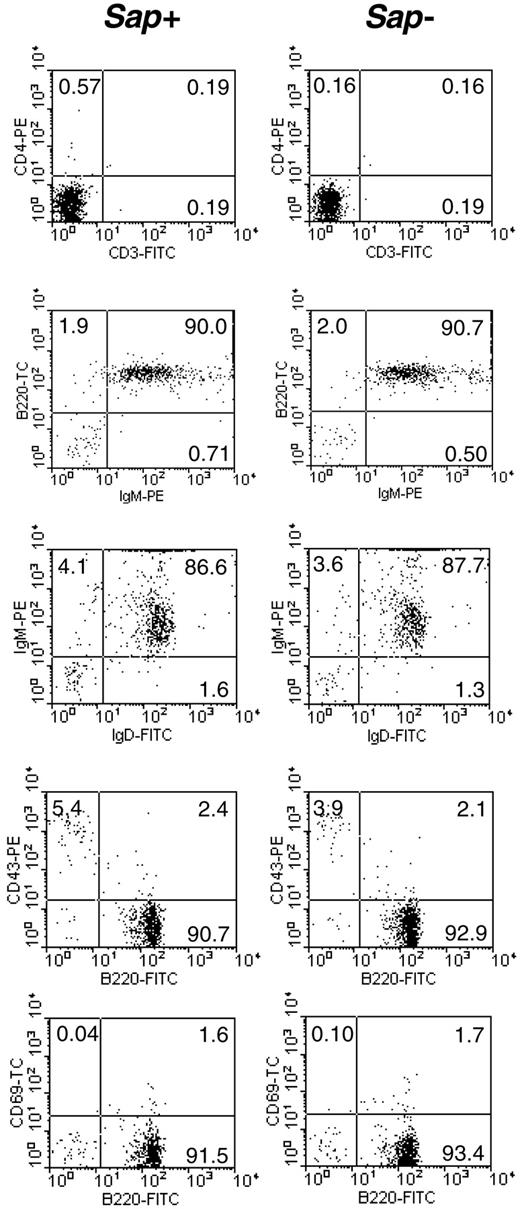
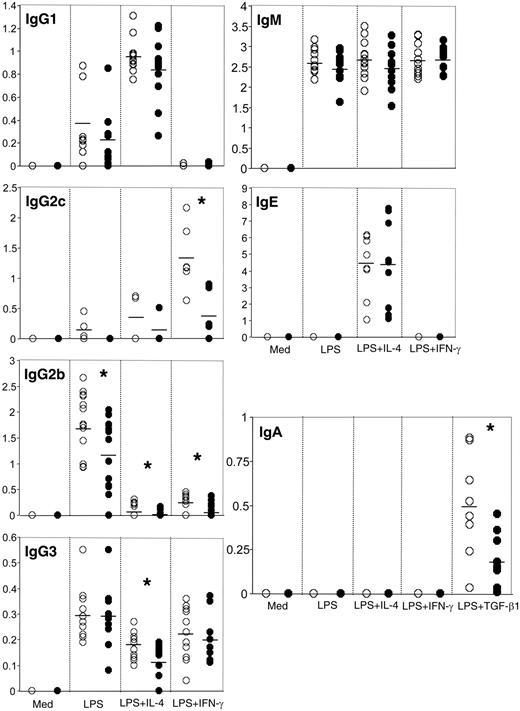
Since SAP was shown to be expressed in wild-type B cells, we investigated the impact of SAP deficiency on B-cell function, in particular, Ig production. In order to eliminate any in vivo preprogramming/interference, we used small resting B cells, which represent the mature naive B-cell population. The small resting B cells were isolated from the spleen by negative selection followed by density centrifugation. The resulting cell population was 92% B220+ and contained fewer than 0.8% CD3+ cells (Figure 3), as assessed by flow cytometry. Of the B220+ cells, 98% were IgM+, and 94% were IgM+IgD+ (Figure 3). The percentages of the B220+CD43+ and B220+CD69+ populations, representing the mature activated B cells, were low in both Sap-deficient and wild-type cell preparations (Figure 3). In order to study Ig production, these isolated small resting B cells were stimulated for 6 days with LPS in combination with IL-4, IFN-γ, or TGF-β1, and the secreted immunoglobulins were measured by ELISA. As shown in Figure 4, the IgM and IgE levels were comparable in both genotypes; however, IgG and IgA production was impaired in Sap-deficient B cells under most conditions. In particular, the levels of secreted IgG2c (equivalent to IgG2a in C57BL/6 background33 ), IgG2b, IgG3, and IgA were significantly lower in Sap-deficient B cells compared with wild-type B cells (Figure 4).
In order to rule out the possibility that the decreased in vitro Ig production was due to poor outgrowth of the Sap-deficient B cells during culture, we assessed the proliferation rates of the cultured cells by measuring the incorporated [3H]-thymidine. As shown in Figure 5, the proliferation rates of Sap-deficient and wild-type B cells were comparable. Moreover, numbers of Sap-deficient and wild-type live B cells, determined by trypan blue exclusion, were similar at various time points (data not shown).
Impaired Ig class switch recombination in Sap-deficient B cells
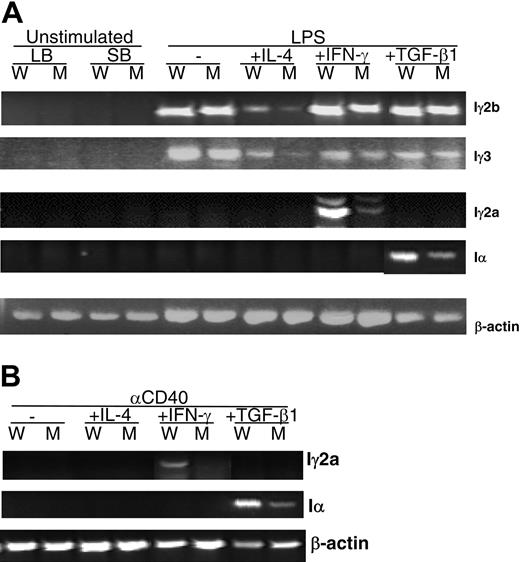
Subsequently, we addressed whether the decreased IgG2c, IgG2b, IgG3, and IgA production is due to a defect of Ig class switch recombination (CSR). Kinoshita et al demonstrated that CSR can be determined by measuring the abundance of RNA transcripts from the looped-out circular DNA.34 We studied the expression level of Iγ2a, Iγ2b, Iγ3, and Iα circular transcripts in Sap-deficient and wild-type B cells activated by LPS in combination with IL-4, IFN-γ, or TGF-β1. All 4 stimulation conditions led to detectable levels of Iγ2b and Iγ3 transcripts after 36 hours of treatment; however, transcript levels were lower in Sap-deficient B cells than in wild-type B cells (Figure 6A). Similarly, Iγ2a and Iα transcripts, which are clearly induced after 3 days of stimulation by IFN-γ and TGF-β1, respectively, were found to be significantly lower in Sap-deficient B cells than in wild-type B cells (Figure 6A). In addition, we analyzed the response of B cells following a more physiologic stimulation, anti-CD40 in combination with the 3 cytokines. All the aforementioned circular transcripts were measured. Iγ2b and Iγ3 were undetectable (not shown). However, Iγ2a and Iα transcripts were dramatically lower in anti-CD40 plus IFN-γ- and anti-CD40 plus TGF-β1-stimulated Sap-deficient B cells as compared with wild-type B cells (Figure 6B).
Expression of SAP in B cells. (A) Expression of the Sap transcript. RT-PCR products with primers 91U and 457L23 from 1μg total RNA extracted from the following cells: thymocytes (T), splenocytes (S), sorted CD3+ cells (CD3), sorted CD19+ cells (B1), negatively selected B cells, unstimulated (B2), and stimulated by LPS+IL-4 for 3 days (D3) and 6 days (D6), respectively. Mouse fibroblast cell line NIH 3T12 (F) and mouse neuroblastoma cell line Neuro 2A (N) were used as negative controls for Sap expression. The transcript of the mouse β-actin gene was used as the amplification control, and the transcript of mouse CD3 as the purity control of the enriched B cells. (B) Expression of the SAP protein. Top panel: SAP protein detected from thymocytes (T), splenocytes (S), T cells selected by Dynal beads (T cell), negatively selected B cells, unstimulated (B), and stimulated with LPS+IL-4 for 6 hours (6 h), 18 hours (18 h), 3 days (D3), and 6 days (D6), respectively; bottom panel: SAP protein detected from negatively selected wild-type B cells (B cells SAP+) and from negatively selected Sap-deficient B cells (B cells SAP-) mixed with wild-type T cells mimicking various percentages of T-cell contamination. The same amount of cell extract (0.5 mg protein) was immunoprecipited by anti-SAP antibody and the Western blot was hybridized with the same anti-SAP antibody.
Expression of SAP in B cells. (A) Expression of the Sap transcript. RT-PCR products with primers 91U and 457L23 from 1μg total RNA extracted from the following cells: thymocytes (T), splenocytes (S), sorted CD3+ cells (CD3), sorted CD19+ cells (B1), negatively selected B cells, unstimulated (B2), and stimulated by LPS+IL-4 for 3 days (D3) and 6 days (D6), respectively. Mouse fibroblast cell line NIH 3T12 (F) and mouse neuroblastoma cell line Neuro 2A (N) were used as negative controls for Sap expression. The transcript of the mouse β-actin gene was used as the amplification control, and the transcript of mouse CD3 as the purity control of the enriched B cells. (B) Expression of the SAP protein. Top panel: SAP protein detected from thymocytes (T), splenocytes (S), T cells selected by Dynal beads (T cell), negatively selected B cells, unstimulated (B), and stimulated with LPS+IL-4 for 6 hours (6 h), 18 hours (18 h), 3 days (D3), and 6 days (D6), respectively; bottom panel: SAP protein detected from negatively selected wild-type B cells (B cells SAP+) and from negatively selected Sap-deficient B cells (B cells SAP-) mixed with wild-type T cells mimicking various percentages of T-cell contamination. The same amount of cell extract (0.5 mg protein) was immunoprecipited by anti-SAP antibody and the Western blot was hybridized with the same anti-SAP antibody.
Purity of the isolated small resting B-cell preparations. Left column: wild-type B cells (Sap+); right column: Sap-deficient B cells (Sap-). Percentage of cells positive for selected surface markers is indicated in each quadrant.
Purity of the isolated small resting B-cell preparations. Left column: wild-type B cells (Sap+); right column: Sap-deficient B cells (Sap-). Percentage of cells positive for selected surface markers is indicated in each quadrant.
Discussion
To study the role of SAP in the immune system, several laboratories including ours have generated Sap knockout mice.20,21,23 Although there are differences in the gene targeting strategies, all 3 Sap-deficient mice have basal Ig deficiencies. Previous investigations20,21 attributed the Ig disregulation in Sap-deficient mice to a defect in Th2 function. Following these studies, Crotty et al22 demonstrated that defective CD4+ T-cell help is responsible for the lack of long-lived plasma cells and memory B cells in Sap-deficient mice, Hron et al27 showed that only the T-dependent antibody responses are impaired in Sap-deficient mice, and more recently, Ma et al36 reported the lack of isotype-switched memory B cells in the peripheral blood of the patients with XLP, which is associated with defective IL-10 production by CD4+ T cells. However, whereas the T-cell defect has been unequivocally demonstrated, none of these studies have excluded the possibility of an intrinsic functional defect in a subset of Sap-deficient B cells. To address this question, we examined the B-cell compartment in Sap-deficient mice. Purified small resting B cells were used to study in vitro Ig production following stimulation with LPS and anti-CD40 in combination with IL-4, IFN-γ, and TGF-β1. We observed impaired IgG and IgA production in Sap-deficient B cells, and we demonstrated that this was not a result of either abnormal growth during the cell culture or altered development of the major B-cell subpopulations. Thus, these data expand the role of SAP in regulating cellular functions to include the B-cell compartment. Consistent with our finding, a recent study provides convincing evidence demonstrating the presence of defective B-cell responses in a Sap-deficient background by adoptive transfer of wild-type and Sap-deficient T and B cells.28
While some studies reported the absence of the SAP protein in B-cell lysates,37,38 others showed that SAP is expressed in B cells of the interfollicular zone of the tonsil and lymph nodes of the humans39 and in follicular germinal center (GC) B cells in the spleen and lymph nodes in humans4 or in the mouse.28 Moreover, it has been shown that the SAP protein is detectable in Hodgkin and non-Hodgkin lymphoma, tonsillar B cells, some B-cell lines, and Burkitt lymphoma cell lines.4,15,38,39 We observed the expression of the Sap mRNA and the SAP protein in B cells from wild-type mice, although at very low levels (Figure 2). We also showed that activation of wild-type, mature naive B cells by LPS in the presence of IL-4 down-regulates the transcript and the protein product of the Sap gene. This result indicates that SAP is a tightly controlled molecule, expressed transiently in a specific stage or stages of B-cell differentiation. The exact subset of B cells expressing SAP and the signaling pathways that are regulated by SAP in B cells remain to be elucidated. One plausible model is that SAP regulates B-cell function by interacting with SLAM, as engagement of SLAM in B cells has been shown to enhance B-cell proliferation and Ig production.40 This is supported by the results of Mikhalap et al, who showed that SAP and SLAM are coexpressed in B cells in the interfollicular zone.39 It is also possible that SAP interferes with B-cell function through CD84 signaling, which is up-regulated in memory B cells and recruits SAP.8,9 Finally, all the SLAM family members with the exception of 2B4 are expressed on B cells, and therefore are good candidates for further investigation.
Aberrant Ig production by Sap-deficient B cells after in vitro activation. Small resting B cells from wild-type (○) and Sap-deficient mice (•) were stimulated for 6 days with LPS plus different cytokines as indicated. Each circle represents the concentration (μg/mL) of secreted Ig in the supernatant of the cells stimulated by the given culture condition. The average value of each isotype is indicated by a horizontal bar. Student t test, *P < .05.
Aberrant Ig production by Sap-deficient B cells after in vitro activation. Small resting B cells from wild-type (○) and Sap-deficient mice (•) were stimulated for 6 days with LPS plus different cytokines as indicated. Each circle represents the concentration (μg/mL) of secreted Ig in the supernatant of the cells stimulated by the given culture condition. The average value of each isotype is indicated by a horizontal bar. Student t test, *P < .05.
Proliferation rates of the stimulated wild-type and Sap-deficient B cells. □ and ▪ columns represent the wild-type and Sap-deficient B cells, respectively. Cells were cultured for 3 days or 6 days. Average values (cpm) of incorporated [3H]-thymidine, ± standard deviations, are shown. A similar pattern was observed in 3 other independent experiments.
Proliferation rates of the stimulated wild-type and Sap-deficient B cells. □ and ▪ columns represent the wild-type and Sap-deficient B cells, respectively. Cells were cultured for 3 days or 6 days. Average values (cpm) of incorporated [3H]-thymidine, ± standard deviations, are shown. A similar pattern was observed in 3 other independent experiments.
Circular transcripts generated by activated B cells. (A) Circular transcripts detected after LPS plus cytokine stimulation. Iγ2b and Iγ3 were analyzed after a 36-hour stimulation; Iγ2a and Iα after 3 days. (B) Iγ2a and Iα production after 3 days of stimulation with anti-CD40 plus cytokine. RT-PCR products were amplified from 1μg total RNA extracted from stimulated cells. The same patterns were observed in at least 3 independent experiments. LB, large activated B cells isolated by centrifugation on the Percoll gradient; SB, small resting B cells purified by Percoll gradient; the remaining samples were from small resting B cells stimulated in vitro. W, wild-type B cells; M, Sap-deficient B cells. The transcript of the mouse β-actin gene was used as the amplification control.
Circular transcripts generated by activated B cells. (A) Circular transcripts detected after LPS plus cytokine stimulation. Iγ2b and Iγ3 were analyzed after a 36-hour stimulation; Iγ2a and Iα after 3 days. (B) Iγ2a and Iα production after 3 days of stimulation with anti-CD40 plus cytokine. RT-PCR products were amplified from 1μg total RNA extracted from stimulated cells. The same patterns were observed in at least 3 independent experiments. LB, large activated B cells isolated by centrifugation on the Percoll gradient; SB, small resting B cells purified by Percoll gradient; the remaining samples were from small resting B cells stimulated in vitro. W, wild-type B cells; M, Sap-deficient B cells. The transcript of the mouse β-actin gene was used as the amplification control.
It is well established that LPS stimulation of B cells in the presence of cytokines can induce in vitro isotype switching.41 Results in Figure 4 demonstrate that Sap-deficient B cells produce lower levels of IgG2c, IgG2b, IgG3, and IgA in vitro than do wild-type B cells. This defect is not caused by abnormal proliferation of Sap-deficient B cells in response to the stimulation (Figure 5); instead, it reflects an impaired function of B cells. To exclude the possibility that the reduction in Ig production of Sap-deficient B cells was a result of a decrease in the number of memory B cells, we used small resting B cells, which were principally B220+IgM+ (98%). The proportion of B220+IgM- cells, which might represent the switched IgG+ or IgA+ cells, was very low and comparable in Sap-deficient and in wild-type B-cell preparations. We also excluded the possibility that decreased Ig production in Sap-deficient B cells is due to a lower percentage of activated B cells before stimulation, since percentages of the B220+CD43+ and B220+CD69+ populations are equally low in Sap-deficient and in wild-type B cells (Figure 3). It is worth noting that the impairment of Ig production in Sap-deficient B cells is only partial. Consistent with our observations, Morra et al observed that whereas the follicular GC B cells are affected in Sap-deficient mice, mature B cells outside the GC and T-independent responses are normal.28 Therefore, one could speculate that the absence of SAP leads to a defect in Ig class switch only in the subset of B cells expressing SAP and that this defect is partially masked by the presence of functionally unaltered B cells in the total B-cell compartment.
To study the mechanism of the impaired Ig secretion, we measured the CSR rates in Sap-deficient and wild-type B cells to study the isotype switching process. After cytokine stimulation, CSR is accompanied by looping-out DNA segments between the Cμ switch region and one of the downstream switch regions of each CH gene, including its I promoter.34 Kinoshita et al have shown that the I promoter is active in the excised circular DNAs and directs the production of circular transcripts, which are sensitive molecular markers of active CSR.34 We observed decreased levels of Iγ2a, Iγ2b, Iγ3, and Iα circular transcripts in Sap-deficient B cells stimulated with either LPS or anti-CD40 in combination with IL-4, IFN-γ or TGF-β1. This result indicates a defect in Ig class switch in Sap-deficient B cells, which is not Th1- or Th2-cytokine dependent. We further tested other components of Ig transcriptional machinery (eg, activation-induced cytidine deaminase [AID] and T-bet) in Sap-deficient mice. The AID gene is expressed in activated B cells and is essential to both CSR and somatic hypermutation (SHM).42-44 The T-box transcription factor T-bet is involved in IgG class switch, especially IgG2a, by regulating germ-line transcription.45,46 Interestingly, no differences in AID and T-bet expression were observed between wild-type and Sap-deficient B cells (data not shown). Thus, another pathway or pathways, which remain to be determined, may be responsible for the impaired Ig class switch in B cells from Sap-deficient mice. In exploring the potential role for SAP in CSR, one must consider the finding of Sylla et al, who demonstrated that the wild-type form of the SAP protein has an affinity for Ku70 and Ku80 proteins,47 which are known to be required for CSR.48,49 Therefore, one possibility is that SAP may be linked to some components of the CSR machinery.
Taken together, the impaired in vitro Ig production in Sap-deficient B cells and the decreased level of CSR suggest a B-cell dysfunction in Sap-deficient mice in addition to the abnormal CD4+ T-cell help previously reported.20-22,27 Its presence in wild-type B cells further supports the concept that the adaptor molecule SAP directly regulates B-cell function. This significant finding may provide novel insights into the pathology and/or mechanism of the XLP disease.
Prepublished online as Blood First Edition Paper, June 7, 2005; DOI 10.1182/blood-2004-07-2731.
Supported in part by grants from the Association for International Cancer Research, Association pour la Recherche sur le Cancer, GIS-Institut des Maladies Rares, Institut Nationale de la Santé et de la Recherche Médicale, and the Swiss Federal Office of Public Health. S.L. is a scientist from the Centre National de la Recherche Scientifique (France).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This work is dedicated to our late colleague Jun Liang. We thank Dr Giovanni Romeo (IARC, Lyon) for his encouragement and support; Mr Jean-Jacques Medard (IARC) for technical assistance; Miss Lily Wang (IARC) and Dr Benoit Pasquier (Unité INSERM 429, Paris) for some of their work; and Mrs Antoinette Trochard (IARC) for assistance in preparing the manuscript. We are grateful to Drs Uzma Hasan and Eric Garcia (Schering-Plough, Dardilly, France) and Mrs Francoise Le Deist (INSERM 429) for help with FACS, and Drs Anne Durandy (INSERM 429), Bao Vuong, and Paul Rothman (Columbia University, New York, NY) for stimulating discussions and critical reading of the manuscript.

![Figure 5. Proliferation rates of the stimulated wild-type and Sap-deficient B cells. □ and ▪ columns represent the wild-type and Sap-deficient B cells, respectively. Cells were cultured for 3 days or 6 days. Average values (cpm) of incorporated [3H]-thymidine, ± standard deviations, are shown. A similar pattern was observed in 3 other independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/6/10.1182_blood-2004-07-2731/6/m_zh80180584310005.jpeg?Expires=1769167267&Signature=5Ka9arWd99--vArasnJS9XWwYaOBg5mbYeVohOdvYlqeIG9A2gyTnlsiFZC~2ijpPNZ5yLdNXCtc~BHaSGTNF4IjPP~Hui~3n~6aNbFqhf~vMZ6yemG-gON3Is9hw84Dztjm0pTyv~EPbrKiHdvShSdbMZFnypUqsicF4VEMqn-rp9~j5aR8x4Nz67yehu-EomPdYLeThkq0PUnv2vhWyOaac74YV-Sb4ld1tPxUnVEd~leeKMF0drHUbdEu5ca109JtVPR43-nr7ln57y27U2XeHdmrn3tegVcz713D92N6Y7RfL4YKvhOxKfYxvd8V7zgvs0OSza325qbTKGz1fA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal