Abstract
Regulation of osteoclast differentiation is key to understanding the pathogenesis and to developing treatments for bone diseases such as osteoporosis. To gain insight into the mechanism of the receptor activator of nuclear factor (NF)–κB ligand (RANKL)–specific induction of the osteoclast differentiation program, we took a suppression-subtractive hybridization screening approach to identify genes specifically induced via the RANKL-Rac1 signaling pathway. Among identified targets, we show that RANKL selectively induces cyclooxygenase (COX) 2 expression via Rac1 that results in turn in production of prostaglandin E2 (PGE2) in RAW 264.7 cells. By using transient transfection assays, we found that the –233/–206 region of the COX-2 promoter gene was critical for RANKL-induced promoter activity. This RANKL-responsive region contained an NF-κB site that, when mutated, completely abolished the induction of NF-κB DNA-binding activity by RANKL. Blockade of COX-2 by celecoxib inhibits differentiation of bone marrow-derived monocyte/macrophage precursor cells (BMMs) into tartrate-resistant acid phosphatase-positive (TRAP+) osteoclastic cells. This inhibition can be rescued by the addition of exogenous PGE2, suggesting that COX-2–dependent PGE2 induction by RANKL in osteoclast precursors is required for osteoclast differentiation.
Introduction
Bone remodeling depends on a delicate balance between bone formation and bone resorption, wherein bone-forming osteoblasts and bone-resorbing osteoclasts play central roles.1 Tipping this balance in favor of osteoclasts leads to pathologic bone resorption, as seen in bone diseases such as autoimmune arthritis and postmenopausal osteoporosis.2 Thus, the investigation of the regulatory mechanisms of osteoclast differentiation is critical to understand the physiology and pathology of the skeletal system.
Receptor activator of nuclear factor (NF)–κB ligand (RANKL; also called TRANCE [tumor necrosis factor-related activation-induced cytokine], ODF [osteoclast differentiation factor], and OPGL [osteoprotegrin ligand]), a member of the tumor necrosis factor (TNF) family,3-6 plays an essential role in osteoclastogenesis, and its signaling mechanism has been extensively studied.7 Briefly, binding of RANKL to its receptor, RANK, results in the recruitment of TNF receptor-associated factor (TRAF) family proteins such as TRAF6, which, in turn, activates the NF-κB and Jun kinase (JNK) pathways. RANKL also induces c-Fos by an as yet unknown mechanism. The essential role of the TRAF6 and c-Fos pathways in osteoclastogenesis determined by gene targeting studies has been well documented.8,9 In addition, several negative regulators of RANKL signaling have been reported.10-12 A typical example is osteoprotegerin (OPG), which functions as a soluble decoy receptor for RANKL, thereby attenuating excessive RANKL signaling.
To gain insight into how osteoclasts differentiate and function to control bone metabolism, we took a suppression-subtractive hybridization (SSH) screening approach to identify genes specifically induced via RANKL-Rac1 signaling pathway. In this screening, we found that cyclooxygenase 2 (COX-2) is one of the strongly induced genes following RANKL stimulation in osteoclast precursors. COX-2 is known to be expressed at very low levels and is strongly induced by proinflammatory stimuli, including lipopolysaccharide (LPS), as well as by several activated oncogenes.13,14 The significance of COX-2 in inflammation is highlighted by the observation that COX-2 inhibitors block the synthesis of prostaglandin (PG) E2 (PGE2) and, as a result, inhibit inflammation and confer analgesia.15 In bone, PGE2 is produced mainly by osteoblasts and acts as a potent stimulator of bone resorption.16 The production of PGE2 by osteoblasts is regulated by several cytokines, including interleukin 1 (IL-1) and IL-6. The addition of indomethacin, an inhibitor of PG synthesis, to mouse bone marrow cultures strikingly suppresses osteoclast formation induced by IL-1, indicating that PGE2 production by osteoblasts is involved in the mechanism of bone resorption induced by IL-1.17,18 In the inflammatory processes, in which the mononuclear phagocyte family plays a crucial part, PGs by themselves have little inflammatory capacity, but in the presence of other mediators they can synergistically amplify the local inflammatory responses.19,20 Because the rate-limiting step in PGE2 synthesis is catalyzed by COX-2,21 the fact that osteoclast precursors express COX-2 by RANKL stimulation led us to the investigation of direct involvement of PGE2 in osteoclast differentiation induced by RANKL signaling.
Here, we provide evidence that COX-2 expression is dependent on RANKL-regulated small guanosine triphosphatase (GTPase) Rac1 pathway. We also show that RANKL stimulation results in the production of PGE2, which contributes to the accelerated osteoclast differentiation. We further demonstrate that the ability of Rac1 to regulate COX-2 expression depends on NF-κB, a key regulator of COX-2 transcription. Finally, we show that inhibition of COX-2 by celecoxib severely suppresses differentiation of bone marrow-derived monocyte/macrophage cells (BMMs) into tartrate-resistant acid phosphatase-positive (TRAP+) osteoclasts. This inhibition can be rescued by the addition of exogenous PGE2. Collectively, our results show that RANKL-Rac1-NF-κB–dependent COX-2 expression plays a key role in the RANKL-induced osteoclast differentiation via PGE2 production.
Materials and methods
Cell culture and reagents
The RAW264.7 monocyte/macrophage cell line was maintained at 37°C in 5% CO2 atmosphere in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). RAW264.7 cells stably transfected with a cDNA encoding a dominant-negative mutant of Rac1, RacN17 (RAW-RacN17), were maintained in DMEM supplemented with 10% FBS and 400 μg/mL G418. Celecoxib, purchased from LKT Laboratories (Minneapolis, MN), was dissolved in dimethyl sulfoxide (DMSO) as a 100-mM stock solution and stored at –20°C. Recombinant macrophage colony-stimulating factor (M-CSF) was purchased from R&D Systems (Minneapolis, MN); RANKL was from PeproTech (London, United Kingdom); and all other chemicals were from Sigma (St Louis, MO). Anti-COX-2 (C-20) was from Santa Cruz Biotechnology (Santa Cruz, CA).
In vitro osteoclastogenesis
Nonadherent bone marrow-derived monocytes/macrophages cells (BMMs) derived from C57BL/6 mice were seeded and cultured in α-minimal essential medium (α-MEM; Gibco, Grand Island, NY) with 10% FBS containing 10 ng/mL M-CSF. After 2 days, nonadherent cells including lymphocytes were washed out, and adherent cells were used as BMMs. These osteoclast precursor cells were further cultured in the presence of 50 ng/mL RANKL and 10 ng/mL M-CSF to generate osteoclasts. After 5 days of culture, cells were fixed and stained for tartrate-resistant acid phosphatase (TRAP). TRAP+ multinucleated (> 3 nuclei) cells were counted as osteoclast-like cells. The cells were observed with a Zeiss Axiovert 200 microscope (Carl Zeiss, Oberkochen, Germany) equipped with a Plan-Neofluor 20×/0.50 objective lens (Carl Zeiss), and images were obtained with an AxioCam HR (Carl Zeiss) using AxioVision 3.1 software (Carl Zeiss).
Subtractive hybridization and differential screening
Total RNA was prepared using TRIzol reagent (Gibco). Then, 1 mg total RNA was subjected into an Oligotex poly A+ RNA column (Qiagen, Valencia, CA) to purify poly A+ RNA. Then, 2 μg poly A+ RNA from RANKL-treated RAW264.7 and RAW-RacN17 cells was used to make tester and driver cDNAs. Subtracted polymerase chain reaction (PCR) was performed using the PCR-select cDNA subtraction kit according to the manufacturer's protocol (Clontech, Palo Alto, CA). Briefly, tester and driver cDNAs were digested with RsaI, and the tester was ligated to adapter DNA. After 2 hybridizations with the tester and driver cDNAs, the resulting mixture was diluted 1:1000 and amplified by PCR using flanking and nested primers to produce a subtracted and normalized PCR fragment library. The efficiency of subtraction was verified via Southern blot analysis of the unsubtracted and subtracted PCR products using a 32P-labeled glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA probe. Subtracted PCR products were cloned into pGEM-T easy vector (Promega, Madison, WI) for DNA sequence analysis.
Northern hybridization and immunoblotting analysis
For the Northern analysis, 20 μg total RNAs extracted from RANKL-treated RAW264.7 and RAW-RacN17 cells was separated and transferred to nylon membranes as described.4 The membranes were hybridized with α-32P]dCTP probe prepared using a ready-to-go labeling kit and a ProbeQuant G-50 purification kit (Amersham Biosciences, Buckinghamshire, United Kingdom). For immunoblotting analysis, 106 cells of RAW264.7 and RAW-RacN17 cells were stimulated with RANKL (50 ng/mL) for the indicated times and then lysed in 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.4, 150 mM NaCl, 1% Triton X-100, 10% glycerol, and proteinase inhibitors mix for 30 minutes on ice. After centrifugation, supernatants were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Amersham Biosciences). The proteins were detected with specific antibodies against COX-2 and tubulin (Santa Cruz Biotechnology).
PGE2 measurements
RAW264.7 cells and RAW-RacN17 cells were cultured for 15 hours in 96-well plates. Indomethacin (10 μM) and celecoxib (100 ng/mL) were applied 30 minutes prior to RANKL (50 ng/mL) stimulation for 8 hours. The PGE2 level was measured in cells plated 104/well, using a commercially available PGE2 enzyme immunoassay kit (Cayman Chemicals, Ann Arbor, MI).
Transient transfections and luciferase assay
Luciferase reporter pGL3 plasmid including 5′-flanking region (from –1838 to +123 bp) of the human COX-2 gene was kindly provided by Dr S. Prescott (University of Utah, Salt Lake). DNA fragments of various lengths of the COX-2 promoter regions (–1838/+123, –518/+123, –361/+123, –200/+123, –96/+123, –78/+123, and –44/+123) were prepared by PCR using the pGL3 plasmid as the template and subsequently subcloned into pGL3 plasmid. For luciferase assay, RAW264.7 and RAW-RacN17 cells (1 × 107), suspended in 400 μL serum-free medium, were mixed with 10 μg pGL3-basic or pGL3-COX-2 promoter-luciferase reporter and 2 μg pCMV-β-galactosidase plasmid and electroporated (BTX, Holliston, MA) using the preset protocol for RAW264.7 cells (155 V, 70 msec, 1 pulse). Thirty hours after electroporation, RANKL (50 ng/mL) was applied for 6 hours. Cells were then harvested and lysed in a reporter lysis buffer (Promega). Luciferase value was normalized to β-galactosidase value. The relative luciferase value (RLV) was defined as the ratio of the COX-2 luciferase activity divided by the activity of pGL3-basic luciferase in cell lysates. The RLV of RANKL-stimulated pGL3-basic transfected cultures was given the arbitrary value of 1. The fold induction was calculated by dividing the RLV of a given culture by the RLV of the pGL3-basic control.
EMSA
RANKL-stimulated or unstimulated cells were lysed and nuclear extracts were prepared as described previously.22 For the electrophoretic mobility shift assay (EMSA), 1 × 105 cpm of a 32P-end-labeled probe was incubated with a mixture of nuclear extract (5 μg) in the binding buffer (10 mM Tris [tris(hydroxymethyl)aminomethane)–HCl, pH 7.5, 50 mM KCl, 1 mM dithiothreitol, 1 mM EDTA [ethylenediaminetetraacetic acid], 5% glycerol, and 2 μg poly (dI-dC) plus 5% NP-40) for 15 minutes at room temperature, and the protein-DNA complexes were resolved in a 5% native polyacrylamide gel and detected by autoradiography. We used the following double-stranded probes: proximal NF-κB site of wild-type probe, 5′-CAGGAGAGTGGGGACTACCCCCTCTGCT-3′ (–233/–206); distal NF-κB site of wild-type probe, 5′-GGCGGGAGAGGGGATTCCCTGCGCCCCCG-3′ (–457/–429); proximal NF-κB site of mutant probe, 5′CAGGAGAGTGGccACTACCCCCTCTGCT-3′; and distal NF-κB site of mutant probe, 5′-GGCGGGAGAGGGGATctCCTGCGCCCCCG-3′. A double-stranded, NF-κB–specific oligonucleotide probe containing the 2 tandemly repeated NF-κB sites derived from the HIV long terminal repeat (5′-ATCAGGGACTTTCCGCTGGGGACTTTCCG-3′) was also used as a control.23 The underlined nucleotides indicate the NF-κB–binding sites, and the lowercase nucleotides indicate the mutations.
Results
Identification of Rac1-dependent genes highly expressed during osteoclastogenesis on RANKL stimulation
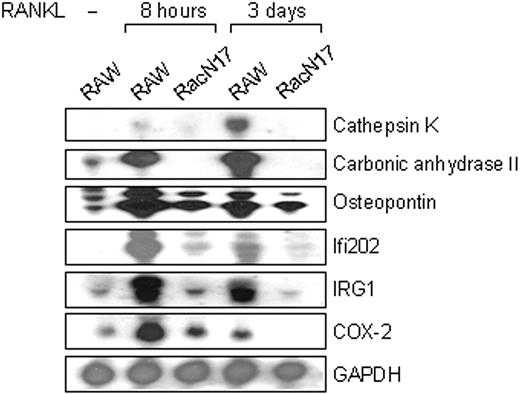
We have recently reported that RANKL activates Rac1 in RAW264.7 cells thereby regulating osteoclast differentiation (Fig7).24 To identify genes selectively activated by RANKL through Rac1-dependent pathway, differentially expressed genes between RANKL-stimulated RAW264.7 cells and RANKL-stimulated RAW-RacN17 cells (denoted as RacN17) expressing a dominant interfering mutant RacN17 were isolated using SSH, a cDNA subtraction technique based on suppression PCR that is sensitive to rare transcripts. The subtractive cDNA libraries were constructed at the 2 different RANKL-stimulated time points (8 hours and 3 days) and were analyzed as described previously.25 The most abundant clones chosen from the subtraction were carbonic anhydrase II, osteopontin, and cathepsin K, all of which were previously known to play critical roles in osteoclast function.26,27 As shown in Figure 1, mRNAs for carbonic anhydrase II and cathepsin K, both of which are known to be strongly expressed in osteoclasts, were specifically induced by RANKL in RAW264.7 cells but not in RAW-RacN17 cells, confirming the validity of our screening protocol. Among genes that have not previously been functionally implicated in osteoclast differentiation were immune response gene (IRG) 1,28 interferon (IFN)–inducible gene (Ifi) 202,29 and COX-2 (Figure 1). In osteoblasts, COX-2 is known to play a critical role in synthesis of PGE221 thereby stimulating osteoclast differentiation indirectly.30 Because RANKL-stimulated COX-2 induction in osteoclast precursors has not been reported previously, COX-2 was chosen for further analysis in this study.
Requirement of the Rac1 pathways in RANKL-induced COX-2 expression and PGE2 induction
To confirm that Rac1 controls the induction of COX-2 by RANKL, we measured COX-2 expression by Northern blotting with RNA preparation from RAW264.7 and RAW-RacN17 cells at various time points after RANKL stimulation. The results showed that COX-2 induction of RAW264.7 cells was seen from 2 hours after RANKL stimulation and decreased toward basal level after 24 hours (Figure 2A). In contrast, COX-2 induction by RANKL was significantly lower in RAW-RacN17 cells. Consistent with this result, immunoblotting analysis revealed that robust induction of COX-2 protein occurred in RAW264.7 cells but not in RAW-RacN17 cells (Figure 2B).
Identification of specifically expressed genes in RANKL-stimulated RAW 264.7 cells. Northern blot analysis of genes expressed during osteoclast differentiation. Total RNA was extracted from RAW264.7 and RAW-RacN17 cells stimulated with RANKL for the indicated periods. Total RNA (20 μg) was applied to each lane. cDNAs for cathepsin K, carbonic anhydrase II, osteopontin, Ifi202, IRG1, and COX-2 were used to prepare probes and hybridized to total RNA. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a control for total RNA loading.
Identification of specifically expressed genes in RANKL-stimulated RAW 264.7 cells. Northern blot analysis of genes expressed during osteoclast differentiation. Total RNA was extracted from RAW264.7 and RAW-RacN17 cells stimulated with RANKL for the indicated periods. Total RNA (20 μg) was applied to each lane. cDNAs for cathepsin K, carbonic anhydrase II, osteopontin, Ifi202, IRG1, and COX-2 were used to prepare probes and hybridized to total RNA. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a control for total RNA loading.
COX-2 is rapidly induced by RANKL stimulation in a Rac1-dependent manner. (A) Expression of COX-2 mRNA in RAW264.7 and RAW-RacN17 cells stimulated with RANKL for the indicated periods was examined by Northern analysis. GAPDH was used as a control for total RNA loading. (B) Expression of COX-2 protein in RAW264.7 and RAW-RacN17 cells stimulated with RANKL for the indicated periods. Whole cell lysates (50 μg each) were immunoblotted with anti–COX-2 antibody and reprobed with antitubulin antibody.
COX-2 is rapidly induced by RANKL stimulation in a Rac1-dependent manner. (A) Expression of COX-2 mRNA in RAW264.7 and RAW-RacN17 cells stimulated with RANKL for the indicated periods was examined by Northern analysis. GAPDH was used as a control for total RNA loading. (B) Expression of COX-2 protein in RAW264.7 and RAW-RacN17 cells stimulated with RANKL for the indicated periods. Whole cell lysates (50 μg each) were immunoblotted with anti–COX-2 antibody and reprobed with antitubulin antibody.
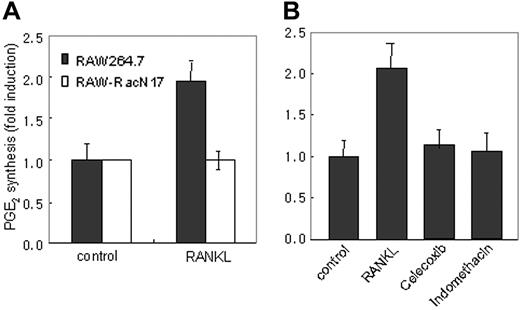
To determine whether PGE2 secretion depends on the RANKL-Rac1–dependent pathway, we measured the level of PGE2 in supernatants of RAW264.7 and RAW-RacN17 cell cultures harvested before and after 8 hours of stimulation with RANKL. As shown in Figure 3A, RANKL stimulation induced PGE2 production in RAW264.7 cells, whereas RAW-RacN17 cells were defective in PGE2 induction by RANKL. Consistently, COX inhibitors, celecoxib and indomethacin, completely inhibited RANKL-induced PGE2 secretion (Figure 3B). These data showed that COX-2 expression by Rac1-transduced signals is required for PGE2 induction by RANKL.
RANKL-stimulated PGE2 secretion is compromised in RAW-RacN17 cells. (A) RAW264.7 cells and RAW-RacN17 cells were stimulated with or without RANKL (50 ng/mL) for 8 hours. Culture supernatants were collected and PGE2 concentration was measured as described in “Materials and methods.” Unstimulated cultures from RAW264.7 and RAW-RacN17 cells secreted similar levels of PGE2. PGE2 levels produced by unstimulated cells were given the arbitrary value of 1. Data are expressed as mean (± SD) of triplicates. Similar results were obtained in 2 additional experiments. (B) RAW264.7 cells were pretreated for 30 minutes with indomethacin (10 μM) and celecoxib (100 ng/mL) and stimulated with RANKL for 8 hours. PGE2 levels were measured as described in panel A. Data are expressed as mean (±SD) of triplicates.
RANKL-stimulated PGE2 secretion is compromised in RAW-RacN17 cells. (A) RAW264.7 cells and RAW-RacN17 cells were stimulated with or without RANKL (50 ng/mL) for 8 hours. Culture supernatants were collected and PGE2 concentration was measured as described in “Materials and methods.” Unstimulated cultures from RAW264.7 and RAW-RacN17 cells secreted similar levels of PGE2. PGE2 levels produced by unstimulated cells were given the arbitrary value of 1. Data are expressed as mean (± SD) of triplicates. Similar results were obtained in 2 additional experiments. (B) RAW264.7 cells were pretreated for 30 minutes with indomethacin (10 μM) and celecoxib (100 ng/mL) and stimulated with RANKL for 8 hours. PGE2 levels were measured as described in panel A. Data are expressed as mean (±SD) of triplicates.
Functional activity of COX-2 promoter in response to RANKL
To explore the transcriptional regulation of COX-2 gene by RANKL stimulation, we studied the COX-2 promoter-driven transcription in RAW264.7 cells transiently transfected with DNAs containing a region spanning from –1838 to +123 bp relative to the transcription start site of the human COX-2 gene in the reporter plasmid pGL3 basic (P1-1838). As shown in Figure 4A, luciferase activity of this construct was strongly up-regulated on activation by RANKL. To map the regions responsible for this induction, subsequent 5′-deletion constructs ranging from –518 down to –44 bp were generated (Figure 4A). An induction in promoter activity on RANKL stimulation was observed only in the reporter construct, P-518, indicating that a critical regulatory region at –518/–362 bp was responsible for this induction. Sequence analysis revealed the existence of a potential distal NF-κB–binding site at –457/–429 bp.31 In contrast, the promoter activity of a shorter construct, P-361, containing a proximal NF-κB–binding site at –233/–206 bp could not be induced by RANKL stimulation, suggesting that the distal NF-κB–binding site but not the proximal one is important for RANKL-induced activation of COX-2 promoter.
To determine whether Rac1 regulates COX-2 promoter activity, RAW264.7 and RAW-RacN17 cells were electroporated with a reporter construct containing the luciferase gene under the control of the COX-2 promoter (P-1838) or reporter basic (pGL3-basic). After 30 hours, cells were untreated or treated with RANKL or LPS for 6 hours. Cells were then lysed and luciferase activity was measured in the lysates. The results showed that LPS increased COX-2 promoter activity in both RAW264.7 and RAW-RacN17 cells. However, RANKL activated the COX-2 promoter in RAW264.7 cells but not RAW-RacN17 cells (Figure 4B). Taken together, these data indicate that the transcriptional induction of COX-2 in RAW264.7 cells on RANKL stimulation occurs via Rac1-NF-κB–signaling cascades.
Analysis of the region responsible for the promoter activity of COX-2 gene in response to RANKL stimulation. (A) Putative consensus sequences in the 5′-flanking region of human COX-2 gene are illustrated in the upper left. Each deleted promoter fragment was inserted into a pGL3-basic luciferase (Luc) vector. Numbers indicate distance in base pairs from the start site of transcription. RAW264.7 cells were transiently transfected with these plasmids along with β-galactosidase plasmid. The transfected cells were stimulated with RANKL (50 ng/mL) for 6 hours, and then the activities of luciferase and β-galactosidase were measured. The normalized luciferase activities in the cells transfected with various reporter plasmids are presented as the fold induction calculated by dividing the relative luciferase value (RLV) of a given culture by the RLV of the pGL3-basic control. Data are expressed as mean (± SD) of triplicates. NFAT indicates nuclear factor of activated T cells; API, activator protein 1; ATF, activating transcription factor; CRE, cyclic adenosine monophosphate (cAMP)–responsive element; TATA, TATA box. (B) As described in panel A, except that RAW264.7 cells and RAW-RacN17 cells were transfected with the reporter plasmid P-1838 and stimulated with RANKL (50 ng/mL) and LPS (1μg/mL), respectively. PBS indicates phosphate-buffered saline used for a control. Data are expressed as mean (±SD) of triplicates.
Analysis of the region responsible for the promoter activity of COX-2 gene in response to RANKL stimulation. (A) Putative consensus sequences in the 5′-flanking region of human COX-2 gene are illustrated in the upper left. Each deleted promoter fragment was inserted into a pGL3-basic luciferase (Luc) vector. Numbers indicate distance in base pairs from the start site of transcription. RAW264.7 cells were transiently transfected with these plasmids along with β-galactosidase plasmid. The transfected cells were stimulated with RANKL (50 ng/mL) for 6 hours, and then the activities of luciferase and β-galactosidase were measured. The normalized luciferase activities in the cells transfected with various reporter plasmids are presented as the fold induction calculated by dividing the relative luciferase value (RLV) of a given culture by the RLV of the pGL3-basic control. Data are expressed as mean (± SD) of triplicates. NFAT indicates nuclear factor of activated T cells; API, activator protein 1; ATF, activating transcription factor; CRE, cyclic adenosine monophosphate (cAMP)–responsive element; TATA, TATA box. (B) As described in panel A, except that RAW264.7 cells and RAW-RacN17 cells were transfected with the reporter plasmid P-1838 and stimulated with RANKL (50 ng/mL) and LPS (1μg/mL), respectively. PBS indicates phosphate-buffered saline used for a control. Data are expressed as mean (±SD) of triplicates.
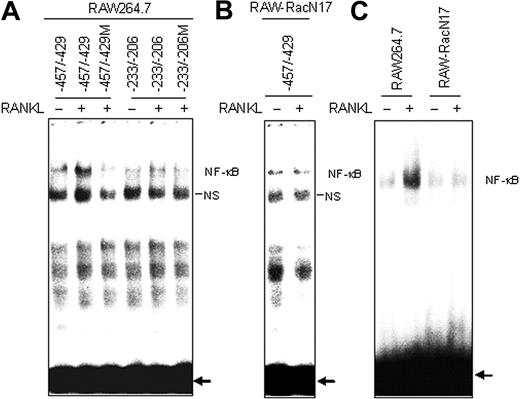
EMSA targeting NF-κB sites. (A) Nuclear extracts from RAW264.7 cells stimulated with RANKL (50 ng/mL) for 15 minutes were analyzed by EMSA. Gel shift assays were performed using oligonucleotides corresponding to the COX-2 distal NF-κB site (nucleotides –457/–429) and the COX-2 proximal site (nucleotides –233/–206). The COX-2 distal site (nucleotides –457/–429) probe containing a mutation in NF-κB element abolished the complex formation. (B) As described in panel A, except that nuclear extracts were prepared from RAW-RacN17 cells. Gel shift assays were performed using oligonucleotides corresponding to the COX-2 distal NF-κB site (nucleotides –457/–429). (C) Gel shift assays were performed using oligonucleotides containing the 2 tandemly repeated NF-κB sites derived from the HIV long terminal repeat. NS indicates a nonspecific protein complex. The arrows point to free probes. Similar results were obtained in 2 additional experiments.
EMSA targeting NF-κB sites. (A) Nuclear extracts from RAW264.7 cells stimulated with RANKL (50 ng/mL) for 15 minutes were analyzed by EMSA. Gel shift assays were performed using oligonucleotides corresponding to the COX-2 distal NF-κB site (nucleotides –457/–429) and the COX-2 proximal site (nucleotides –233/–206). The COX-2 distal site (nucleotides –457/–429) probe containing a mutation in NF-κB element abolished the complex formation. (B) As described in panel A, except that nuclear extracts were prepared from RAW-RacN17 cells. Gel shift assays were performed using oligonucleotides corresponding to the COX-2 distal NF-κB site (nucleotides –457/–429). (C) Gel shift assays were performed using oligonucleotides containing the 2 tandemly repeated NF-κB sites derived from the HIV long terminal repeat. NS indicates a nonspecific protein complex. The arrows point to free probes. Similar results were obtained in 2 additional experiments.
To further analyze the functionality of these sequences as NF-κB–binding elements, we performed EMSA with nuclear extracts of RAW264.7 cells using DNA probes of the 2 putative NF-κB–binding sites. A retarded complex was resolved in nuclear extracts from RAW264.7 cells stimulated by RANKL with the distal NF-κB probe (–457/–429) but not with the proximal probe (–233/–206; Figure 5A). This band was not seen with a distal NF-κB probe (–457/–429M) containing a mutation in the NF-κB–binding element, confirming the specificity of the reaction. As expected, the retarded complex could not be seen in RAW-RacN17 cells on RANKL stimulation (Figure 5B). We also performed EMSA with an oligonucleotide probe containing the 2 NF-κB sites derived from the HIV long terminal repeat. Consistent with the results in RAW264.7 cells, RANKL-induced NF-κB activation was severely affected in RAW-RacN17 cells (Figure 5C).
Inhibition of osteoclast differentiation by COX-2 inhibitors
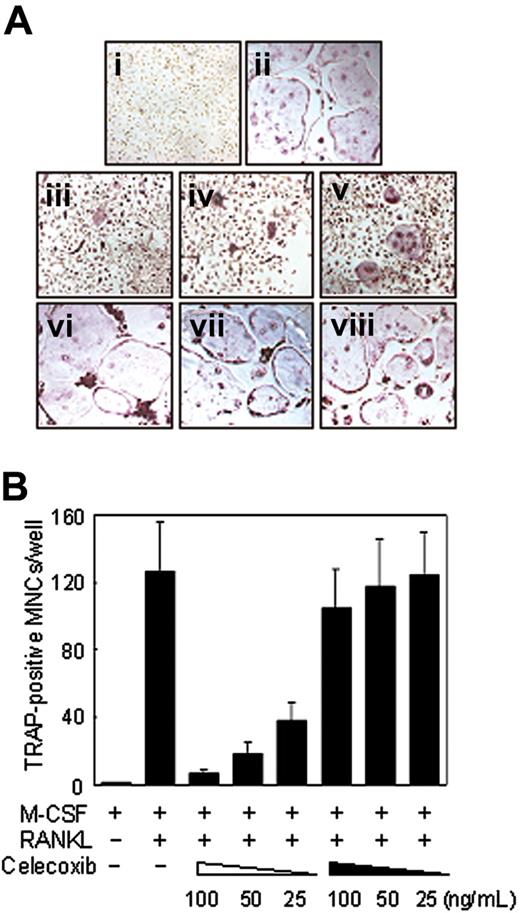
To investigate the role of RANKL-stimulated COX-2 expression in osteoclast precursors, we examined the effect of a specific inhibitor of COX-2, celecoxib, on osteoclast differentiation of BMMs in the absence of stromal cells. Celecoxib inhibited RANKL-induced osteoclast differentiation from BMMs in a dose-dependent manner (Figure 6Aiii-v). The similar inhibitory effect was observed with indomethacin, COX-1 and COX-2 inhibitors in BMMs (data not shown).
Because the expression of COX-2 is clearly seen within 15 hours after RANKL stimulation (Figure 2), we next examined whether celecoxib affects the early stage or the late stage of osteoclast differentiation by performing temporal treatment with celecoxib. Addition of celecoxib 2 days after RANKL stimulation had no effect on osteoclast formation (Figure 6Avi-viii), consistent with the fact that the early time induction of PGE2 by RANKL stimulation is necessary for osteoclast differentiation.
Exogenous PGE2 reversed the inhibitory effects of celecoxib
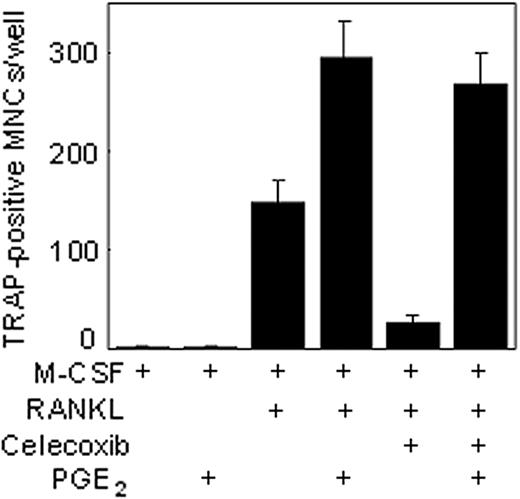
Celecoxib is known to inhibit COX-2,32 the enzyme responsible for catalyzing the formation of PGE2 from arachidonic acid. Whether the inhibitory effects of celecoxib on osteoclastogenesis are mediated through the suppression of COX-2 was investigated. To determine this, BMMs were exposed to exogenous PGE2 together with celecoxib and then examined for RANKL-induced osteoclast differentiation. The results in Figure 7 show that suppressive effects of osteoclast differentiation by celecoxib could be reversed by PGE2. The latter alone had no effect on osteoclast differentiation. Thus, these results suggest that celecoxib inhibits osteoclast differentiation through the suppression of COX-2 activity, thereby resulting in the inhibition of PGE2 induction.
Suppressive effects of COX-2 inhibitor, celecoxib, on RANKL-induced osteoclastogenesis in BMMs. (A) Demonstration of TRAP+ cells differentiating from BMMs unstimulated (i) or stimulated (ii) with RANKL. On day 5 with RANKL stimulation, the cells were fixed and stained for TRAP. TRAP+ cells appear as red cells. (iii-viii) Dose-dependent effects of celecoxib on the formation of TRAP+ multinucleated osteoclasts (MNCs) with RANKL stimulation. BMMs were pretreated (iii, 100 ng/mL; iv, 50 ng/mL; v, 25 ng/mL) with different concentrations of celecoxib or treated (vi, 100 ng/mL; vii, 50 ng/mL; viii, 25 ng/mL) 2 days after RANKL stimulation. On day 5 with RANKL stimulation, the cells were fixed and stained for TRAP (original magnification, × 100). (B) Total numbers of TRAP+ MNCs are shown. Note that formation of TRAP+ MNCs was inhibited by pretreatment of celecoxib in a dose-dependent manner. The open and filled arrowheads at bottom indicate pretreatment and posttreatment with celecoxib, respectively. Similar results were obtained in 2 additional experiments. Data are expressed as means ± SDs of triplicates.
Suppressive effects of COX-2 inhibitor, celecoxib, on RANKL-induced osteoclastogenesis in BMMs. (A) Demonstration of TRAP+ cells differentiating from BMMs unstimulated (i) or stimulated (ii) with RANKL. On day 5 with RANKL stimulation, the cells were fixed and stained for TRAP. TRAP+ cells appear as red cells. (iii-viii) Dose-dependent effects of celecoxib on the formation of TRAP+ multinucleated osteoclasts (MNCs) with RANKL stimulation. BMMs were pretreated (iii, 100 ng/mL; iv, 50 ng/mL; v, 25 ng/mL) with different concentrations of celecoxib or treated (vi, 100 ng/mL; vii, 50 ng/mL; viii, 25 ng/mL) 2 days after RANKL stimulation. On day 5 with RANKL stimulation, the cells were fixed and stained for TRAP (original magnification, × 100). (B) Total numbers of TRAP+ MNCs are shown. Note that formation of TRAP+ MNCs was inhibited by pretreatment of celecoxib in a dose-dependent manner. The open and filled arrowheads at bottom indicate pretreatment and posttreatment with celecoxib, respectively. Similar results were obtained in 2 additional experiments. Data are expressed as means ± SDs of triplicates.
Discussion
RANKL signaling is known to be essential for osteoclast differentiation, at the stage after the commitment of the bone marrow progenitor cells into the myelomonocytic lineage.7 Although our understanding of signaling pathways associated with osteoclast differentiation advanced considerably recently, the regulatory factors of RANKL-mediated osteoclastogenesis, involved in transmission and modulation of RANKL signaling, are still unknown. By using PCR-based SSH, we have identified a COX-2 gene whose expression is highly enriched during RANKL stimulation and whose induction is impaired in RAW264.7 cells expressing a dominant-negative form of Rac1.
Our study clearly demonstrates a pathway uniquely linked to RANKL signaling, culminating in the expression of COX-2 through RANK-Rac1-NF-κB–dependent pathway. Osteoclast formation induced by RANKL was accompanied by PGE2 production, which was blocked by a selective COX-2 inhibitor celecoxib. We also showed that suppression of osteoclastogenesis by celecoxib could be reversed by PGE2. Moreover, simultaneous treatment with PGE2 in the presence of RANKL cooperatively induced osteoclast differentiation. Taken together, these data show RANKL-induced COX-2 is responsible for PGE2 production in osteoclast precursors for regulating osteoclast differentiation.
PGE2 plays a crucial role in regulation of bone resorption.33 Agonists reported to stimulate PGE2-dependent osteoclast formation include IL-1,34 TNF-α,35 and parathyroid hormone.36 Most of these agonists induce PGE2 production in osteoblasts and stromal cells, and osteoclast formation induced by these agonists is dependent on PGE2 synthesis in mouse bone marrow cultures.37,38 It has also been reported that COX-2 and PGE2 play important roles in bone formation of rat long bones as shown in an experiment using systemic injection of PGE239 and are required for mesenchymal cell differentiation into osteoblasts.40 Because of the complexity of responses to PGE2 in bone cells including osteoclasts and osteoblasts, it is difficult to define precisely the net effect of PGE2 on bone resorption and formation. However, our data suggest that RANKL-induced PGE2 synthesis in osteoclast precursors is cell autonomous and that PGE2 acts directly on the precursors for osteoclast formation in a paracrine or an autocrine manner. This is consistent with the previous finding of a synergistic effect of PGE2 on the ability of RANKL to induce osteoclast differentiation.41 In addition, Okada et al42 showed a reduction in RANKL-stimulated osteoclast formation in spleen cell cultures in the absence of osteoblasts from COX-2–/– mice, suggesting that PGE2 originates from multiple sources during osteoclastogenesis in vivo.
PGE2 exerts its effects through interaction with specific cell surface receptors.43 The PGE receptors, divided into 4 subtypes (EP1, EP2, EP3, and EP4), are important in regulating PGE2 action in the target cells. Among them, EP4 and EP2 are coupled to activation of adenyl cyclase and are implicated in PGE2-induced osteoclastogenesis.44-46 It is tempting to speculate that the PGE2-EP signaling works in osteoclast precursors to induce a maximal osteoclast formation. Evidence for expression of both EP2 and EP4 receptors in the osteoclast precursors including RAW264.7 cells and BMMs47 supports this possibility. It remains to be clarified under what physiologic or pathologic contexts the PGE2-EP signaling is mobilized for bone resorption.
Exogenous PGE2 reversed the inhibitory effects of celecoxib. BMMs were pretreated with celecoxib (50 ng/mL) and PGE2 either alone or in combination as indicated and then stimulated with RANKL. On day 5, the cells were fixed and stained for TRAP as described in “Materials and methods.” Similar results were obtained in 2 additional experiments.
Exogenous PGE2 reversed the inhibitory effects of celecoxib. BMMs were pretreated with celecoxib (50 ng/mL) and PGE2 either alone or in combination as indicated and then stimulated with RANKL. On day 5, the cells were fixed and stained for TRAP as described in “Materials and methods.” Similar results were obtained in 2 additional experiments.
Whatever the detailed mechanism underlying this process, our results indicate that RANKL induces COX-2 expression in osteoclast precursors through Rac1-dependent NF-κB activation pathway, thereby resulting in PGE2 production. Although PGE2 itself is not sufficient for regulating osteoclast formation, PGE2 likely acts in an autocrine or a paracrine loop to enhance the early phase differentiation of osteoclasts. These findings suggest that signaling molecules involved in COX-2–PGE2 pathway in osteoclast precursors could potentially be novel targets for drugs to inhibit osteoclast function.
Prepublished online as Blood First Edition Paper, April 28, 2005; DOI 10.1182/blood-2004-12-4975.
Supported in part by the Molecular and Cellular BioDiscovery Research Grant (M1-0401-00-0045) from the Ministry of Science and Technology (S.Y.L), by the Korea Science and Engineering Foundation (KOSEF) through the Center for Cell Signaling Research at Ewha Womans University, and by grant no. R01-2005-000-10305 from the Basic Research Program of the KOSEF. S.Y.H., N.K.L. and K.H.K. were supported by BK21 fellowship from the Ministry of Education. S.Y.H. and N.K.L. have contributed equally to this work.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Jaesang Kim for critical reading of the manuscript and S. Prescott for providing the COX-2 promoter vector.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal