Abstract
Jak (Janus kinase) family nonreceptor tyrosine kinases are central mediators of cytokine signaling. The Jak kinases exhibit distinct cytokine receptor association profiles and so transduce different signals. Jak3 expression is limited to the immune system, where it plays a key role in signal transduction from cytokine receptors containing the common gamma-chain, γc. Patients unable to signal via γc present with severe combined immunodeficiency (SCID). The finding that Jak3 mutations result in SCID has made it a target for development of lymphocyte-specific immunosuppressants. Here, we present the crystal structure of the Jak3 kinase domain in complex with staurosporine analog AFN941. The kinase domain is in the active conformation, with both activation loop tyrosine residues phosphorylated. The phosphate group on pTyr981 in the activation loop is in part coordinated by an arginine residue in the regulatory C-helix, suggesting a direct mechanism by which the active position of the C-helix is induced by phosphorylation of the activation loop. Such a direct coupling has not been previously observed in tyrosine kinases and may be unique to Jak kinases. The crystal structure provides a detailed view of the Jak3 active site and will facilitate computational and structure-directed approaches to development of Jak3-specific inhibitors.
Introduction
The Janus kinase (Jak) family of cytoplasmic tyrosine kinases are essential for signal transduction from a wide variety of cell-surface receptors. There are 4 members of the family in vertebrates: Jak1, Jak2, Jak3, and tyrosine kinase 2 (Tyk2).1,2 Jak kinases share a characteristic domain architecture, which includes an amino-terminal FERM domain (Band 4.1, Ezrin, Radixin, Moesin homology domain), an src homology 2 (SH2)–like region, a pseudokinase domain, and a carboxy-terminal kinase domain. Parts of these structural domains have historically been termed Jak homology (JH) domains 1 through 7, based on primary sequence alignments. The FERM domain mediates association with the cytoplasmic region of cytokine receptors and may also participate in catalytic regulation. The function and activity of the SH2-like region is unclear. The pseudokinase (or JH2) domain is unique to Jak kinases. This domain is thought to have a protein kinase fold but to lack catalytic activity, as residues critical for phosphotransfer are absent. The pseudokinase domain has been shown to be intrinsic to the autoregulation of Jak kinases via a direct interaction with the kinase domain.3 The JH1 kinase domain lies at the C-terminus and is a functional tyrosine kinase. To date, no 3-dimensional structure has been reported for any portion of any of the Jak kinases.
A wide variety of cytokine receptor superfamily members signal via the Jak/Stat (signal transducer and activator of transcription) pathway, including granulocyte colony-stimulating factor (G-CSF), thrombopoietin, the interferons, erythropoietin, and the interleukins. The Jak/Stat pathway is consequently involved in regulation of diverse cell processes, including proliferation, differentiation, migration, and apoptosis.1,2
In a significant number of patients with severe combined immunodeficiency (SCID), the disease arises from mutations either in the cytokine receptor common gamma-chain, γc, or in the interleukin receptor IL-7R which uses γc, or in Jak3 (accounting for ∼ 50%, ∼ 10%, and ∼ 7%-14% of human SCIDs, respectively).4 The phenotype of patients with γc and Jak3 mutations is virtually identical; they present with no T or natural killer cells and a normal number of poorly functioning B cells (T-B+NK-SCID).5-8 Human SCID patients do not produce specific antibodies in response to in vivo antigenic challenge, and the disease usually presents in infants as an array of opportunistic infections and mortality in the first 2 years of life. Human SCID is currently treated by reconstitution of the immune defenses with hematopoietic stem cell transplantation. The γc/Jak3 SCID phenotype is limited and specific to the immune system, and patients with SCID are otherwise healthy and display almost no symptoms following stem cell transplantation.8 The Jak3 mutations that give rise to SCID have been reviewed recently in O'Shea et al.4
The profound immune-specific effects of disrupted Jak3 signaling highlight the possibility of therapeutic targeting of Jak3 as a highly specific mode of immune system suppression.9 Potentially, a Jak3-specific inhibitor would target the immune system by depleting natural killer and T cells through down-regulation of cell proliferation. Jak3-specific inhibitors are being studied as supplements to current organ transplant rejection therapies and to treat T-cell–specific autoimmune diseases, including psoriasis, multiple sclerosis, inflammatory bowel disease, and rheumatoid arthritis.10 Jak-specific inhibitors may also be useful for treatment of hematologic and other malignancies that involve pathologic Jak activation.11 Activated translocated ets leukemia (TEL) protein/Jak2 fusions are produced by chromosomal translocations in some human leukemias,12 and Jak activation has been shown to be transforming when activated by other tyrosine kinase oncogenes. For example v- and breakpoint cluster region-abelson (BCR-Abl) activate the Jak/Stat signaling pathway.13-15 Thus Jak-specific inhibitors may be useful to augment kinase-specific treatments such as imatinib mesylate (Novartis Pharma AG, Basel, Switzerland) for CML. A number of Jak-specific inhibitors are currently in clinical and preclinical trials,9 with CP-690550 (Pfizer, New York, NY) showing some selectivity for Jak3 (IC50 [inhibitory concentration 50%] Jak1, 112 nM; Jak2, 20 nM; Jak3, 1 nM) and demonstrating efficacy against organ allograft rejection.16
We describe here the crystal structure of the Jak3 kinase domain in complex with a staurosporine-based inhibitor. The kinase is in an active, doubly phosphorylated state. The structure provides mechanistic insights into Jak activation and provides a detailed view of the kinase that will facilitate development of Jak-specific inhibitors.
Materials and methods
Synthesis of AFN941
2,3,10,11,12,13,15,16,17,18-Decahydro-10-methoxy-9-methyl-11-(methylamino)-(9S,10R,11R,13R)-9,13-epoxy-1H,9H-di-indolo[1,2,3-gh:3′,2′,1′-lm]pyrrolo[3,4-j][1,7]benzodiazonin-1-one (5, 1,2,3,4-tetrahydrostaurosporin). A solution of staurosporine was incubated (20 g, 42.9 mmol) in Tetrahydrofuran (THF; 1 L) containing Pd/C (30 g of 10%) for 120 hours at 65°C and 30 bar H2; the mixture was filtered, and the residue was washed with THF. The combined filtrate and washings were evaporated to dryness under reduced pressure to give the crude product which was chromatographed (silica gel, eluent 5% MeOH, 95% CH2Cl2) and recrystallized from MeOH to give AFN941 as a white powder: melting point 252°C to 255°C (tartrate 238-245°C).
Cloning, expression, and purification of Jak3 kinase
A cDNA encoding the kinase domain (residues 811-1124) of human Jak317-19 was obtained from a human spleen-liver-placenta library (Stratagene, La Jolla, CA) using primers 5′-3′ caagaccccacgatcttcgagg and 3′-5′ tgaaaaggacagggagtggtgtttgccc. The cDNA was subcloned into transfer vector pAcG2T (Pharmingen, San Diego, CA) using BamHI (5′) and EcoRI (3′) restriction enzyme sites. Recombinant baculovirus was generated using the Baculogold expression system (Pharmingen). The kinase domain was expressed as a glutathione S transferase (GST) fusion protein in High 5 cells (Invitrogen, Carlsbad, CA).
High 5 cells infected with the recombinant baculovirus were grown in shaker flasks and Express 5 serum-free medium (Invitrogen). Cells were infected at a density of 1.6 × 106 cells/mL with 20 mL virus and harvested 72 hours after infection. The cells were resuspended in 20 mM Tris (tris(hydroxymethyl)aminomethane)–HCl, pH 6.8, 150 mM NaCl, 5 mM DTT (dithiothreitol), 10% glycerol, supplemented with complete protease inhibitor mixture (Roche Diagnostics GmbH, Mannheim, Germany) and flash-frozen in liquid nitrogen and stored overnight at -80°C. On thawing the cells were disrupted by sonication. Cell debris was removed by centrifugation (45 000 g, 1 hour, 4°C) and the supernatant was incubated for 1 hour with glutathione Sepharose beads (Amersham Biosciences AB, Uppsala, Sweden). The beads were washed with Tris-buffered saline (TBS) supplemented with 5 mM DTT and 10% glycerol and then incubated overnight with 5 μL of 3 mg/mL human α-thrombin (Enzyme Research, South Bend, IN) at 4°C. Jak3 kinase domain was eluted; dialyzed against 20 mM Tris-HCl, pH 8, 50 mM NaCl, 5 mM DTT, 10% glycerol; and loaded onto a Mono Q column (Amersham Biosciences AB). Jak3 kinase domain eluted at approximately 150 mM NaCl. For crystallization, the protein was concentrated to 5 mg/mL. Aliquots were flash-frozen in liquid nitrogen and stored at -80°C. The kinase domain was shown to be catalytically active in a kinase assay using the T-cell receptor (TCR) ζ-chain as a substrate (T.J.B., unpublished observations, September 1, 2004).
Crystallization and diffraction data collection
Following incubation with inhibitor AFN941 for 1 hour on ice (5 μL protein at 5 mg/mL, 0.25 μL AFN941 at 50 mM, and 0.2 μL DTT at 1 M) and centrifugation at 16 000 g for 30 minutes, crystallization trays were set up. Crystallization trays were set up at 4°C using hanging-drop vapor diffusion methodology with reservoirs containing 50 mM Pipes (1,4-piperazinedi-ethanesulfonic acid) pH 6, 960 mM Na malonate pH 6, 1.6% glycerol, and 10 mM DTT. Crystallization drops contained 0.3 μL precipitant buffer and 0.5 μL protein solution. A crystal appeared after 2 weeks and grew to its final dimension (100 × 100 × 200 μm) in 4 weeks. The primitive orthorhombic cell had dimensions a = 46.3 Å, b = 54.2 Å, and c = 118.6 Å. For data collection the mother liquor in the drop was substituted with 2M Na malonate, pH 6. The crystal was flash frozen to 100K using an in-house cryostream. Data were collected at 100K in house on a Rigaku (The Woodlands, TX) RU300-RC rotating anode X-ray generator with Osmic Confocal Max-Flux (CMF12-38Cu6) optics and a Mar345dtb image plate area detector (Mar Research, Norderstedt, Germany). The data were processed in space group P212121 with DENZO and scaled using SCALE-PACK.20 Data are summarized in Table 1.
Data collection and refinement statistics
. | Value . |
|---|---|
| No. crystals | 1 |
| Space group | P212121 |
| Crystal size, μm | 100 × 100 × 200 |
| Cell, Å | |
| a | 46.3 |
| b | 54.2 |
| c | 118.6 |
| Wavelength, Å | 1.5418 |
| Resolution, Å (in highest resolution range) | 25.0-2.55 (2.64-2.55) |
| Reflections, no. total/no. unique | 136 320/9 949 |
| Rsym, % (in highest resolution range) | 11.3 (56.9) |
| Mn/Mn, I/σI (in highest resolution range) | 9.2 (1.9) |
| Completeness, % (in highest resolution range) | 97.9 (97.4) |
| Redundancy (in highest resolution range) | 3.1 (3.1) |
| Refinement | |
| Resolution range, Å (in highest resolution range) | 25.0-2.55 (2.64-2.55) |
| Free R reflections, % (in highest resolution range) | 4.9 (5.0) |
| Free R reflections, no. (in highest resolution range) | 481 (43) |
| rmsd bond length, Å* | 0.011 |
| rmsd bond angles, °* | 2.3 |
| Average B value | |
| Overall, Å2 | 31.1 |
| Protein atoms | 30.7 |
| N-lobe protein atoms | 33.4 |
| C-lobe protein atoms | 29.6 |
| AFN941 | 22.6 |
| DTT | 51.9 |
| Water | 36.6 |
| Rfactor, %† (in highest resolution range) | 20.4 (33.2) |
| Free Rfactor, %†‡ (in highest resolution range) | 25.4 (47.1) |
| No. nonhydrogen protein atoms | 2358 |
| No. water molecules | 166 |
| No. AFN941 atoms | 35 |
| No. DTT atoms | 16 |
| Ramachandran plot§ | |
| Most favorable region, % (no.) | 85.6 (214) |
| Disallowed regions, % (no.) | 0.8 (2) |
. | Value . |
|---|---|
| No. crystals | 1 |
| Space group | P212121 |
| Crystal size, μm | 100 × 100 × 200 |
| Cell, Å | |
| a | 46.3 |
| b | 54.2 |
| c | 118.6 |
| Wavelength, Å | 1.5418 |
| Resolution, Å (in highest resolution range) | 25.0-2.55 (2.64-2.55) |
| Reflections, no. total/no. unique | 136 320/9 949 |
| Rsym, % (in highest resolution range) | 11.3 (56.9) |
| Mn/Mn, I/σI (in highest resolution range) | 9.2 (1.9) |
| Completeness, % (in highest resolution range) | 97.9 (97.4) |
| Redundancy (in highest resolution range) | 3.1 (3.1) |
| Refinement | |
| Resolution range, Å (in highest resolution range) | 25.0-2.55 (2.64-2.55) |
| Free R reflections, % (in highest resolution range) | 4.9 (5.0) |
| Free R reflections, no. (in highest resolution range) | 481 (43) |
| rmsd bond length, Å* | 0.011 |
| rmsd bond angles, °* | 2.3 |
| Average B value | |
| Overall, Å2 | 31.1 |
| Protein atoms | 30.7 |
| N-lobe protein atoms | 33.4 |
| C-lobe protein atoms | 29.6 |
| AFN941 | 22.6 |
| DTT | 51.9 |
| Water | 36.6 |
| Rfactor, %† (in highest resolution range) | 20.4 (33.2) |
| Free Rfactor, %†‡ (in highest resolution range) | 25.4 (47.1) |
| No. nonhydrogen protein atoms | 2358 |
| No. water molecules | 166 |
| No. AFN941 atoms | 35 |
| No. DTT atoms | 16 |
| Ramachandran plot§ | |
| Most favorable region, % (no.) | 85.6 (214) |
| Disallowed regions, % (no.) | 0.8 (2) |
Cell: a = 46.3 Å, b = 54.2 Å, c = 118.6 Å, α = β = γ = 90°.
Rsym = ΣII - <I>/Σi <I>, where Ii is the intensity of the i th term and <I> is the mean observed intensity
RMS (root mean square) deviations from ideal values calculated with CNS21
Rfactor = Σ Fobs - Fcalc/Σ Fobs; free-Rfactor is the Rfactor for reflections excluded from the refinement
Values were calculated using Procheck22
Structure determination and refinement
Structure determination by molecular replacement was implemented with PHASER23,24 using the EGFR kinase domain (residues 672 to 960 of Protein Data Bank [PDB] submission code 1M17; http://www.rscb.org/pdb/) as the search model. The correct solution yielded a log(likelihood) gain of 150 in the 25 to 2.55 Å resolution range. The program Arp-Warp was then used to build from the molecular replacement model in an automated fashion.25 Next, repeated rounds of manual refitting and crystallographic refinement were then performed in O v9.0.726 and CNS v1.1.21 Staurosporine analog AFN941 was modeled into a region of closely fitting positive Fobs - Fcalc electron density in the catalytic cleft of the crystal structure. Stereochemical parameter files for the ligand were generated using PRODRG.27 The final crystallographic model was evaluated using Procheck,22 and waters were added using the program O. The final crystallographic model has an R-factor of 20.4% and an R-free of 25.4%.
Crystallographic data deposition
Refined crystallographic coordinates for the kinase domain of Jak3 in complex with staurosporine analog AFN941 are deposited in the Protein Data Bank and are assigned an accession code 1YVJ.
Results and discussion
Structure determination
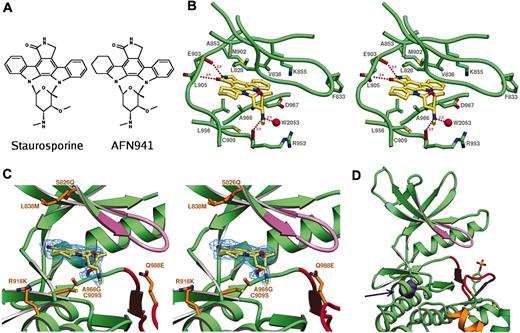
The crystal structure of the tyrosine kinase domain of human Jak3 has been determined at 2.55 Å resolution and refined to an R factor of 20.4% (Table 1; “Materials and methods”). The crystallographic model includes residues Pro814 to Arg1103 of Jak3, the tetrahydrostaurosporine inhibitor AFN941, 2 dithiothreitol molecules, and 166 water molecules (Figure 1). The first 5 and last 21 residues in the expressed protein are not visible, and the electron density is weak for 3 presumably flexible loops (those connecting β3-αC and β4-β5 in the N-lobe and αFG-αG in the C-lobe). Otherwise, clear electron density is observed throughout the structure, and the position and conformation of the inhibitor is well defined in the enzyme active site (Figure 2C).
Overall structure
The Jak3 kinase domain exhibits the classic bilobed architecture conserved among the catalytic domains of all protein kinases28 (Figure 1). The N-terminal lobe contains a 6-stranded antiparallel β-sheet and a single helix (αC) which is involved in catalytic regulation in many protein kinases. The glycine-rich loop, or P-loop, which participates in coordination of the phosphate groups of adenosine triphosphate (ATP), is also found in the N-lobe. The C-terminal lobe is predominantly helical, but contains 2 short strands of β-sheet within the activation loop. The 2 lobes are connected by a short linker termed the “hinge” region. The ATP-binding site is located between the 2 lobes and abutting the hinge. The tetrahydro-staurosporine analog AFN941 is located in this catalytic cleft, where it makes extensive interactions with residues in both the N- and C-lobes.
Jak3 is crystallized in a catalytically active state and is tyrosine-phosphorylated on both of the autophosphorylation sites in the activation loop. The overall conformation of the kinase, including the position of the C-helix, the interlobe orientation, and the conformation of the activation loop, closely resembles that found in other active tyrosine kinases (Figure 1A-B). Among tyrosine kinases for which structures are available the present structure superimposes most closely with that of the Abl kinase domain in complex with inhibitor Pd17395529 (PDB entry 1M52, Chain A). Corresponding residues from Jak3 and this Abl structure superimpose with an RMS deviation of 1.4 Å for 234 matching carbon-α atoms (the sequence identity is 37% for the 234 matched residues).
The architecture of the Jak3 kinase domain diverges significantly in 2 respects from other tyrosine kinases studied to date. Jak3 contains an additional helix formed by residues 1029 through 1038, which is inserted between helices αF and αG in the C-lobe. We label this helix αFG (Figure 1). Although it is not found in other tyrosine kinases, it is well conserved in the catalytic domain of vertebrate Jak family members (Figure 1C). The function of this additional helix is unclear, but given that it appears to be present in all vertebrate Jak kinases, it is tempting to speculate that it could participate in intramolecular regulatory interactions with the pseudokinase or FERM regions. Additionally, the conformation of the loop connecting the β2 and β3 sheets is dramatically divergent. The loop folds “forward” (to the right as viewed in Figure 1A), leaving a small, mostly hydrophobic cleft exposed on the back of the N-lobe. The conformation of the loop is stabilized by an extensive set of hydrogen bonds. Most prominently, the side chain of Asp842 bridges across the turn to make hydrogen bonds with the backbone amides of Asn847 and Gly849 (not pictured). The conformation of the loop requires glycine at position 849, and Gly849 as well as most of the other residues that define the conformation of the loop are conserved among all vertebrate Jak kinases. The cleft formed by the altered loop conformation is roughly defined by Tyr841, Pro843, Ala850, and Phe817. As with the novel αFG helix, this cleft may represent a site of interdomain contact, or of interaction with another regulator. Interestingly, residues involved in formation of this cleft and the FG helix are respectively not conserved or are deleted in insect Jaks; this may reflect differences in Jak regulation in invertebrates. Structural analysis of an intact Jak kinase will afford a better understanding of a possible role of these Jak-specific architectural features in intramolecular regulation.
SCID-related Jak3 mutations
The majority of Jak3 point mutations found in human SCIDs are either nonsense mutations or frameshift mutations that lead to truncation of the translated protein and hence loss of function. A single missense mutation has been documented in the fragment of Jak3 studied here; this mutation leads to substitution of Leu910 with serine. Examination of the structure reveals that Leu910 is located proximal to the catalytic site (Figure 2D), and is largely buried in a hydrophobic environment. Substitution of a hydrophobic leucine residue at this position with the polar serine residue would be expected to produce at least a local structural disruption and hence a loss of catalytic function.
Ribbon diagram of the Jak3 kinase domain and sequence alignment of Jak family members. (A) Overview of the crystal structure of the catalytic domain of Jak3 in complex with tetrahydro-staurosporine AFN941. β-Strands are shown as arrows and α-helices as coils. The glycine-rich loop is colored purple and the activation-loop is blue. The novel αFG helix is colored orange. The N-lobe, C-lobe, and C-helix are indicated. Phosphorylated tyrosines pTyr980 and pTyr981 are shown in stick form. The staurosporine analog is shown as space-filling spheres with carbon atoms colored yellow. (B) Ribbon diagram as in panel A but with the view rotated by 90°. (C) Sequence alignment for the kinase (JH1) domains of Jak3, Jak1, Jak2, and Tyk2; the JH2 domain of Jak3; and the kinase domains of c-Src and epidermal growth factor receptor (EGFR). The numbering and secondary structure assignments for Jak3 crystal structure are indicated. β-Strands are indicated as arrows, α-helices as cylinders, and loops as lines. The activation loop is indicated in red, and the novel helix αFG in orange. The N-lobe is denoted by green secondary structure assignments and the C-lobe with blue. Residues that interact with the staurosporine analog AFN941 are indicated by green boxes. The catalytically important residues Lys855, Glu871 are boxed in red as are the phosphorylated tyrosine residues pTyr980 and pTyr981. GenBank accession codes for Jak1, Jak2, Jak3, Tyk2, c-Src, and EGFR are NP_002218, NP_004963, NP_000206, AAS37680, AAH11566, and NP_005219, respectively.
Ribbon diagram of the Jak3 kinase domain and sequence alignment of Jak family members. (A) Overview of the crystal structure of the catalytic domain of Jak3 in complex with tetrahydro-staurosporine AFN941. β-Strands are shown as arrows and α-helices as coils. The glycine-rich loop is colored purple and the activation-loop is blue. The novel αFG helix is colored orange. The N-lobe, C-lobe, and C-helix are indicated. Phosphorylated tyrosines pTyr980 and pTyr981 are shown in stick form. The staurosporine analog is shown as space-filling spheres with carbon atoms colored yellow. (B) Ribbon diagram as in panel A but with the view rotated by 90°. (C) Sequence alignment for the kinase (JH1) domains of Jak3, Jak1, Jak2, and Tyk2; the JH2 domain of Jak3; and the kinase domains of c-Src and epidermal growth factor receptor (EGFR). The numbering and secondary structure assignments for Jak3 crystal structure are indicated. β-Strands are indicated as arrows, α-helices as cylinders, and loops as lines. The activation loop is indicated in red, and the novel helix αFG in orange. The N-lobe is denoted by green secondary structure assignments and the C-lobe with blue. Residues that interact with the staurosporine analog AFN941 are indicated by green boxes. The catalytically important residues Lys855, Glu871 are boxed in red as are the phosphorylated tyrosine residues pTyr980 and pTyr981. GenBank accession codes for Jak1, Jak2, Jak3, Tyk2, c-Src, and EGFR are NP_002218, NP_004963, NP_000206, AAS37680, AAH11566, and NP_005219, respectively.
The activation loop (and active site)
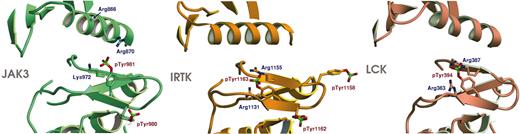
Activation of Jak kinases involves phosphorylation of 2 tyrosines within the activation loop, Tyrosines 980 and 981 in Jak3. Although the structures of many tyrosine kinases have been determined in “active” conformations, surprisingly few have been studied with the activation loop actually phosphorylated. To date, only lymphocyte-specific kinase (Lck),30 the insulin receptor tyrosine kinase (IRTK),31 and insulin-like growth factor receptor-1 (IGFR-1)32 (which is very closely related to the IRTK) have been studied in the phosphorylated state (vascular endothelial growth factor 2 [VEGF2] is phosphorylated on the activation loop but is not in an active conformation33 ). Analysis of the Jak3 activation loop reveals features in common with these kinases but also important differences which suggest a unique mechanism by which phosphorylation promotes coalescence of the active site in Jak kinases.
The Jak activation loop adopts an extended conformation with 2 β-strands. This general β hairpin-like conformation was first observed in the cyclic adenosine monophosphate (cAMP)–dependent kinase,34 and appears to be a general characteristic of active protein kinases. The activation loop of Jak3 is shown in a similar orientation with those of Lck and IRTK in Figure 3. The phosphate group of the primary activating tyrosine phosphorylation site, pTyr981, is coordinated directly by residue Lys972, and by Arg870 in the C-helix via a water-mediated hydrogen bond. Additionally, the side chain of Arg866 in the C-helix extends toward pTyr 981 but is not well ordered in the present structure. An interaction of pTyr981 with arginines 870 and 866 in the C-helix provides favorable electrostatic and hydrogen bond interactions that may directly “pull” the C-helix into the active conformation. The ability of the C-helix to hydrogen bond with pTyr981 is conserved in all Jak kinases, both vertebrate and invertebrate. Residue 870 is conserved as an arginine or lysine in all Jaks except human Tyk2, where it is a glutamine, which can also hydrogen bond with pTyr981. Thus, the structure suggests a direct mechanism by which activation loop phosphorylation may promote the active position of helix C. By comparison, the equivalent phosphotyrosines in Lck (pTyr394) and IRTK (pTyr1163) are similarly positioned but are coordinated by arginines that lie nearby on the large lobe of the kinase; there is no interaction with basic residues in the C-helix or elsewhere in the N-lobe. In Lck and other Src-family kinases, as well as in IRTK, phosphorylation of the activation loop is thought to indirectly promote the active conformation of the C-helix, by removing steric constraints imposed by an autoinhibitory conformation of the unphosphorylated activation loop. It is important to note that such a “release of steric constraints” mechanism may also apply in the case of Jak kinases, but no autoinhibited structure is yet available for comparison with the active form described here.
Binding of AFN941 to Jak3 active site. (A) Structure of AFN941 juxtaposed with that of staurosporine. (B) Stereoview of the catalytic cleft of Jak3 bound to AFN941. The protein backbone trace is shown in the region of the active site with residues Leu828, Phe833, Val836, Ala853, Lys855, Met902, Glu903, Cys909, Arg953, Leu956, Ala966, and Asp967 depicted in stick representation. Staurosporine analog AFN941 is shown in stick representation with carbon atoms colored yellow. Carbonyl oxygens of residues Arg953 and Glu903 are shown in red, and water 2053 is depicted as a red sphere. Residue numbers are indicated. Hydrogen bonds to backbone atoms and water are indicated and their distances are noted. (C) Stereoview of the catalytic cleft of Jak3 bound to the staurosporine analog AFN941. The experimental Fobs - Fcalc electron density omit map contoured at 2.5σ in blue is shown for the ligand AFN941. The glycine-rich loop is shown in purple and the activation loop in red. Residues proximal to the active site and divergent between Jak3 and Jak2 are shown in orange stick format and are labeled by Jak3 residue type, Jak3 residue number, and Jak2 residue type. These residues are Ser828Gln, Leu838Met, Cys909Ser, Arg916Lys, Ala966Gly, and Gln988Glu. (D) Location of point mutation Leu910Ser noted in human SCID patients.4 Leu910 is colored purple and indicated by the arrow.
Binding of AFN941 to Jak3 active site. (A) Structure of AFN941 juxtaposed with that of staurosporine. (B) Stereoview of the catalytic cleft of Jak3 bound to AFN941. The protein backbone trace is shown in the region of the active site with residues Leu828, Phe833, Val836, Ala853, Lys855, Met902, Glu903, Cys909, Arg953, Leu956, Ala966, and Asp967 depicted in stick representation. Staurosporine analog AFN941 is shown in stick representation with carbon atoms colored yellow. Carbonyl oxygens of residues Arg953 and Glu903 are shown in red, and water 2053 is depicted as a red sphere. Residue numbers are indicated. Hydrogen bonds to backbone atoms and water are indicated and their distances are noted. (C) Stereoview of the catalytic cleft of Jak3 bound to the staurosporine analog AFN941. The experimental Fobs - Fcalc electron density omit map contoured at 2.5σ in blue is shown for the ligand AFN941. The glycine-rich loop is shown in purple and the activation loop in red. Residues proximal to the active site and divergent between Jak3 and Jak2 are shown in orange stick format and are labeled by Jak3 residue type, Jak3 residue number, and Jak2 residue type. These residues are Ser828Gln, Leu838Met, Cys909Ser, Arg916Lys, Ala966Gly, and Gln988Glu. (D) Location of point mutation Leu910Ser noted in human SCID patients.4 Leu910 is colored purple and indicated by the arrow.
The second phosphorylated tyrosine in the Jak3 activation loop, pTyr980, is in an equivalent position to pTyr1162 in IRTK. This phosphotyrosine points outward, away from the protein core toward the solvent, and it appears to play a lesser role in stabilizing the active conformation of the activation loop. Its phosphate group contacts Lys978 and 3 ordered water molecules. Interestingly, in a study of Jak3 regulation phosphorylation of Tyr980 was observed to play a positive role in activation and phosphorylation of Tyr981 a negative regulatory role.36 The structural basis for these findings is unclear. It is possible that phosphorylation at the 2 sites differently affects binding of peptide substrates, or that the substitution of these tyrosines with phenylalanine,36 does not accurately mimic the nonphosphorylated tyrosine. Such a confounding effect would not be unexpected if Tyr981 plays a key role in maintaining an autoinhibited conformation in Jak3, as the corresponding Tyr1163 does in IRTK.37 Structural analysis of Jak3 in complex with substrate peptides and in the inactive state may further address these issues.
Comparison of activation loop conformation for Jak3, Lck, and IRTK. The 3 panels show ribbon representation diagrams for the activation loops of Jak3, IRTK,31 and Lck.30 The Jak3 activation loop is phosphorylated on 2 adjacent tyrosines, Tyr980 and Tyr981. pTyr981 is in a similar location to phosphorylated tyrosines in the previously determined activated crystal structures of Lck30 (PDB entry 3LCK) and insulin receptor tyrosine kinase31 (PDB entry 1IR3). Lck is phosphorylated on a single tyrosine, in the equivalent position to Tyr981; however, insulin receptor is phosphorylated on 3 tyrosines on the activation loop, 2 of which are the equivalent residues to Tyr980 and Tyr981 in Jak3. pTyr980 in Jak3 is in a similar conformation to that of pTyr1162 in IRTK. In Jak3, Arg870 of the C-helix coordinated pTyr981. Arg866 also extends toward pTyr981 but is poorly ordered; therefore, its side chain is not illustrated. Figures were prepared using the program SETOR.35
Comparison of activation loop conformation for Jak3, Lck, and IRTK. The 3 panels show ribbon representation diagrams for the activation loops of Jak3, IRTK,31 and Lck.30 The Jak3 activation loop is phosphorylated on 2 adjacent tyrosines, Tyr980 and Tyr981. pTyr981 is in a similar location to phosphorylated tyrosines in the previously determined activated crystal structures of Lck30 (PDB entry 3LCK) and insulin receptor tyrosine kinase31 (PDB entry 1IR3). Lck is phosphorylated on a single tyrosine, in the equivalent position to Tyr981; however, insulin receptor is phosphorylated on 3 tyrosines on the activation loop, 2 of which are the equivalent residues to Tyr980 and Tyr981 in Jak3. pTyr980 in Jak3 is in a similar conformation to that of pTyr1162 in IRTK. In Jak3, Arg870 of the C-helix coordinated pTyr981. Arg866 also extends toward pTyr981 but is poorly ordered; therefore, its side chain is not illustrated. Figures were prepared using the program SETOR.35
AFN941 binding mode and the active site cleft
Ligand AFN941 binds Jak3 in the catalytic cleft between the N- and C-lobes of the kinase. The inhibitor sits in a pocket bounded by the glycine-rich P-loop, the hinge segment, and the extreme N-terminus of the activation-loop (Figure 2B). The N-terminal portion of the activation loop, which bears the highly conserved sequence Asp-Phe-Gly (DFG), adopts the “inward” conformation characteristic of active kinases. The electron density for the ligand is well defined (Figure 2C), and it refines with an overall B-factor of 22.6 Å2 , comparable to that of the structure as a whole. The ligand binds in approximately the position occupied by the adenine ring of ATP in previously solved kinase crystal structures, and in a fashion analogous to that seen in previously reported kinase-staurosporine complexes. AFN941 is derived from staurosporine and differs only in that one indole ring system has been hydrogenated to break its planarity, thereby increasing the solubility of the compound (Figure 2A).
The deepest part of the binding pocket is defined by residue Met902, the so-called gate-keeper residue, with 2 atoms of the indole ring of AFN941 abutting this residue (Figure 2B). The ligand binding pocket is bounded in the back by hinge region residues Met902 through Cys909. Two hydrogen bonding interactions take place between the lactam ring of the staurosporine analog and backbone atoms of the hinge. The carbonyl oxygen of residue Glu903 interacts with the lactam ring amide with a distance of 2.9 Å, and the backbone nitrogen of Leu905 bonds to the keto-oxygen of the lactam ring with distance of 2.9 Å. The tetrahydro-indole ring adopts a chair conformation and sits in the region of the catalytic cleft termed the “lipophilic plug” (Figure 2A). The glycosidic group of AFN941 is in a boat conformation and is perpendicular to the plane of the indolocarbazole ring system. A hydrogen bond is formed between the carbonyl oxygen of residue Arg953 and the methylamino nitrogen of the glycosidic ring of AFN941 with a distance of 2.9 Å. The methylamino nitrogen also makes a hydrogen bond to a water molecule. The ligand is also in van der Waals contact with a number of side chains, including between Leu828, Val836, and Ala853 in the roof of the binding cleft and between Leu956 and Gly908 in the floor of the cleft.
Comparison of the Jak3/AFN941 complex with available tyrosine kinase/staurosporine complexes reveals that the closest similarity is with Lck bound to staurosporine38 (PDB entry 1QPJ, with 229 carbon-α atoms superimposing with an RMS distance of 1.3 Å). The mode of binding of the AFN941 in Jak3 is also extremely similar to that seen for staurosporine in ζ-associated protein 70 (Zap-70),39 but the conformation of the kinase immediately surrounding the inhibitor differs somewhat. The conformation differs locally around residue Cys909, which is a proline in Zap-70 (Pro421). Also the P-loop differs, especially at residue Phe833. In contrast to the corresponding residue in Zap-70 (Phe349), in the Jak3 complex Phe833 is oriented away from the inhibitor, toward the C-helix. This conformation is similar to that seen for the corresponding Phe256 in the Lck complex.38
In addition to the clear density for AFN941, additional electron density is visible for 2 similarly shaped nonwater ligands. One is in a solvent channel proximal to residues Leu973, Ala877, Leu878, and Arg943; the second is seen in the triphosphate region of the active site cleft, abutting the staurosporine analog. The electron density for both of these ligands is well-fit by DTT, which was a component of the crystallization buffer. Neither appears to be covalently coordinated to the protein. The putative DTT molecule in the active site region is proximal to residues Gly831 through Gly834 of the P-loop, and to Asp967, Gln988, Lys855, and Leu970. The interactions made by this moiety in the active site cleft may be exploited in structure-directed design of Jak3 inhibitors.
Insights for inhibitor development
Inhibitor specificity is a particularly critical issue for development of therapeutically useful inhibitors of Jak3 for use in transplantation. For this indication, treatment is typically long term, and inhibition of the very closely related Jak2 is undesirable. Jak2 activity is required for erythropoietin, thrombopoietin, and G-CSF signaling, so cross-inhibition of Jak2 might be expected to result in therapeutic side effects, including anemia, thrombocytopenia, and generalized leukopenia. Jak3 shares sequence identity of 62% with Jak2, and almost all residues in the inhibitor binding cleft are conserved (Figure 1C). Sequence differences between Jak3 and Jak2 that are proximal to the catalytic cleft are illustrated in Figure 2C. Two notable differences in residues abutting the staurosporine analog are at residues Cys909 and Ala966, which in Jak2 are replaced by serine and glycine, respectively. Thiol-based interactions with the cysteine in Jak3 might be exploited to achieve specificity for Jak3 over Jak2 and many other tyrosine kinases that do not have a cysteine at this position. Similarly, steric clash with the methyl side chain of Ala966 in Jak3 might be used as a design strategy in development of compounds with enhanced selectivity for Jak2 over Jak3. A Jak2-specific inhibitor could be useful for treatment of leukemias that result from translocations that fuse the dimerization domain of the Tel transcription factor with the Jak2 kinase.11,12
Although staurosporine and its analogues are not therapeutically useful because of their pan-specificity and toxicity, the present structure provides the first view of the Jak3 kinase domain and will be useful for computational chemistry approaches to inhibitor discovery and optimization. Furthermore, we expect that the Jak3 expression, purification, and crystallization procedures described here will be useful for cocrystallization of Jak3 with specific inhibitors as they are discovered and developed.
Prepublished online as Blood First Edition Paper, April 14, 2005; DOI 10.1182/blood-2005-02-0707.
Supported by an American Society of Hematology Basic Research Scholar Award (T.J.B.), by the National Institutes of Health (NIH; grant CA080942) (M.J.E.), and by a Scholar Award from the Leukemia and Lymphoma Society (M.J.E.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Florence Poy and Yingwu Xu for help designing cloning and purification protocols.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal