Abstract
Both deregulated growth and blocks in differentiation cooperate in the multistage process of leukemogenesis. Thus, understanding functional interactions between genes that regulate normal blood cell development, including cell growth and differentiation, and how their altered expression contributes to leukemia, is important for rational drug design. Previously, we have shown that the zinc finger transcription factor Egr-1 plays a role in monocytic differentiation. Ectopic expression of Egr-1 in M1 myeloblastic leukemia cells was observed to activate the macrophage differentiation program in the absence of the differentiation inducer interleukin 6 (IL-6) and to promote terminal differentiation in its presence. In addition, we have shown that deregulated expression of the proto-oncogene c-myc blocks the myeloid terminal differentiation program. Here we show that restoring expression of Egr-1 in M1 cells that express deregulated c-Myc abrogates the c-Myc block in terminal differentiation, resulting in cells that undergo functional macrophage maturation. However, there is an absence of both growth arrest and cell adhesion. In addition, Egr-1 expression diminished M1myc leukemogenicity in vivo. These findings indicate that Egr-1 can act as a tumor suppressor gene and suggest that Egr-1 or Egr-1 targets may provide important tools for differentiation therapy in certain leukemic phenotypes.
Introduction
Deregulated growth and blocks in differentiation, which result in impaired blood cell homeostasis cooperate in the multistage process of leukemogenesis.1 Understanding functional interactions between genes that regulate normal blood cell development, including cell growth and differentiation, and how their altered expression may contribute to leukemia is important for rational drug design because these proteins represent excellent targets for chemotherapeutic drug intervention.
One of many genes that participate in regulating hematopoietic homeostasis is the proto-oncogene c-myc. c-Myc has been shown to play a pivotal role in the regulation of proliferation, differentiation, and apoptosis.2-6 As expected for a gene product that participates in these different processes, alterations in c-Myc expression are associated with hematologic malignancies.7,8 Therefore, it is important to understand how c-Myc carries out its multiple functions, both with regard to normal hematopoiesis, the establishment and maintenance of hematologic malignancies, and the responsiveness of these leukemias to different therapeutic regimens.
The murine M1 myeloid leukemic cell line proliferates autonomously and can be induced with the physiologic inducer interleukin 6 (IL-6) to undergo terminal differentiation and growth arrest. As is the case for many cell types, c-Myc is highly expressed in proliferating M1 myeloblasts and is suppressed on induction of terminal differentiation.3,9 Deregulated and continued expression of c-Myc in M1 cells blocked terminal myeloid differentiation induced by IL-6 at an intermediate stage in the progression from immature blasts to mature macrophages and prevented exit from the cell cycle.9,10 Similar results were also observed in normal bone marrow (BM).11
Because deregulated c-Myc both blocks differentiation and its associated growth arrest, it can be asked if positive regulators of differentiation can act as tumor suppressors when coexpressed with c-Myc. This type of study should yield new insights into the regulation of terminal differentiation and progression to the tumorigenic state, and provide additional tools to design drugs for differentiation therapy.
Early growth response gene (Egr-1), a member of the egr family of genes that encode for zinc finger transcription factors, plays a role in the development, growth control, and survival of several cell types, including T and B cells, neuronal cells, and myeloid cells.12-17 Previously, using both differentiation-inducible myeloid cell lines and normal BM cells, we have shown that Egr-1 plays a role in monocytic differentiation of myeloid cells.14-17 It has been demonstrated that Egr-1 stimulates development of hematopoietic progenitor cells along the macrophage lineage at the expense of the granulocyte and erythroid lineages.17 Egr-1 was found to be a macrophage differentiation primary response gene. Its ectopic expression in M1 cells activated the macrophage differentiation program in the absence of differentiation inducer; this included the appearance of morphologically differentiated cells, decreased growth rate, and down-regulation of c-Myc mRNA. In addition, ectopic expression of Egr-1 in M1 cells dramatically increased the sensitivity of the cells for IL-6–induced differentiation, allowed a higher proportion of M1 cells to become terminally differentiated, and decreased M1 leukemogenicity in vivo.16
These observations have raised the interesting question of whether the positive regulator of differentiation, Egr-1, can act as a tumor suppressor when coexpressed with c-Myc. In this work we have shown that ectopic expression of Egr-1 abrogated the c-Myc block in terminal differentiation. Interestingly, it was shown that the ability of Egr-1 to override the c-Myc block in terminal differentiation occurred in the absence of growth arrest and cell adhesion, resulting in cells with the characteristics of functionally mature macrophages. As with normal macrophages, the cells also underwent programmed cell death. It was also observed that Egr-1 diminished the aggressiveness of M1myc leukemias and abrogated the leukemic potential of IL-6–treated M1myc cells. Taken together, these observations demonstrate that Egr-1 behaves like a tumor suppressor, suggesting that, on the one hand, genetic lesions that alter Egr-1 expression can cooperate with deregulated c-Myc in exacerbating the leukemic phenotype and, on the other hand, that enhancement of Egr-1 expression, or its downstream targets, can overcome blocks in differentiation due to deregulated c-Myc, as well as its associated leukemias.
Materials and methods
Cells, cell culture, cytokines, and chemicals
The M1 murine myeloblastic leukemia cell line, competent for induction of terminal differentiation on addition of physiologic factors such as IL-6 or leukemia inhibitory factor (LIF), has been described previously.9,16 M1 cells were cultured in Dulbecco modified Eagle medium (DMEM; Gibco, Gaithersburg, MD) supplemented with 10% heat-inactivated horse serum (Gibco) and 1% penicillin and streptomycin (Gibco). M1myc and the M1Egr-19,16 cell lines were similarly cultured, except that the culture medium was supplemented with 400 mg/mL G418 (Gibco) to maintain selection of the transgenes. Because the c-myc transgene carried a puromycin-resistance selection marker, the M1Egr-1/myc cells were grown in medium that was supplemented with 3.5 mg/mL puromycin, dissolved in 10% ethanol and 1 × phosphate-buffered saline (PBS; Sigma, St Louis, MO), in addition to G418. All cells were maintained in a humidified atmosphere with 10% CO2 at 37°C. Purified recombinant human IL-6 (rhuIL-6) was a generous gift from L. Souza (Amgen, Thousand Oaks, CA).
Establishment of genetically engineered cell lines
Freshly selected PA317 (American Type Culture Collection, Rockville MD) viral packaging cells were transiently transfected overnight with 10 μg DNA using the CaPO4 method.10,11 Supernatant containing infectious retroviral pseudotypes was harvested 48 hours after transfection and filtered through a 0.45-μm membrane. Infection of exponentially growing M1, M1myc, and M1Egr-1 cells was performed in 10% CO2 atmosphere with 5 mL virus-enriched culture supernatant and 10 μg/mL Polybrene (Sigma) per 100-mm dish. After 18 hours, the medium was changed to remove the Polybrene, and the cells were allowed to recuperate for 24 hours. For selection, the infected cells were distributed into 24-well plates at 100 cells/well, 500 cells/well, and 1000 cells/well, in DMEM supplemented with the appropriate antibiotic (400 μg/mL G418 or 3.5 μg/mL puromycin or both). Individual clones were isolated and established as cell lines. For each cell type, at least 3 clones were used for analysis and found to behave in a similar manner. Characteristics of control cell lines (M1-MSCV-puro, M1-MSCV-neo, M1-MSCV-puro/neo, and M1Egr-1-MSCV-puro) were indistinguishable from that of their respective parental cells.
Assay for leukemia
Four- to 6-week old CD-1 nu/nu mice obtained from Charles River Laboratories (Wilmington, MA) were given intravenous injections (tail vein) of 105 cells prepared in 200 μL 1 × PBS. Control animals were injected with the same volume of 1 × PBS. Cells were treated for 5 days with or without IL-6 (100 ng/mL) prior to inoculation into mice. The number of mice that were leukemia-bearing or leukemia-free was evaluated statistically using the standard Kaplan-Meier survival analysis method. Three weeks following inoculation, one mouse injected with each cell type was humanely killed and the BM analyzed for the presence of myeloid leukemic cells. This analysis included factor-independent growth and the ability to differentiate in response to IL-6.
General recombinant DNA techniques and expression vectors
Plasmids and DNA probes were prepared as previously described.9,16 The retroviral plasmid expression vectors, MSCV-neomycin (neo) and MSCV-puromycin (puro), were a gift from Dr Robert G. Hawley18 ; University of Toronto, Toronto, ON, Canada). To generate the M1Egr-1/myc cells, MSCV-puro/myc plasmid was infected into the M1Egr-1 cells.16 As a control M1myc cells were infected with MSCV-puro/Egr-1 retrovirus.
Analysis of cell morphology and cell viability
Cells were collected at indicated time points and, following cytocentrifugation, were stained with May-Grünwald-Giemsa. Morphologic differentiation was determined by counting 250 to 300 cells and scoring the proportion of immature blast cells, cells at intermediate stages of differentiation, and mature macrophages.9,16,19 Cell viability was determined using the trypan blue dye exclusion method. Results of all experiments represent the mean of at least 3 independent determinations.
Phagocytosis analysis
Analysis of phagocytotic activity was performed with latex beads (obtained from Polysciences, Warrington, PA), according to manufacturer's protocol. Cells were seeded in 5-mL tissue culture plates at 0.1 × 106 cells/mL without or with IL-6 (50 ng/mL), without or with latex beads (150/cell). At indicated time points cells were harvested to pellet extra beads. Cells were resuspended, acridine orange was added (10 μg/mL), and cells were incubated 15 minutes at 37°C. Following incubation, cells were washed 5 times in 1 × PBS. Cells were mounted on slides with small cover slips sealed with nail polish. Cells were visualized and photomicrographs (original magnification × 400; 40×/0.7 NA objective) were taken with a Wild Leitz (Wetzlar, Germany) fluorescent microscope equipped with an MPS52 camera.
Analysis of apoptotic DNA fragmentation
Cells (5 × 106 cells/0.5 mL) were harvested, washed in 1 × PBS, and incubated overnight in TNE lysis buffer (10 mM Tris[tris(hydroxymethyl) aminomethane]–HCL, pH 8.0, 150 mM NaCl, and 10 mM EDTA [ethylenediaminetetraacetic acid], pH8.0, supplemented with 200 mg/mL Proteinase K and 0.5% Triton-X 100) at 55°C. Total high molecular DNA was isolated by extracting twice with phenol (equilibrated in Tris and H2O saturated), followed by 2 extractions with equal volumes of 1:1 phenol/1:24 chloroform/isoamyl alcohol, followed by 2 extractions with 1:24 chloroform/isoamyl alcohol. The DNA was then precipitated, resuspended in Tris-EDTA buffer, and subjected to electrophoretic analysis in a 2% agarose gel, run overnight at 20 V. The gel was then stained with ethidium bromide for analysis.20
Assay of caspase-8 activity
Caspase-8 (IETDase) activity was assayed using the ApoAlert Flice/Caspase-8 fluorescent assay kit (Clontech, Palo Alto, CA; catalogue no. K2028-1) according to the manufacturer's recommended protocol. Briefly, 106 cells/sample were lysed and the supernatant collected. The IETD–7-amino-4-trifluoromethyl coumarin (AFC) substrate (50 μM) was added together with reaction buffer and the samples were incubated at 37°C for 1 hour. Fluorescent emission of the AFC product was detected with a fluorometer equipped with a 400-nm excitation filter and a 505-nm emission filter. To confirm the specificity of the protease reaction, control samples were preincubated with IETD-fmk inhibitor prior to the addition of IETD-AFC substrate. The amount of substrate cleaved as a function of time was determined using a standard calibration curve relative to free AFC as per the manufacturer's instructions (Clontech).
RNA extraction, Northern blots, and probes
Total RNA was prepared from 5 to 10 × 106 cells using TRIzol reagent (Gibco) as described in the manufacturer's specifications. Total RNA (10 mg/lane) was electrophoresed on a 1% agarose gel containing formaldehyde (0.7%), and the loading of equal amounts was confirmed by comparing intensity of ethidium bromide staining of ribosomal RNA bands. Northern blot analysis and stripping blots of probe to rehybridize were done as described previously.11,21 Probes used include the lysozyme EcoR1 fragment (600 base pair [bp]) cut from the pBS vector9 and a ferritin fragment excised from pBS.9 Probes were labeled by random priming (Gibco, RadPrime DNA labeling kit, catalogue no. 18428-011) to a specific activity equal to or more than 109 cpm/mg.
Protein extraction and immunoblotting
Preparation of cell extracts, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, and probing with antibody were as previously described.21 Primary antibodies against c-Myc and Egr-1 were from Santa Cruz Biotechnology (Santa Cruz, CA). The antibody against poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) was from Boehringer Mannheim (Mannheim, Germany), and the antibody used to detect caspase 8 cleavage was from Stressgen (Victoria, BC, Canada).
Flow cytometry analysis
For cell cycle analysis 2 × 106 cells were harvested by centrifugation, washed 2 times in 1 × PBS, and fixed in 70% ice-cold ethanol. The cells were subsequently treated for 30 minutes with RNAse A (180 mg/mL) and stained with propidium iodide (34 mg/mL in 7.6 mM sodium citrate; Sigma) prior to analysis using the Coulter Epics Elite system (Miami, FL). An appropriate window was chosen in the fluorescence-activated cell sorter (FACS) such that both living and apoptotic cells were analyzed. Cell cycle analysis was performed at least 3 times yielding similar results.
Results
Establishment of M1Egr-1/myc cell lines
M1Egr-1 cells16 were infected with an MSCV-puro retroviral vector encoding for a c-myc transgene. Northern blot analysis was used to screen for positive clones expressing exogenous c-Myc and Egr-1 transcripts (data not shown). Selected M1Egr-1/myc clones, as well as parental M1Egr-1 and M1myc clones, were then analyzed for Myc and Egr-1 protein levels by Western blotting (Figure 1). M1Egr-1/myc6 and M1Egr-1/myc7 clones, which were observed to express Egr-1 and c-Myc proteins at levels comparable to parental M1 clones, were used for further studies. In all experiments, similar data were obtained for both M1Egr-1/myc clones; data are presented for M1Egr-1/myc6. M1Egr-1/myc clones established using M1myc as the parental cell line behaved similarly to clones established using M1Egr-1 as the parental cell line.
Establishment of M1Egr1-myc cell lines. M1Egr-1 cells were transduced with an MSCV-puro retroviral vector, encoding for a c-myc transgene. Following selection in puromycin (3.5 mg/mL) surviving clones were expanded, as described in “Materials and methods.” Indicated M1 clones were cultured without or with IL-6 (50 ng/mL) for 48 hours and were assessed for c-Myc (A) and Egr-1 (B) protein expression by Western blotting, using appropriate antibodies, as described in “Materials and methods.”
Establishment of M1Egr1-myc cell lines. M1Egr-1 cells were transduced with an MSCV-puro retroviral vector, encoding for a c-myc transgene. Following selection in puromycin (3.5 mg/mL) surviving clones were expanded, as described in “Materials and methods.” Indicated M1 clones were cultured without or with IL-6 (50 ng/mL) for 48 hours and were assessed for c-Myc (A) and Egr-1 (B) protein expression by Western blotting, using appropriate antibodies, as described in “Materials and methods.”
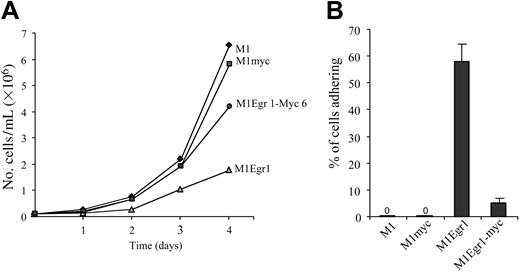
Growth kinetics and adherence of M1, M1myc, M1Egr-1, and M1Egr-1/myc cells. (A) Cells were seeded in DMEM at 0.05 × 106 cells/mL. On the second and third day of the experiment cells were diluted 1:4 to prevent overgrowing. At indicated time points cells were tested for viable cell numbers. Each time point represents the average of 3 experiments, with an SD up to 10% (ie, 40 ± 4). Using the Student t test to compare the different cell types 2 × 2, the difference between M1Egr-1/myc and M1Egr-1 at 4 days was P < .05. (B) Percent cells adhering to the surface of the tissue culture dish was determined 3 days following seeding 0.1 × 106 cells/mL, as previously described.8 Results are the average of 3 experiments done in duplicate, with an SD up to 15%. The difference between M1Egr-1/myc and M1Egr-1 was P < .01.
Growth kinetics and adherence of M1, M1myc, M1Egr-1, and M1Egr-1/myc cells. (A) Cells were seeded in DMEM at 0.05 × 106 cells/mL. On the second and third day of the experiment cells were diluted 1:4 to prevent overgrowing. At indicated time points cells were tested for viable cell numbers. Each time point represents the average of 3 experiments, with an SD up to 10% (ie, 40 ± 4). Using the Student t test to compare the different cell types 2 × 2, the difference between M1Egr-1/myc and M1Egr-1 at 4 days was P < .05. (B) Percent cells adhering to the surface of the tissue culture dish was determined 3 days following seeding 0.1 × 106 cells/mL, as previously described.8 Results are the average of 3 experiments done in duplicate, with an SD up to 15%. The difference between M1Egr-1/myc and M1Egr-1 was P < .01.
M1Egr-1/myc cells were compared to M1Egr-1 and M1myc cells, both without and following treatment with IL-6.
Expression of macrophage-associated differentiation markers is reduced in untreated M1Egr-1/myc cells compared to untreated M1Egr-1 cells
It has been shown that M1Egr-1 cells proliferate more slowly than M1 cells, and a high percentage of M1Egr-1 cells adhere to the culture plate and express markers associated with differentiated macrophages.16 In addition, over 70% of the cell population appears differentiated.16 Consistent with these observations, the level of c-Myc expression is reduced16 (Figure 1). By simultaneously expressing c-Myc and Egr-1 in M1 cells, it can be determined if restoring c-Myc expression will alter the differentiation-associated changes observed in untreated M1Egr-1 cells.
As seen in Figure 2A, although the growth rate of M1Egr-1 cells was retarded, the growth of M1Egr-1/myc cells was comparable to that of parental M1 and M1myc cells, indicating that restoring c-Myc expression was sufficient to override the Egr-1–mediated inhibition of cell proliferation. It was also readily apparent that there were very few M1Egr-1/myc cells adhering to the culture dish, in contrast to M1Egr-1 cells; fewer than 10% of the M1Egr-1/myc cells adhered, compared to 60% of the M1Egr-1 cells (Figure 2B). Therefore, c-Myc expression prevented the adherence of M1Egr-1 cells to the surface of the culture dish.
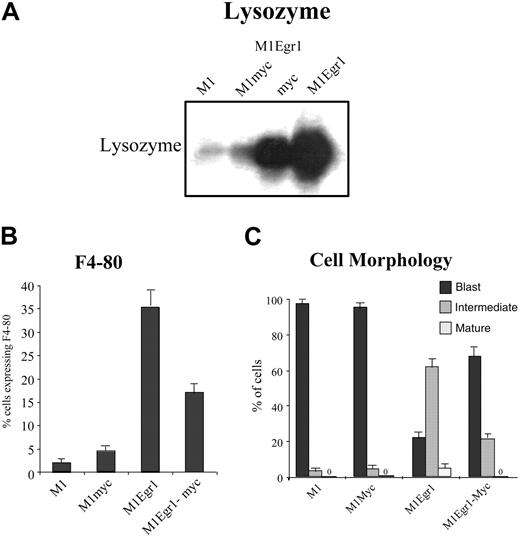
Interestingly, despite expression of the c-myc transgene, untreated M1Egr-1/myc cells, like M1Egr-1 cells, expressed elevated levels of macrophage differentiation markers, including lysozyme and F4-80, assessed by Northern blot and FACS analysis, respectively (Figure 3A-B). Furthermore, as seen by May-Grünwald-Giemsa staining, almost one third of the population of untreated M1Egr-1/myc cells is at an intermediate stage of differentiation. This is in stark contrast to the untreated M1 and M1Myc cell populations, which are composed predominantly (98%) of blast cells (Figure 3C). However, all these differentiation-associated characteristics were expressed at a lower level by M1Egr-1/myc cells than by M1Egr-1 cells. These data indicate that deregulated expression of c-Myc significantly, but not completely, reduced the ability of Egr-1 to promote spontaneous macrophage differentiation.
Analysis of differentiation markers in untreated M1, M1myc, M1Egr-1, and M1Egr-1/myc6 cells. (A) Lysozyme expression was determined by Northern blot analysis. Total RNA was extracted from 0.8 × 106 cells, loaded on formaldehyde-agarose gels (10 mg/lane), and transferred to Duralon nylon membranes for analysis, as described in “Materials and methods.” Blots were hybridized with 32P-labeled lysozyme cDNA probe (see “Materials and methods”). This is a representative experiment that was carried out 3 times. (B) F4-80 macrophage-specific antigen expression was assayed by FACS analysis. Cells (2.0 × 106) were incubated with a fluorescein isothiocyanate (FITC)–linked anti–F4-80 antibody, and fluorescence was measured using flow cytometry, as described in “Materials and methods.” Results are the average of 3 experiments, with an SD of up to 15%. The differences between M1 and M1myc were P > .05 (not significant), between M1 and M1Egr-1 P < .01 (significant), between M1 and M1Egr-1/myc P < .05 (significant), and between M1Egr-1 and M1Egr-1/myc P < .05 (significant). (C) Morphologic characteristics of cells were determined counting at least 300 cells from cytospin smears stained with May-Grünwald-Giemsa, scoring the proportion of immature blasts, cells at intermediate stages of differentiation, and mature macrophages (see “Materials and methods”). Results are the average of 3 experiments, with an SD for each cell clone and morphologic type of up to 10%. Using the intermediate stage of differentiation as a measure of differentiation of the untreated population of cells, the differences between M1 and M1myc were P > .1 (not significant), between M1 and M1Egr-1 P < .01 (significant), between M1 and M1Egr-1/myc P < .05 (significant), and between M1Egr-1 and M1Egr-1/myc P < .05 (significant).
Analysis of differentiation markers in untreated M1, M1myc, M1Egr-1, and M1Egr-1/myc6 cells. (A) Lysozyme expression was determined by Northern blot analysis. Total RNA was extracted from 0.8 × 106 cells, loaded on formaldehyde-agarose gels (10 mg/lane), and transferred to Duralon nylon membranes for analysis, as described in “Materials and methods.” Blots were hybridized with 32P-labeled lysozyme cDNA probe (see “Materials and methods”). This is a representative experiment that was carried out 3 times. (B) F4-80 macrophage-specific antigen expression was assayed by FACS analysis. Cells (2.0 × 106) were incubated with a fluorescein isothiocyanate (FITC)–linked anti–F4-80 antibody, and fluorescence was measured using flow cytometry, as described in “Materials and methods.” Results are the average of 3 experiments, with an SD of up to 15%. The differences between M1 and M1myc were P > .05 (not significant), between M1 and M1Egr-1 P < .01 (significant), between M1 and M1Egr-1/myc P < .05 (significant), and between M1Egr-1 and M1Egr-1/myc P < .05 (significant). (C) Morphologic characteristics of cells were determined counting at least 300 cells from cytospin smears stained with May-Grünwald-Giemsa, scoring the proportion of immature blasts, cells at intermediate stages of differentiation, and mature macrophages (see “Materials and methods”). Results are the average of 3 experiments, with an SD for each cell clone and morphologic type of up to 10%. Using the intermediate stage of differentiation as a measure of differentiation of the untreated population of cells, the differences between M1 and M1myc were P > .1 (not significant), between M1 and M1Egr-1 P < .01 (significant), between M1 and M1Egr-1/myc P < .05 (significant), and between M1Egr-1 and M1Egr-1/myc P < .05 (significant).
IL-6–treated M1Egr-1/myc cells do not undergo G0/G1 arrest and cell surface adherence
To further dissect the regulation of terminal differentiation, the effect of constitutive coexpression of the positive regulator Egr-1 and the negative regulator c-Myc on the terminal differentiation program of M1 myeloid leukemic cells following treatment with IL-6 was studied.
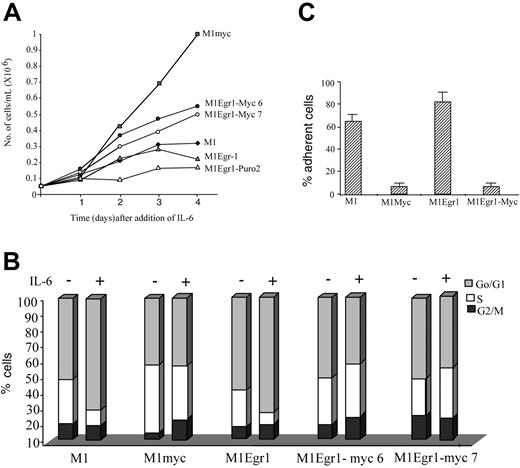
As shown in Figure 4A, in response to the optimal concentration of IL-6 (50 ng/mL) M1Egr-1/myc cells continued to proliferate, albeit at a slower rate than similarly treated M1myc cells, for 4 days following addition of IL-6. FACS analysis performed on these cells following 3 days of treatment with IL-6 showed that the distribution of cells in the different phases of the cell cycle resembled similarly treated M1myc cells, as well as untreated cells, where there was no increase in the percentage of cells in the G0/G1 phase of the cell cycle. In contrast, both M1 and M1Egr-1 cells treated with IL-6 accumulated in the G0/G1 phase, mostly at the expense of the S phase (Figure 4B). In addition, it was readily apparent that IL-6–treated M1Egr-1/myc cells, similar to M1myc cells, did not adhere to the culture dish (Figure 4C). The effect of the proto-oncogene c-myc appeared to be dominant to egr-1 with regard to cell cycle regulation and cell surface adhesion in IL-6–treated M1Egr-1/myc cells. It should be noted that also in untreated M1Egr-1/myc cells, the effect of Myc expression was to override the growth inhibitory and cell adhesion effects mediated by Egr-1.
IL-6–treated M1Egr-1/myc cells undergo terminal myeloid differentiation in the absence of both growth arrest and cell adhesion
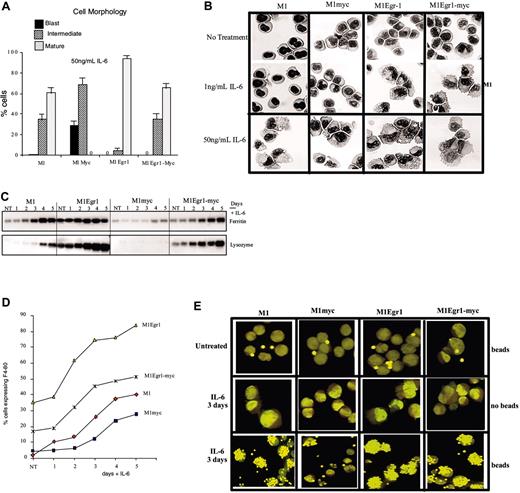
Despite the fact that constitutive Egr-1 expression had no effect on the ability of constitutive c-Myc to prevent G0/G1 arrest and cell adherence following treatment with the differentiation inducer IL-6, it was still necessary to ascertain the morphology of the cells and the expression of different markers associated with the differentiated phenotype. Cytospin smears stained with May-Grünwald-Giemsa were examined to assess the cell population for any morphologic differentiation. Interestingly, following 3 days of treatment with IL-6 (50 ng/mL), 67% of the M1Egr-1/myc cells appeared mature and 33% appeared to be at an intermediate stage of differentiation, similar to IL-6–treated M1 cells (Figure 5A-B). M1Egr-1 cells exhibited a higher percentage of mature cells, as previously reported.16 Ferritin and lysozyme expression, assessed by Northern blot analysis (Figure 5C), and F4-80 expression, assessed by flow cytometry (Figure 5D) were similar or somewhat higher for M1Egr-1/myc than M1 cells following IL-6 treatment, but always reduced relative to M1Egr-1 cells. Finally, to ascertain if the morphologically differentiated M1Egr-1/myc cells were functional, the ability of the cells to phagocytize latex beads was determined. Figure 5E clearly demonstrates that IL-6–treated M1Egr-1/myc cells phagocytize latex beads comparable to similarly treated M1 and M1Egr-1 cells. IL-6–treated M1myc cells, where no mature cells were detectable, did not phagocytize latex beads (Figure 5E). Thus, constitutive Egr-1 expression overrode the c-Myc–mediated block in myeloid differentiation resulting in terminal myeloid differentiation, yet the cells neither adhered to the culture dish nor underwent G0/G1 arrest.
Growth kinetics and cell cycle analysis of M1, M1myc, M1Egr-1, and M1Egr-1/myc (clones 6 and 7) cells treated with IL-6. (A) Cells were seeded at 0.05 × 106 cells/mL with IL-6 (50 ng/mL). At indicated time points viable cell numbers were determined using a hemocytometer (see “Materials and methods”). Results are the average of 3 independent experiments with an SD up to 15%. Applying the Student t test to the day 4 data, the differences between M1myc and either M1Egr-1/myc6 or M1Egr-1/myc7 were P < .05 (significant), between M1Egr-1 and either M1Egr-1/myc6 or M1Egr-1/myc7 P < .05 (significant), and between M1 and M1Egr-1 P > .05 (not significant). (B) Cells were seeded at 0.05 × 106 cells/mL with or without IL-6 (50 ng/mL). After 3 days cell cycle analysis was performed using FACS analysis, as described in “Materials and methods.” This is a representative experiment, which was repeated a total of 4 times. (C) Percent cells adhering to the surface of the tissue culture dish was determined 3 days following seeding 0.05 × 106 cells/mL in the presence of IL-6 (50 ng/mL). Results are the average of 3 experiments, with an SD up to 15%. The differences between M1myc and M1Egr-1/myc were P > .1 (not significant), between M1 and M1myc P < .05 (significant), between M1 and M1Egr-1/myc P < .05 (significant), and between M1 and M1Egr-1 P > .05 (not significant).
Growth kinetics and cell cycle analysis of M1, M1myc, M1Egr-1, and M1Egr-1/myc (clones 6 and 7) cells treated with IL-6. (A) Cells were seeded at 0.05 × 106 cells/mL with IL-6 (50 ng/mL). At indicated time points viable cell numbers were determined using a hemocytometer (see “Materials and methods”). Results are the average of 3 independent experiments with an SD up to 15%. Applying the Student t test to the day 4 data, the differences between M1myc and either M1Egr-1/myc6 or M1Egr-1/myc7 were P < .05 (significant), between M1Egr-1 and either M1Egr-1/myc6 or M1Egr-1/myc7 P < .05 (significant), and between M1 and M1Egr-1 P > .05 (not significant). (B) Cells were seeded at 0.05 × 106 cells/mL with or without IL-6 (50 ng/mL). After 3 days cell cycle analysis was performed using FACS analysis, as described in “Materials and methods.” This is a representative experiment, which was repeated a total of 4 times. (C) Percent cells adhering to the surface of the tissue culture dish was determined 3 days following seeding 0.05 × 106 cells/mL in the presence of IL-6 (50 ng/mL). Results are the average of 3 experiments, with an SD up to 15%. The differences between M1myc and M1Egr-1/myc were P > .1 (not significant), between M1 and M1myc P < .05 (significant), between M1 and M1Egr-1/myc P < .05 (significant), and between M1 and M1Egr-1 P > .05 (not significant).
Analysis of differentiation-associated characteristics of M1, M1myc, M1Egr-1, and M1Egr-1/myc cells. Cells were seeded at 0.05 × 106 cells/mL with or without IL-6 (50 ng/mL). (A) After 3 days cells were harvested and cell morphology was determined, as described in Figure 3, and “Materials and methods.” Results are the average of 3 independent experiments yielding similar results with an SD up to 10%. Using mature cells as a measure of cell differentiation, the differences between M1myc and each of the other 3 cell lines used were P < .01 (significant), between M1 and M1Egr-1/myc P > 0.1 (not significant), and between either M1 or M1Egr-1/myc and M1Egr-1 P < .05 (significant). (B) Photomicrographs of M1, M1myc, M1Egr1, and M1Egr1-myc cells from May-Grünwald-Giemsa–stained cytospin smears untreated and after 3 days of treatment with either 1 ng/mL or 50 ng/ml IL-6. Slides were analyzed and photographed (original magnification, × 400). Images were cpatured with a Zeiss Axioplan microscope using a 40×/0.75 NA Neofluar lens and a SenSys camera (Roper Scientific, Tucson, AZ) and SmartCapture 2 software (Digital Scientific, Cambridge, United Kingdom). (C) Expression of ferritin and lysozyme mRNA was determined by Northern blot analysis using of total RNA as described in “Materials and methods.” This is a representative experiment that was carried out 3 times. (D) F4-80 expression was determined using FITC-conjugated anti–F4-80 primary antibody and flow cytometry, as described in Figure 3 and “Materials and methods.” (E) Photomicrographs of phagocytosis analysis showing cells phagocytosing latex beads. Cells were seeded in the absence or presence of IL-6 and incubated with or without latex beads as described in “Materials and methods.” Images were captured using a Nikon dissectin microscope (SMZ-U, zoom 1:10) and an ED Plan APO lense (Nikon, Surrey, United Kingdom) and a Nikon CoolPix 990 camera and were processed with Adobe Photoshop (Adobe Systems, San Jose, CA). Photomicrographs are representative of at least 30 fields examined per cell line. Results are shown for M1Egr-1/myc6. Similar data were obtained with M1Egr-1/myc7.
Analysis of differentiation-associated characteristics of M1, M1myc, M1Egr-1, and M1Egr-1/myc cells. Cells were seeded at 0.05 × 106 cells/mL with or without IL-6 (50 ng/mL). (A) After 3 days cells were harvested and cell morphology was determined, as described in Figure 3, and “Materials and methods.” Results are the average of 3 independent experiments yielding similar results with an SD up to 10%. Using mature cells as a measure of cell differentiation, the differences between M1myc and each of the other 3 cell lines used were P < .01 (significant), between M1 and M1Egr-1/myc P > 0.1 (not significant), and between either M1 or M1Egr-1/myc and M1Egr-1 P < .05 (significant). (B) Photomicrographs of M1, M1myc, M1Egr1, and M1Egr1-myc cells from May-Grünwald-Giemsa–stained cytospin smears untreated and after 3 days of treatment with either 1 ng/mL or 50 ng/ml IL-6. Slides were analyzed and photographed (original magnification, × 400). Images were cpatured with a Zeiss Axioplan microscope using a 40×/0.75 NA Neofluar lens and a SenSys camera (Roper Scientific, Tucson, AZ) and SmartCapture 2 software (Digital Scientific, Cambridge, United Kingdom). (C) Expression of ferritin and lysozyme mRNA was determined by Northern blot analysis using of total RNA as described in “Materials and methods.” This is a representative experiment that was carried out 3 times. (D) F4-80 expression was determined using FITC-conjugated anti–F4-80 primary antibody and flow cytometry, as described in Figure 3 and “Materials and methods.” (E) Photomicrographs of phagocytosis analysis showing cells phagocytosing latex beads. Cells were seeded in the absence or presence of IL-6 and incubated with or without latex beads as described in “Materials and methods.” Images were captured using a Nikon dissectin microscope (SMZ-U, zoom 1:10) and an ED Plan APO lense (Nikon, Surrey, United Kingdom) and a Nikon CoolPix 990 camera and were processed with Adobe Photoshop (Adobe Systems, San Jose, CA). Photomicrographs are representative of at least 30 fields examined per cell line. Results are shown for M1Egr-1/myc6. Similar data were obtained with M1Egr-1/myc7.
It has been previously shown by us that ectopic expression of Egr-1 increases the sensitivity of M1 cells to be induced for macrophage differentiation by IL-6.16 About 5% to 10% of M1 and M1myc cells assumed intermediate-stage morphology, the remainder of the population appearing blastlike. In contrast, most of the M1Egr-1 cells underwent morphologic differentiation, 33% appeared to be at the intermediate stage and 64% appeared mature. M1Egr-1/myc cells also responded to the suboptimal concentration of IL-6 (1 ng/mL; Figure 5B); 50% of the cells exhibited an intermediate morphology and 38% appeared mature (data not shown). In conclusion, coexpression of Egr-1 and c-Myc in M1 cells maintained Egr-1–mediated sensitivity to IL-6.
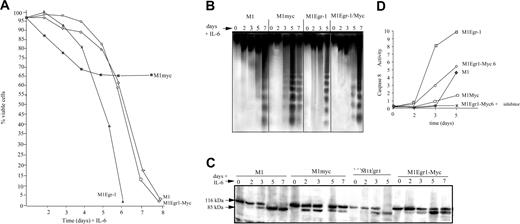
Subsequent to completion of the myeloid differentiation program, M1 and normal myeloid cells undergo programmed cell death. It was asked if IL-6–treated M1Egr-1/myc cells undergo apoptosis following macrophage differentiation. As a first step, viability of the IL-6–treated cells was ascertained (Figure 6A). The kinetics of loss of viability was very similar to IL-6–treated M1 cells and delayed relative to M1Egr-1 cells, which differentiated more rapidly. In each case the loss of viability followed terminal differentiation and eventually the entire population succumbed (Figure 6A). The DNA ladders were consistent with the loss of viability (Figure 6B). Deregulated expression of c-Myc in M1 cells blocked differentiation and its associated growth arrest and prematurely induced an incompletely penetrant apoptotic pathway.11 Both the viability curve and the DNA ladders (Figure 6A-B) illustrate these previously reported observations.11 M1Egr-1 cells differentiated so rapidly that the apoptotic response also occurred prior to M1 cells; however, it was complete and by 6 days all the cells had lost viability.
It has been shown that IL-6 induces Fas receptor in M1 and M1Myc cells, and data have been reported that are consistent with terminally differentiating M1 cells undergoing apoptosis via the Fas/CD95 pathway.20 Fas receptors were also induced by IL-6 in M1Egr-1 and M1Egr-1/myc cells (data not shown). The kinetics of both PARP cleavage and activation of caspase 8 were consistent with the apoptotic response of these variant M1 cell lines using the death receptor pathway (Figure 6C-D).
Analysis of differentiation associated apoptosis of M1, M1myc, M1Egr-1, and M1Egr-1/myc cells following treatment with IL-6. Cells were seeded at 0.05 × 106 cells/mL with or without IL-6 (50 ng/mL). At indicated time points cells were analyzed as indicated. Because M1myc cells treated with IL-6 continue to proliferate, it was necessary to split the cell culture on days 3, 5, and 7. (A) Cell viability was assessed by trypan blue dye exclusion using a hemocytometer, as described in “Materials and methods.” Results are the average of 3 independent experiments yielding similar results, with an SD up to 10%. (B) DNA fragmentation was determined as indicated in “Materials and methods.” DNA samples were resolved on a 2% agarose gel and assessed for fragmentation. Results shown are representative of 3 independent experiments. (C) Analysis of PARP cleavage was determined as indicated in “Materials and methods.” This is a representative experiment that was carried out 3 times. (D) Caspase 8 activity was determined using the ApoAlert FLICE assay (Clontech), as described in “Materials and methods.” Results are the average of 3 independent experiments, with SDs less than 10% for each sample.
Analysis of differentiation associated apoptosis of M1, M1myc, M1Egr-1, and M1Egr-1/myc cells following treatment with IL-6. Cells were seeded at 0.05 × 106 cells/mL with or without IL-6 (50 ng/mL). At indicated time points cells were analyzed as indicated. Because M1myc cells treated with IL-6 continue to proliferate, it was necessary to split the cell culture on days 3, 5, and 7. (A) Cell viability was assessed by trypan blue dye exclusion using a hemocytometer, as described in “Materials and methods.” Results are the average of 3 independent experiments yielding similar results, with an SD up to 10%. (B) DNA fragmentation was determined as indicated in “Materials and methods.” DNA samples were resolved on a 2% agarose gel and assessed for fragmentation. Results shown are representative of 3 independent experiments. (C) Analysis of PARP cleavage was determined as indicated in “Materials and methods.” This is a representative experiment that was carried out 3 times. (D) Caspase 8 activity was determined using the ApoAlert FLICE assay (Clontech), as described in “Materials and methods.” Results are the average of 3 independent experiments, with SDs less than 10% for each sample.
These data demonstrate that in M1Egr-1/myc cells, ectopic expression of Egr-1 abrogated the c-Myc–mediated block in IL-6–induced terminal myeloid differentiation. The resulting cells had characteristics of functionally mature macrophages that underwent programmed cell death following maturation, in the absence of both G0/G1 arrest and cell adhesion.
M1Egr-1/myc cells are less leukemogenic than M1myc cells
M1 cells are leukemogenic when injected into syngeneic or nude mice, and their ability to cause leukemia is lost after induction of differentiation in vitro or in vivo. It has been shown that constitutive expression of Egr-1 in M1 cells decreased the leukemogenicity of the cells in vivo.8 M1myc cells treated with differentiation factors fail to undergo differentiation and continue to proliferate, whereas M1Egr-1/myc cells treated with differentiation factors undergo terminal differentiation followed by apoptosis and are responsive to low doses of the differentiation factor IL-6. To correlate the effect of Egr-1 expression on the behavior of M1myc cells in vitro and in vivo, assays for leukemogenicity of the various M1 cell line derivatives were carried out.
Consistent with previous observations, M1Egr-1 cells were less aggressive than M1 cells in causing leukemia, whereas M1myc cells were more aggressive than M1 cells8,21 (Table 1). M1Egr-1/myc cells behaved similarly to M1 cells and were less aggressive than M1myc cells (Table 1, compare the effect of all untreated cells on nude mice). M1myc cells did not lose the ability to cause leukemia following treatment in vitro with IL-6, which was in accord with the observation that M1myc cells continued to proliferate in the presence of differentiation factors. M1Egr-1/myc cells behaved similarly to M1 cells with regard to loss of the ability to cause leukemia following treatment with differentiation factor IL-6 (Table 1, compare the effect of IL-6–treated cells on nude mice). Furthermore, examination of the BM from a single mouse from each group 3 weeks after injection revealed the presence of myeloid leukemic cells, as determined by growth and differentiation characteristics, in BM from mice that had received injections of each of the untreated M1 cell variants, as well as of IL-6–treated M1myc cells. Leukemic cells were undetectable in BM from mice that had been given IL-6–treated M1, M1Egr-1 or M1Egr-1/myc cells. In conclusion, Egr-1 diminished the aggressiveness of M1myc leukemias and abrogated the leukemic potential of IL-6–treated M1myc cells.
Effect of constitutive Egr-1 expression on M1myc leukemogenicity
. | Mice dead, % . | . | . | Myeloid leukemic cells recovered from BM† . | ||
|---|---|---|---|---|---|---|
| Cells* . | 4 wk . | 8 wk . | 12 wk . | . | ||
| M1 | ||||||
| No IL-6 | 63 | 100 | — | + | ||
| With IL-6 | 0 | 0 | 0 | - | ||
| M1Egr-1 | ||||||
| No IL-6 | 0 | 23 | 100 | + | ||
| With IL-6 | 0 | 0 | 0 | - | ||
| M1myc | ||||||
| No IL-6 | 100 | — | — | + | ||
| With IL-6 | 87 | 100 | — | + | ||
| M1Egr-1/myc | ||||||
| No IL-6 | 53 | 100 | — | + | ||
| With IL-6 | 0 | 0 | 0 | - | ||
. | Mice dead, % . | . | . | Myeloid leukemic cells recovered from BM† . | ||
|---|---|---|---|---|---|---|
| Cells* . | 4 wk . | 8 wk . | 12 wk . | . | ||
| M1 | ||||||
| No IL-6 | 63 | 100 | — | + | ||
| With IL-6 | 0 | 0 | 0 | - | ||
| M1Egr-1 | ||||||
| No IL-6 | 0 | 23 | 100 | + | ||
| With IL-6 | 0 | 0 | 0 | - | ||
| M1myc | ||||||
| No IL-6 | 100 | — | — | + | ||
| With IL-6 | 87 | 100 | — | + | ||
| M1Egr-1/myc | ||||||
| No IL-6 | 53 | 100 | — | + | ||
| With IL-6 | 0 | 0 | 0 | - | ||
Each cell type was intravenously injected into 13 nude mice. Cells were treated for 5 days with or without IL-6 (100 ng/mL) prior to inoculation into mice.
Three weeks following inoculation, one mouse injected with each cell type was humanely killed and the BM analyzed for the presence of myeloid leukemic cells.
Discussion
Ectopic expression of Egr-1 in IL-6–treated M1myc cells relieved the block in terminal differentiation imparted by ectopic c-Myc, resulting in cells that have the characteristics of functionally mature macrophages. However, the cells failed to adhere to the culture dish, did not become G0/G1 arrested, and continued to cycle prior to undergoing programmed cell death. The ability of Egr-1 to promote terminal myeloid differentiation appears to be dominant to the effect of c-Myc being a negative regulator of differentiation. Although c-Myc prevented growth arrest, the terminally differentiated M1Egr-1/myc cells succumbed to apoptosis. In addition, Egr-1 abrogated the leukemic potential of M1myc cells treated with IL-6 and diminished the aggressiveness of M1myc leukemias. This is consistent with Egr-1 behaving like a tumor suppressor, at least within the context of this experimental system. Interestingly, the human EGR1 gene was localized to chromosome 5, a region often deleted in patients suffering from therapy-induced acute myeloid leukemia.22,23
In the presence of differentiation factors, M1 cells constitutively expressing c-Myc do not differentiate, a subpopulation of cells continue to undergo apoptosis, and the population proliferates indefinitely9-11 (Figure 6). This response to differentiation factors accounts for the failure of M1myc cells to lose leukemogenicity following treatment with IL-6. The increased leukemic potential of untreated M1myc cells relative to M1 cells is probably due to the lower response of the M1myc cells to in vivo factors (Table 1). However, no detectable difference in the rate of proliferation is detected between untreated M1myc and M1 cells in vitro (Figure 2).
Ectopic expression of Egr-1 in M1 cells slows proliferation and causes cell adherence, and 70% of the cell population appears differentiated; these cells also express significantly reduced levels of c-Myc compared to parental M1 cells.8 Restoring c-Myc expression was shown to be sufficient to override the Egr-1–mediated inhibition of cell proliferation and prevented the adherence of M1Egr-1 cells to the surface of the culture dish. In addition, all differentiation-associated characteristics assessed were expressed at much lower levels by M1Egr-1/myc cells than by M1Egr-1 cells. These data indicate that restored expression of c-Myc significantly inhibited the ability of Egr-1 to promote spontaneous macrophage differentiation.
This work revealed that cycling cells can undergo terminal myeloid differentiation and, furthermore, that deregulated c-Myc does not interfere with differentiation by virtue of preventing growth arrest. M1 cells ectopically expressing the Bcr-Abl transgene also undergo macrophage differentiation without arresting proliferation; interestingly, these cells continued to express high levels of c-Myc mRNA.24 The Bcr-Abl transgene supports differentiation and proliferation because the M1Bcr-Abl cells differentiate in the absence of any exogenous differentiation stimuli, and the addition of IL-6 or LIF has no further effect on the cells. M1Egr-1/myc cells require exogenous stimuli to activate the differentiation program, and then the cells undergo differentiation in the absence of cell cycle arrest. However, in the case of M1Egr-1/myc cells the differentiated cells undergo apoptosis, the final step in terminal myeloid differentiation. It is interesting that Bcr-Abl promotes the myeloid differentiation program to the point that the cells become functional macrophages, yet they are blocked from undergoing apoptosis. It would be informative to assess the expression of Egr-1 in M1Bcr-Abl cells, as well as the effect of an egr1 transgene on the proliferating ability of these cells. In addition, gene array studies comparing IL-6–treated M1Egr-1/myc cells to untreated M1Bcr-Abl cells should give important information on the regulation of apoptosis following myeloid differentiation and the block in the apoptotic response following differentiation, which may be relevant to understanding the chronic phase of chronic myeloid leukemia.
Fibronectin (FN) is an Egr-1 target gene that promotes Egr-1–mediated cell adhesion.25,26 It plays an important role in organizing the extracellular matrix and facilitates cell adhesion, migration, wound healing, and tumor metastasis.25 The matrix is not formed spontaneously. Cells use their β-integrin to capture secreted FN and convert it into fibrils that are deposited into the matrix. Enhanced expression of either FN or β-integrin reduces both the motility and tumorigenicity of transformed cells.27,28 These facts raise the possibility that ectopic expression of Egr-1 in M1 cells causes adhesion in the absence of differentiation inducer and decreased leukemogenicity8 by modulating FN expression. Furthermore, it has been demonstrated that FN is a c-Myc target gene that is repressed on activation of a MycER transgene, resulting in decreased cell adhesion.29 On induction of differentiation of the myeloid cell lines HL60 and U937 by tissue plasminogen activator, c-Myc is rapidly down regulated and FN was observed to be one of the earliest up-regulated genes.29 These findings may provide an explanation as to why M1Egr-1/myc cells no longer adhere to the culture dish, both in the absence or presence of differentiation inducer. Current work is targeted at addressing this hypothesis, as well as using the more general approach of gene array analysis to elucidate the gene targets relevant to adhesion.
In this laboratory it was also shown that c-Fos expression partially alleviated the block in differentiation imparted by c-Myc in M1 cells. Fos/Jun have been implicated as positive regulators of both apoptosis and terminal differentiation in hematopoietic progenitor cells of the myeloid lineage.30 Activation of the foser transgene product partially abrogated the block imparted by deregulated c-Myc on myeloid differentiation and also increased the sensitivity of the cells to respond to differentiation factors.21 Similar to the effect of Egr-1, c-Fos expression diminished the effect of constitutive c-Myc on the aggressiveness of M1 leukemias. These findings predict that genetic lesions that abolish either c-Fos or Egr-1 expression would cooperate with deregulated c-Myc in leukemogenesis.
Although c-Myc expression prevents M1Egr-1/myc cells from undergoing growth arrest when treated with differentiating cytokines, Egr-1 promotes terminal differentiation followed by loss of viability. Thus, Egr-1 or Egr-1 target genes may be important tools for differentiation therapy in some leukemias. Analysis of gene expression for the different established M1 variant cell lines should give an understanding of how Egr-1 overrides the c-Myc–mediated block in myeloid differentiation, which c-Myc target genes participate in blocking differentiation, and which promote proliferation. It is also important to ascertain if Egr-1 can behave as a tumor suppressor when myeloid differentiation is blocked by other oncogenes, and studies are currently in progress to accomplish this. These lines of investigation should provide insights regarding how Egr-1 or Egr-1 targets may be used to treat and suppress hematologic malignancies.
Prepublished online as Blood First Edition Paper, April 19, 2005; DOI 10.1182/blood-2004-08-3056.
Supported by National Institutes of Health grants 1 RO1 CA81168 (B.H.) and 1 RO1 CA59774 (D.A.L.) and the shared Resources for Cancer Research CA88261-03.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal