Abstract
Although the zeta-associated protein of 70 kDa (ZAP-70) is overexpressed in patients with chronic lymphocytic leukemia (CLL) displaying unmutated IGVH genes and poor prognosis, a previous microarray study from our group identified overexpression of LPL and ADAM29 genes among unmutated and mutated CLL, respectively. To assess the prognostic value of these genes, we quantified their expression by real-time quantitative polymerase chain reaction (PCR) in a cohort of 127 patients with CLL and correlated this with clinical outcome, IGVH mutational status, and ZAP-70 protein expression. IGVH mutational status, ZAP-70, and the LPL and ADAM29 mRNA ratios (L/A ratio) were predictive of event-free survival for the whole cohort and for patients with stage A disease. In patients in stage B and C, the L/A ratio was an independent prognostic factor, whereas ZAP-70 did not predict survival. Simultaneous usage of the L/A ratio and ZAP-70 expression allowed an almost perfect (99%) assessment of the IGVH status in the 80% of patients with concordant results (L/A+, ZAP-70+ or L/A-, ZAP-70-). LPL and ADAM29 gene expression could also be determined by a simple competitive multiplex reverse transcription PCR assay. Overall, quantification of LPL and ADAM29 gene expression is a strong prognostic indicator in CLL, providing better prognostic assessment than ZAP-70 in advanced stages of the disease. (Blood. 2005;106:650-657)
Introduction
Chronic lymphocytic leukemia (CLL) displays a variable outcome. The classical Rai1 or Binet2 staging systems provided a basis for therapeutic stratification by allocating CLL cases into 3 major risk groups (low, intermediate, and high), according to tumor burden and the presence of anemia and thrombocytopenia. Asymptomatic patients with a low tumor burden (Binet stage A) do not benefit from treatment with chlorambucil.3 However, the disease in half of these patients will progress and both staging systems fail to initially identify such patients. The advent of new treatments such as purine analogues and monoclonal antibodies directed against CD20 and CD52 are able to induce complete remissions and may allow early treatment for asymptomatic patients whose disease is likely to progress.3 Accurate identification of these patients is therefore increasingly important.
Serologic markers such as lactic dehydrogenase, β2-microglobulin,4 soluble CD23,5 and thymidine kinase4,6 are essentially indicators of disease activity or tumor load or both, although some can anticipate disease progression.7 Phenotypic expression of CD38 has been associated with aggressive disease,8 but the threshold level for positive cases, if it exists at all, remains a matter of debate.9-11 Genomic aberrations correlate well with either good (isolated 13q-) or poor (17p-; 11q-) prognosis in CLL,11,12 though their occurrence as a second malignant hit cannot be definitely excluded. The mutational status of immunoglobulin heavy chain variable (IGVH) genes has been considered as the best prognostic marker in CLL.13 In an initial study, we observed that at least half of CLL cases carried mutations using a cut-off of 98% germline homology.14 This was further confirmed by others,15 some of whom also correlated the IGVH mutational status to clinical behavior.8,16,17 Mutated (MT) cases usually demonstrate a favorable evolution when compared to unmutated (UM) cases (> 98% germline homology), which are characterized by progressive disease, continuing treatment needs, and a high proportion of CLL-related deaths.
Unexpectedly, recent reports indicated that ZAP70, normally expressed in T and natural killer (NK) lymphocytes, is also transcribed in CLL B cells lacking IGVH mutations and is associated with poor prognosis.18,19 Further series have confirmed these findings at the protein level,20-23 suggesting a pivotal role for the zeta-associated protein of 70 kDa (ZAP-70) in prognosis prediction in CLL.
In a study of gene expression profiling performed on 18 CLL cases, we identified a limited set of genes (n = 85), which were expressed differentially between progressive UM and stable MT CLLs.24 We validated these results by real-time quantitative polymerase chain reaction (RQ-PCR) for 18 genes on the same cDNAs that were hybridized on the DNA chips. From these RQ-PCR experiments, 4 genes in addition to ZAP70 appeared to provide a better segregation of the 2 groups of CLL. These included the lipoprotein lipase (LPL) and spartin (SPG20) genes, whose expression was higher in UM cases, whereas disintegrin and metalloproteinase 29 (ADAM29) and nuclear receptor-interacting protein 1 (NRIP1) genes were found at higher levels in MT cases. These findings led us to investigate, in an independent and larger CLL series, whether these 4 genes (isolated or in combination) could provide significant prognostic information and compared their value with that of the IGVH mutational status and ZAP-70 protein expression.
Patients, materials, and methods
Patients and samples
This multicenter retrospective study was undertaken on samples from 127 patients whose diagnosis was made between October 1979 and February 2003, and identified from the registries of the Pasteur Institute and the Pitié-Salpêtrière hospitals, Paris, France (n = 92), and the University Hospital of São Paulo, Brazil (n = 35). Inclusion criteria consisted of: (1) diagnosis of typical CLL based on morphologic and phenotypic analyses25 ; (2) availability of frozen samples; (3) previous determination of IgVH mutational status; and (4) patient's informed consent according to French and Brazilian regulations. Approval was obtained from the Pitié-Salpêtrière Hospital, the Pasteur Institute, and the University Hospital of São Paulo for these studies. This series included 87 patients with stage A, 29 with stage B, and 11 with stage C disease. Median follow-up time was 73 months for the whole series (range, 1-291 months), 87 months for patients in stage A and 50 months for patients in stages B and C. Progression was defined as change of clinical stage or need for treatment or both.
All analyses were carried out on frozen peripheral blood samples. For 113 patients, samples had been taken at the time of diagnosis. In 14 stage A cases with a very stable lymphocytosis over time, they were obtained at distance from diagnosis. Mononuclear cells had been separated by Ficoll-Hypaque gradient centrifugation and stored in liquid nitrogen. On thawing, cell viability was first assessed by trypan blue exclusion and only samples displaying more than 80% viability were further analyzed. In 5 cases, leukemic B-cell populations were purified by negative magnetic selection using anti-CD3, anti-CD14, anti-CD16, and anti-CD56 monoclonal antibodies (Dynal, Oslo, Norway). Final purity was evaluated by flow cytometry to be over 98%. Control samples from 20 healthy adult volunteers were obtained using the same procedures and included peripheral mononuclear blood cells (PBMCs, n = 14) and purified peripheral blood B cells (n = 6). In addition the T-cell line Jurkat was cultured in RPMI 1640 medium containing 10% fetal calf serum, 2 mM glutamine, 1% sodium pyruvate, and penicillin-streptomycin.
IGVH mutational status
The IGVH gene sequences were determined as previously described.26 Briefly, amplification of immunoglobulin heavy chain variable regions by PCR was performed on DNA from leukemic cells with consensus primers for the VH framework region 1 and JH genes as previously described26 or following the BIOMED-2 protocols.27 Purified PCR products were sequenced either directly or after a cloning procedure using an automated DNA sequencer. Sequence data were analyzed using IgBLAST (http://www.ncbi.nlm.nih.gov/igblast), the VBASE (http://vbase.mrc-cpe.cam.ac.uk/), and the ImMunoGeneTics database (IMGT; http://imgt.cines.fr). IGVH sequences were considered as mutated if their homology with the closest germline counterpart was less than 98%.
RNA isolation and cDNA synthesis
Total cellular RNA was extracted using the RNeasy kit (Qiagen, Courtaboeuf, France) following the supplier's instructions. The integrity of RNA was assessed by visualization of the 18S and 28S RNA species on electrophoresis in agarose gel after ethidium bromide staining. First-strand cDNA was synthesized from 2 μg total RNA, using Superscript II reverse transcriptase (RT; Invitrogen, Cergy-Pontoise, France) and oligodT or random hexamer primers.
Quantitative RT-PCR
For gene expression analyses of LPL, SPG20, ADAM29, and NRIP1, we performed RQ-PCR using the Light Cycler System (Roche Molecular Biochemicals, Mannheim, Germany) and the SYBR Green I dye. Primers used in this study (Table 1) were designed with the GeneRunner software (Hastings Software, Hastings, NY). RQ-PCRs were carried out on 100 ng reverse transcribed total RNA (cDNA) with the following parameters: 10 minutes at 95°C for initial denaturation, then 40 cycles of 10 seconds at 95°C, 5 seconds at 62°C, and 17 seconds at 72°C. The specificity of the amplified products was verified by analysis of their respective melting curves as provided by the Light Cycler software. All reactions were done in duplicate and each PCR run also included the 5 points of the calibration curve and a no template control. Estimation of the quality of cDNA for each sample was done by quantification of an endogenous reference, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The gene copy number was calculated with a standard curve generated from serially diluted (10-fold dilutions from 106 to 102 copies) plasmids containing the respective sequence-verified insert. Results were expressed as the ratio of mean of gene copy number/mean GAPDH copy number × 100.
Sequences of primers used in RQ-PCR and multiplex PCR
Primer . | Sequence (5′ → 3′) . |
|---|---|
| GAPDH forward | GGTGCTGAGTATGTCGTGGA |
| GAPDH reverse | ATGCCAGTGAGCTTCCGTT |
| LPL forward | GGAATGTATGAGAGTTGGGTGC |
| LPL reverse | CAATGCTTCGACCAGGGGACC |
| ADAM29 forward | TCTTATGTGGGCTGGTGGATCC |
| ADAM29 reverse | GACCTAGATGATGAGCCACTGC |
| SPG20 forward | CTGAAATGTACTGCGGGAGCC |
| SPG20 reverse | CCAACTCACCCAGGAAGCACC |
| NRIP1 forward | GGATAGCACATTACTGGCCTCT |
| NRIP1 reverse | AGGTTTAGGTGAGGTGGCAGG |
Primer . | Sequence (5′ → 3′) . |
|---|---|
| GAPDH forward | GGTGCTGAGTATGTCGTGGA |
| GAPDH reverse | ATGCCAGTGAGCTTCCGTT |
| LPL forward | GGAATGTATGAGAGTTGGGTGC |
| LPL reverse | CAATGCTTCGACCAGGGGACC |
| ADAM29 forward | TCTTATGTGGGCTGGTGGATCC |
| ADAM29 reverse | GACCTAGATGATGAGCCACTGC |
| SPG20 forward | CTGAAATGTACTGCGGGAGCC |
| SPG20 reverse | CCAACTCACCCAGGAAGCACC |
| NRIP1 forward | GGATAGCACATTACTGGCCTCT |
| NRIP1 reverse | AGGTTTAGGTGAGGTGGCAGG |
Multiplex RT-PCR
We also evaluated the relative expression of LPL and ADAM29 by multiplex RT-PCR, using the same primers than those for RQ-PCR (Table 1). Different concentrations of primers, MgCl2, and deoxyribonucleoside triphosphate (dNTP) were first evaluated. Optimized PCR conditions were obtained with primers at the final concentrations of 0.5 μM for LPL and 0.25 μM for ADAM29, 1.5 mM MgCl2, and 200 μM dNTP. Amplifications were performed on 100 ng cDNA and included an initial denaturation step at 94°C for 5 minutes, followed by 29 cycles of 30 seconds at 95°C, 20 seconds at 62°C, and 30 seconds at 72°C. Finally, the reaction was completed with a final elongation step at 72°C for 5 minutes. PCR products were analyzed on 2% agarose gel electrophoresis stained with ethidium bromide, where they appeared as a 445-base pair (bp) band for ADAM29 and a 410-bp band for LPL. Amplification of GAPDH was performed in parallel to ensure cDNA integrity.
Multiparametric flow cytometry
Flow cytometric analysis of ZAP-70 intracellular expression was performed using the method described by Crespo et al20 with some minor modifications. Thawed mononuclear cells were fixed in 2% paraformaldehyde and were then permeabilized by incubation with phosphate-buffered saline containing 0.1% saponin (Sigma, Saint-Quentin Favallier, France) and 0.5% bovine serum albumin. One million cells were first incubated with 2.5 μg anti-ZAP-70 antibody (clone 2F3.2; Upstate Biotechnology, Lake Placid, NY) or irrelevant isotype-matched anti-CD14 monoclonal antibodies (Dako Cytomation, Trappes, France). After washing, they were incubated with 1.5 μg F(ab′)2 fluorescein isothiocyanate (FITC)-conjugated goat antimouse antibody (Immunotech, Marseille, France). Cells were then washed and incubated with phycoerythrin-cyanin 5 (PC5)-conjugated mouse anti-CD19 (Immunotech), allophycocyanin (APC)-conjugated mouse anti-CD3, and APC-conjugated mouse anti-CD56 (BD Biosciences, San Jose, CA). Samples including at least 104 cells were further analyzed with a flow cytometer (FACSCalibur; BD Biosciences) and the use of Cell Quest Pro software (BD Biosciences). Lymphocyte cells were first selected on size structure characteristics and then gated on B cells (CD19+) and T and NK cells (CD3+CD56+). Bi-parametric dot-plot graphs were obtained separately for cells that were stained for, respectively, CD3, CD56, and ZAP-70, or CD19 and ZAP-70, or CD3, CD56, and CD14 (negative control). In those plots as well as in mono-parametric histograms, ZAP-70 expression on CD3+CD56+ cells served to determine the percentage of CLL cells that were positive for ZAP-70.
Statistical analyses
Expression levels of the 4 tested genes and of ZAP-70 protein were compared with the IGVH mutational status as a reference. Threshold values that could best discriminate MT from UM cases were first determined by plotting expression values against IGVH percentage of germline homology, and then further refined by calculating the Youden index28 and validity index (the percentage of correctly classified cases). Thereafter, the performance indexes including sensitivity, specificity, and positive and negative predictive values were determined. Distributions of patients, according to Binet staging, sex, and IgVH mutational status were compared using χ2 test. Median follow-up periods were calculated for each Binet stage group. Because CLL-related deaths were mainly observed in stages B and C (only 4 cases in stage A), whereas disease progression was the most frequent event for patients in stage A, we evaluated event-free survival (EFS), from diagnosis to date of disease progression or CLL-related death or last follow-up visit, for the whole cohort and patients in stage A. Overall survival was calculated only for patients in stage B and C. Survival analyses were performed using the Kaplan-Meier method. Statistical significance of associations between individual variables and survival was calculated by the log-rank test. Univariate and multivariate regression analyses were done according to the Cox proportional hazards regression model. Because the biologic factors studied as prognostic indicators were strongly correlated with IGVH mutational status, it was inappropriate to test them simultaneously by Cox regression. Consequently, 2 Cox regression analyses were performed, with the first one testing IGVH mutational status, and the second one testing the other selected factors, in both circumstances with adjustment for age, sex, and when appropriate the Binet staging. Variables with 2-tailed P values of less than .05 were considered to be significant. All analyses were done using SPSS Statistical Software, version 11.5 (SPSS, Chicago, IL).
Results
Patient characteristics
Table 2 summarizes the characteristics of the 127 patients. Eighty-seven patients were in stage A, 29 in stage B, and 11 in stage C. Twenty disease-related deaths were recorded and 4 additional deaths, unrelated to CLL, occurred among patients in stage A. Fifty-three patients (42%) were allocated to the UM IGVH group, whereas 74 displayed a MT IGVH profile. The MT and UM cases were heterogeneously distributed within Binet stages because 69% of patients in stage A displayed a MT IGVH profile, whereas 65% were UM among B and C cases (P < .001). Sex distribution was also heterogeneous with a high proportion of male patients among UM cases (70%) and a slight female predominance (53%) in the MT group (P = .05). Based on French Cooperative Group guidelines, almost all patients with stage B and C disease received early treatment, whereas treatment was deferred for those with stage A disease until disease progression. This was the case for 22 patients (25%) in stage A, 4 of whom died.
Clinical and biologic characteristics of patients
. | Stage A . | Stage B . | Stage C . | Stages B+C . | Stages A+B+C . |
|---|---|---|---|---|---|
| No. of patients | 87 | 29 | 11 | 40 | 127 |
| Age, y* | 63.0 | 58.0 | 59.0 | 58.5 | 61.0 |
| Male sex, no. (%) | 51 (59) | 24 (83) | 5 (45) | 29 (72) | 80 (63) |
| Lymphocyte count, × 109/L* | 15.2 | 37.2 | 151.8 | 45.0 | 19.0 |
| Hemoglobin level, g/dL* | 14.1 | 13.4 | 7.2 | 13.0 | 13.8 |
| Platelet count, × 109/L* | 212.5 | 160.0 | 187.5 | 172.0 | 198.0 |
| Lymphocyte doubling time, no (%) | |||||
| More than 12 mo | 64 (84) | NA | NA | NA | NA |
| Less than 12 mo | 12 (16) | NA | NA | NA | NA |
| IGVH genes, no (%) | |||||
| UM | 27 (31) | 17 (59) | 9 (82) | 26 (65) | 53 (42) |
| MT | 60 (69) | 12 (41) | 2 (18) | 14 (35) | 74 (58) |
| ZAP-70, no (%) | |||||
| Less than 20% | 45 (65) | 7 (30) | 3 (33) | 10 (31) | 55 (54) |
| 20% or greater | 24 (35) | 16 (70) | 6 (67) | 22 (69) | 46 (46) |
| L/A ratio, no (%) | |||||
| Less than 1 | 56 (69) | 13 (46) | 2 (20) | 15 (39) | 71 (60) |
| 1 or greater | 25 (31) | 15 (54) | 8 (80) | 23 (61) | 48 (40) |
| Progression, no (%) | 22 (25) | NA | NA | NA | NA |
| CLL-related death, no (%) | 4 (5) | 10 (34) | 6 (55) | 16 (40) | 20 (18) |
. | Stage A . | Stage B . | Stage C . | Stages B+C . | Stages A+B+C . |
|---|---|---|---|---|---|
| No. of patients | 87 | 29 | 11 | 40 | 127 |
| Age, y* | 63.0 | 58.0 | 59.0 | 58.5 | 61.0 |
| Male sex, no. (%) | 51 (59) | 24 (83) | 5 (45) | 29 (72) | 80 (63) |
| Lymphocyte count, × 109/L* | 15.2 | 37.2 | 151.8 | 45.0 | 19.0 |
| Hemoglobin level, g/dL* | 14.1 | 13.4 | 7.2 | 13.0 | 13.8 |
| Platelet count, × 109/L* | 212.5 | 160.0 | 187.5 | 172.0 | 198.0 |
| Lymphocyte doubling time, no (%) | |||||
| More than 12 mo | 64 (84) | NA | NA | NA | NA |
| Less than 12 mo | 12 (16) | NA | NA | NA | NA |
| IGVH genes, no (%) | |||||
| UM | 27 (31) | 17 (59) | 9 (82) | 26 (65) | 53 (42) |
| MT | 60 (69) | 12 (41) | 2 (18) | 14 (35) | 74 (58) |
| ZAP-70, no (%) | |||||
| Less than 20% | 45 (65) | 7 (30) | 3 (33) | 10 (31) | 55 (54) |
| 20% or greater | 24 (35) | 16 (70) | 6 (67) | 22 (69) | 46 (46) |
| L/A ratio, no (%) | |||||
| Less than 1 | 56 (69) | 13 (46) | 2 (20) | 15 (39) | 71 (60) |
| 1 or greater | 25 (31) | 15 (54) | 8 (80) | 23 (61) | 48 (40) |
| Progression, no (%) | 22 (25) | NA | NA | NA | NA |
| CLL-related death, no (%) | 4 (5) | 10 (34) | 6 (55) | 16 (40) | 20 (18) |
NA indicates not applicable.
Median values
Selection of LPL and ADAM29 as potential prognostic markers in CLL
Because our microarray study showed that LPL and SPG20 were overexpressed among UM CLLs, whereas ADAM29 and NRIP1 predominated in MT CLLs, we first evaluated expression of these 4 genes in an initial series of 71 patients. Quantification was performed on total lymphocytes because preliminary experiments in 5 patients showed similar results when compared to purified leukemic cells (data not shown). For each gene, we determined which expression levels could best segregate UM from MT cases using the Youden and validity indexes. Results showed that overall LPL and ADAM29 performed better than SPG20 and NRIP1 to predict UM and MT IGVH profiles, respectively (Tables 3 and 4). The concordance rate was 80% for ADAM29, 77% for LPL, 65% for SPG20, and 55% for NRIP1. Next, we investigated whether a combination of the most discriminating parameters by a simple 1:1 LPL/ADAM29 (L/A) ratio could improve their individual performance. With a calculated threshold of 1, the L/A ratio displayed better sensitivity and specificity than each marker taken individually. Positive predictive value (PPV) was 91% for UM cases and negative predictive value (NPV) was 86% for MT patients, providing a better performance than each individual marker (Tables 3 and 4). Thus the L/A ratio constituted the best marker reflecting the mutational status of IGVH genes in this cohort of 71 patients with CLL, with a concordance rate of 89%.
Correlation of gene expression with IGVH mutational status
. | LPL . | . | ADAM29 . | . | SPG20 . | . | NRIP1 . | . | LPL/ADAM29 . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | 1 or more . | Less than 1 . | 3 or more . | Less than 3 . | 3.5 or more . | Less than 3.5 . | 4 or more . | Less than 4 . | 1 or more . | Less than 1 . | |||||
| UM, n = 37 | 30 | 7 | 8 | 29 | 20 | 17 | 17 | 20 | 32 | 5 | |||||
| MT, n =34 | 9 | 25 | 28 | 6 | 8 | 26 | 19 | 15 | 3 | 31 | |||||
. | LPL . | . | ADAM29 . | . | SPG20 . | . | NRIP1 . | . | LPL/ADAM29 . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | 1 or more . | Less than 1 . | 3 or more . | Less than 3 . | 3.5 or more . | Less than 3.5 . | 4 or more . | Less than 4 . | 1 or more . | Less than 1 . | |||||
| UM, n = 37 | 30 | 7 | 8 | 29 | 20 | 17 | 17 | 20 | 32 | 5 | |||||
| MT, n =34 | 9 | 25 | 28 | 6 | 8 | 26 | 19 | 15 | 3 | 31 | |||||
Parameters calculated in relation to unmutated IGVH genes for LPL, SPG20, and LPL/ADAM29 and to mutated IGVH genes for ADAM29 and NRIP1
. | LPL . | ADAM29 . | SPG20 . | NRIP1 . | LPL/ADAM29 . |
|---|---|---|---|---|---|
| Sensitivity, % | 81 | 82 | 54 | 56 | 86 |
| Specificity, % | 74 | 78 | 76 | 54 | 91 |
| PPV, % | 77 | 78 | 71 | 53 | 91 |
| NPV, % | 78 | 83 | 60 | 57 | 86 |
. | LPL . | ADAM29 . | SPG20 . | NRIP1 . | LPL/ADAM29 . |
|---|---|---|---|---|---|
| Sensitivity, % | 81 | 82 | 54 | 56 | 86 |
| Specificity, % | 74 | 78 | 76 | 54 | 91 |
| PPV, % | 77 | 78 | 71 | 53 | 91 |
| NPV, % | 78 | 83 | 60 | 57 | 86 |
Reproducibility of LPL and ADAM29 quantification
Next, we tested the reproducibility of LPL and ADAM29 quantification by RQ-PCR. This was done by comparing results obtained from replicate samples for 4 patients, 2 overexpressing LPL and 2 overexpressing ADAM29. For each patient, 4 replicates were analyzed during the same reaction run (intrarun variability), and this was repeated on 3 different days (interrun variability). The overall variability of these 12 replicates is shown in Table 5. Of note, intrarun variability was always smaller than overall variability, with the coefficient of variation (CV) being less than 0.5% for LPL and less than 1.1% for ADAM29 (data not shown).
Reproducibility of RQ-PCR
. | LPL . | . | ADAM29 . | . | ||
|---|---|---|---|---|---|---|
| Patient . | Ct ± SD . | CV, % . | Ct ± SD . | CV, % . | ||
| CLL-73 | 23.54 ± 0.46 | 1.97 | NA | NA | ||
| CLL-105 | 23.96 ± 0.38 | 1.59 | NA | NA | ||
| CLL-67 | NA | NA | 27.33 ± 1.18 | 4.31 | ||
| CLL-101 | NA | NA | 23.32 | 2.21 | ||
. | LPL . | . | ADAM29 . | . | ||
|---|---|---|---|---|---|---|
| Patient . | Ct ± SD . | CV, % . | Ct ± SD . | CV, % . | ||
| CLL-73 | 23.54 ± 0.46 | 1.97 | NA | NA | ||
| CLL-105 | 23.96 ± 0.38 | 1.59 | NA | NA | ||
| CLL-67 | NA | NA | 27.33 ± 1.18 | 4.31 | ||
| CLL-101 | NA | NA | 23.32 | 2.21 | ||
Overall variability of RQ-PCR quantification of LPL and ADAM29 gene expression was evaluated on 12 replicates performed on 4 different CLL patients. This was done by testing each sample in 4 replicates during the same reaction (intrarun variability), this being repeated on 3 different days (interrun variability).
Ct indicates threshold cycle; SD, standard deviation; CV, coefficient of variation; NA, not applicable.
The L/A ratio is a strong predictor of prognosis in CLL
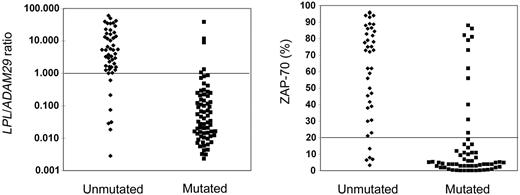
We evaluated the prognostic value of the L/A ratio in an extended cohort of 127 patients. This included the 71 initially studied patients and 56 additional cases selected because of their prolonged follow-up and the availability of frozen samples. In addition we compared its value to that of the mutational profile of IGVH genes and ZAP-70 protein expression. LPL and ADAM29 were available for 119 patients, whereas ZAP-70 could be determined for 101 patients and all 3 parameters for 93 patients. On this larger series, the L/A ratio once again provided a better concordance (90%) with IGVH mutational status than LPL (76%) or ADAM29 (82%) taken individually (data not shown), and threshold values were found to be identical to those of the first cohort (Figure 1). ZAP-70 expression was measured by flow cytometry in leukemic B cells in comparison with that of the patients' T and NK cells. A cutoff value at 20% of cell positivity was found to provide the best correlation with IGVH genes (Figure 1).
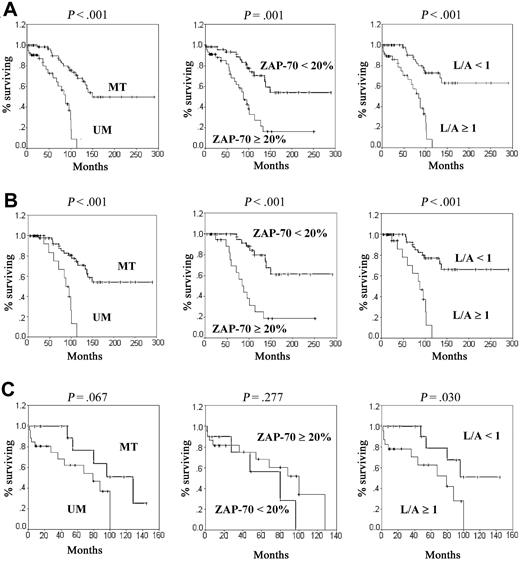
Median EFS for the entire cohort was 87 months in CLLs displaying UM IGVH genes as compared to 149 months in MT patients (P < .001). It was 84 months for patients with an L/A ratio above 1 and 88 months for patients expressing ZAP-70, whereas median EFS was not achieved for those with an L/A ratio below 1 (P < .001) or ZAP-70- (P < .001; Figure 2A). Multivariate Cox regression showed that, with adjustment for age and sex, UM IGVH and stage B or C were independently associated with disease progression or CLL-related death with hazard ratios of, respectively, 5.0 (P < .001) and 2.6 (P = .01; Table 6). Similarly, an L/A ratio above 1 and stage B or C were independently and significantly associated with shorter EFS. ZAP-70, however, was not found to be an independent prognosis factor.
Prognostic factors for disease progression and CLL-related death in multivariate Cox regression
Factor . | Hazard ratio (95% CI) . | P . |
|---|---|---|
| All stages, EFS for all stages | ||
| Model 1 including IGVH | ||
| Age | 1.1 (1.0-1.2) | .20* |
| Sex, male | 1.4 (1.0-2.0) | .29 |
| Binet stage B/C | 2.6 (1.8-3.7) | .01 |
| UM IGVH | 5.0 (3.4-7.2) | <.001 |
| Model 2 including L/A ratib† | ||
| Age | 1.1 (1.0-1.2) | .50* |
| Sex, male | 1.4 (1.2-2.7) | .12 |
| Binet stage B/C | 2.5 (1.7-3.7) | .02 |
| L/A ratio 1 or greater | 5.6 (3.8-8.1) | <.001 |
| EFS for Binet stage A | ||
| Model 1 including IGVH | ||
| Age | 1.0 (0.9-1.1) | .86* |
| Sex, male | 1.6 (1.0-2.3) | .28 |
| UM IGVH | 5.7 (3.6-9.1) | <.001 |
| Model 2 including L/A ratio and ZAP-70 | ||
| Age | 0.9 (0.8-1.0) | .29* |
| Sex, male | 1.7 (1.0-2.7) | .32 |
| ZAP-70 greater than 20% | 4.2 (2.3-7.7) | .02 |
| L/A ratio 1 or greater | 3.9 (2.1-7.2) | .03 |
| OS for stages B and C | ||
| Model 1 including IGVH | ||
| Age | 1.5 (1.2-1.8) | .03* |
| Sex, male | 1.0 (0.5-2.1) | .99 |
| UM IGVH | 7.2 (3.4-15.1) | .01 |
| Model 2 including L/A ratio† | ||
| Age | 1.6 (1.3-2.0) | .05* |
| Sex, male | 0.5 (0.3-1.3) | .46 |
| L/A ratio 1 or greater | 6.8 (3.3-14) | .01 |
Factor . | Hazard ratio (95% CI) . | P . |
|---|---|---|
| All stages, EFS for all stages | ||
| Model 1 including IGVH | ||
| Age | 1.1 (1.0-1.2) | .20* |
| Sex, male | 1.4 (1.0-2.0) | .29 |
| Binet stage B/C | 2.6 (1.8-3.7) | .01 |
| UM IGVH | 5.0 (3.4-7.2) | <.001 |
| Model 2 including L/A ratib† | ||
| Age | 1.1 (1.0-1.2) | .50* |
| Sex, male | 1.4 (1.2-2.7) | .12 |
| Binet stage B/C | 2.5 (1.7-3.7) | .02 |
| L/A ratio 1 or greater | 5.6 (3.8-8.1) | <.001 |
| EFS for Binet stage A | ||
| Model 1 including IGVH | ||
| Age | 1.0 (0.9-1.1) | .86* |
| Sex, male | 1.6 (1.0-2.3) | .28 |
| UM IGVH | 5.7 (3.6-9.1) | <.001 |
| Model 2 including L/A ratio and ZAP-70 | ||
| Age | 0.9 (0.8-1.0) | .29* |
| Sex, male | 1.7 (1.0-2.7) | .32 |
| ZAP-70 greater than 20% | 4.2 (2.3-7.7) | .02 |
| L/A ratio 1 or greater | 3.9 (2.1-7.2) | .03 |
| OS for stages B and C | ||
| Model 1 including IGVH | ||
| Age | 1.5 (1.2-1.8) | .03* |
| Sex, male | 1.0 (0.5-2.1) | .99 |
| UM IGVH | 7.2 (3.4-15.1) | .01 |
| Model 2 including L/A ratio† | ||
| Age | 1.6 (1.3-2.0) | .05* |
| Sex, male | 0.5 (0.3-1.3) | .46 |
| L/A ratio 1 or greater | 6.8 (3.3-14) | .01 |
Multivariate analyses were done separately for IGVH or ZAP-70 and L/A ratio due to the high concordance between the latter 2 parameters and the former.
P for trend test.
ZAP-70 was not independently associated with disease progression or CLL-related death and was then eliminated in the final Cox model.
In patients with stage A disease, an identical median EFS of 87 months was observed for patients with UM IGVH genes, an L/A ratio above 1, and expressing ZAP-70, whereas it was not achieved for cases with MT IGVH genes, an L/A ratio below 1, and ZAP-70- (all P < .001; Figure 2B). In multivariate Cox analyses, after adjustment for age and sex, both ZAP-70 and L/A ratio were independent significant prognostic factors. This was also true for the IGVH mutational status (Table 6).
The 40 patients in stage B and C were analyzed together for evaluation of overall survival (OS; Figure 2C). There was a trend for longer OS in patients with MT than in those with UM IGVH genes (128 versus 79 months; P = .067). The L/A ratio was predictive of survival because patients with a ratio above 1 had a median OS of 79 months, whereas it was not yet reached at time of analysis for those with a ratio below 1 (P = .03). In contrast, ZAP-70 did not correlate with survival in this group (100 versus 80 months; P = .28). By multivariate Cox analysis, an L/A ratio below 1 was found as a significant prognostic factor, whereas ZAP-70 was not independently associated with survival. In a separate Cox analysis, IGVH mutational status became a significant prognosis factor, after adjustment for age and sex (Table 6).
Correlation between L/A ratio, ZAP-70 expression, and IGVH mutational status
Given the strong prognostic value of the 3 markers, we next assessed the correlation between them in the 93 cases for whom all 3 parameters had been determined. Cases were scored as “positive” or “negative” for a given marker based on expression values above or below the thresholds. Concordance rate with IGVH mutational status was 85% for ZAP-70, thus slightly lower than that obtained with the L/A ratio (92%; Table 7). As depicted in Table 7, all double-positive patients (ZAP-70+L/A+; n = 30) except one expressed UM IGVH genes, and all double-negative patients (ZAP-70-L/A-; n = 44) had MT IGVH genes. Although an almost perfect correlation for all 3 markers was found for these cases (73 of 74, 99% accuracy), discordant profiles still accounted for 20% of CLL patients. They included 13 patients expressing a ZAP-70+L/A- profile, whereas 6 patients were ZAP-70-L/A+. Among the 13 ZAP-70+L/A- cases, 4 had UM IGVH genes (1 stage A and 3 stage B), whereas 9 had MT IGVH genes (7 stage A and 2 stage B). Four of the 6 patients with a ZAP-70-L/A+ profile displayed UM IGVH genes (3 stage A and 1 stage C), whereas they were MT in the remaining 2 cases (1 stage A and 1 stage B). Of note, the mutational status of these 19 discordant cases would have been predicted correctly more often by the L/A ratio alone (13 cases) than by ZAP-70 expression alone (6 cases). The small number of patients with discordant markers did not allow prognostic evaluation.
Groups of CLL patients according to L/A ratio and ZAP-70 expression
. | ZAP-70+L/A+ . | ZAP-70+L/A− . | ZAP-70−L/A+ . | ZAP-70−L/A− . |
|---|---|---|---|---|
| UM | 29 | 4 | 4 | 0 |
| MT | 1 | 9 | 2 | 44 |
| Total, no. (%) | 30 (32) | 13 (14) | 6 (6) | 44 (47) |
. | ZAP-70+L/A+ . | ZAP-70+L/A− . | ZAP-70−L/A+ . | ZAP-70−L/A− . |
|---|---|---|---|---|
| UM | 29 | 4 | 4 | 0 |
| MT | 1 | 9 | 2 | 44 |
| Total, no. (%) | 30 (32) | 13 (14) | 6 (6) | 44 (47) |
Positivity or negativity for ZAP-70 refers to expression values ≥20% or <20%, respectively. Positivity or negativity for L/A ratio refers to expression values ≥ 1 or < 1, respectively.
Correlations between LPL/ADAM29 ratio or ZAP-70 expression and the IgVH gene mutational status. The threshold values calculated for the L/A ratio (= 1) and ZAP-70 (= 20%) showing the best concordance rate with the IGVH mutational status, as determined by using the Youden index, are indicated by a horizontal line.
Correlations between LPL/ADAM29 ratio or ZAP-70 expression and the IgVH gene mutational status. The threshold values calculated for the L/A ratio (= 1) and ZAP-70 (= 20%) showing the best concordance rate with the IGVH mutational status, as determined by using the Youden index, are indicated by a horizontal line.
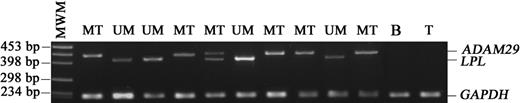
Determination of the LPL and ADAM29 gene expression by a simple qualitative multiplex RT-PCR assay
To simplify the assessment of LPL and ADAM29 gene expression, we developed a multiplex RT-PCR assay, where both genes were simultaneously amplified generating PCR products of different size, respectively, 410 bp and 445 bp (Figure 3). This assay was then evaluated on 95 patients of our series. Experiments were done in duplicate on separate PCRs for 25 cases and showed a perfect reproducibility. Using our defined PCR conditions, the results were unambiguous in 89 cases with production of a single band or 2 bands but with one clearly more intense than the other. They correlated with the RQ-PCR results because 34 of the 36 patients with an L/A ratio above 1.0 expressed predominantly LPL in the multiplex assay, whereas 2 displayed products of both size with roughly similar intensities (doublets). Alternatively, 55 of the 59 patients exhibiting a L/A ratio less than 1.0 expressed predominantly ADAM29, whereas 4 displayed doublets (Table 8). Thus, inconclusive results (doublets) were observed for 6 patients. In 5 of these 6 cases, the L/A ratio values were in the range of 0.7 to 1.5. There was, therefore, a very good concordance between the results obtained by RQ-PCR and qualitative multiplex PCR. Prognostic evaluation performed with LPL and ADAM29 gene expression determined by multiplex PCR produced identical results to those obtained by RQ-PCR (data not shown).
Comparison of multiplex RT-PCR with RQ-PCR
Multiplex RT-PCR . | L/A ratio less than 1 . | L/A ratio greater than 1 . | Total . |
|---|---|---|---|
| ADAM29 | 55 | 0 | 55 |
| LPL | 0 | 34 | 34 |
| Doublet | 4 | 2 | 6 |
| Total | 59 | 36 | 95 |
Multiplex RT-PCR . | L/A ratio less than 1 . | L/A ratio greater than 1 . | Total . |
|---|---|---|---|
| ADAM29 | 55 | 0 | 55 |
| LPL | 0 | 34 | 34 |
| Doublet | 4 | 2 | 6 |
| Total | 59 | 36 | 95 |
Cases were scored as ADAM29 or LPL according to the type of the predominant PCR product obtained by multiplex RT-PCR. When 2 bands of roughly similar intensity were observed, they were qualified as doublet.
Kaplan-Meier survival curves in CLL according to IGVH mutational status, L/A ratio, or ZAP-70 expression. (A) EFS probabilities for the total population. (B) EFS probabilities for patients in stage A. (C) OS probabilities for patients with stages B and C disease.
Kaplan-Meier survival curves in CLL according to IGVH mutational status, L/A ratio, or ZAP-70 expression. (A) EFS probabilities for the total population. (B) EFS probabilities for patients in stage A. (C) OS probabilities for patients with stages B and C disease.
Expression of LPL and ADAM29 genes in normal circulating cells
Although experiments on purified and unpurified CLL cells showed similar results, we wanted to evaluate a possible expression of LPL and ADAM29 genes in normal cells, which might contaminate patient samples. For this, we assessed their expression by multiplex PCR in the peripheral blood cells of 20 healthy individuals. Very low level of expression of LPL was found in 3 of the 14 PBMCs tested and in 1 of the 6 purified B-cell samples. ADAM29 was not detected in any of the PBMCs and at a very low level in one of the purified B-cell samples. In addition the T-cell line Jurkat failed to express either of these 2 genes. Thus, all or most of the LPL and ADAM29 transcripts that we measured in patient samples originated from leukemic cells and not from background normal mononuclear cells.
Multiplex PCR determination of LPL and ADAM29 expression.LPL and ADAM29 transcripts were amplified simultaneously; the PCR products were then separated by electrophoresis on agarose gel and visualized under UV illumination after ethidium bromide staining. Amplification of the GAPDH gene from the same transcripts served as control of cDNA integrity. MWM indicates molecular weight marker; MT, mutated; UM, unmutated; B, purified B cells from a healthy individual; T, Jurkat T-cell line.
Multiplex PCR determination of LPL and ADAM29 expression.LPL and ADAM29 transcripts were amplified simultaneously; the PCR products were then separated by electrophoresis on agarose gel and visualized under UV illumination after ethidium bromide staining. Amplification of the GAPDH gene from the same transcripts served as control of cDNA integrity. MWM indicates molecular weight marker; MT, mutated; UM, unmutated; B, purified B cells from a healthy individual; T, Jurkat T-cell line.
Discussion
The recent development of microarray technology has allowed the discovery of genes, which may have prognostic significance in human tumors. A previous gene expression profiling study in CLL led us to identify a set of genes that appeared to segregate stable MT from progressive UM forms.24 In the present work, we show that the combined expression of 2 of them, LPL and ADAM29, constitutes a new prognostic marker in CLL.
LPL is a heparin-releasable enzyme bound to glycosaminoglycan components of the capillary endothelium and is particularly abundant in muscle, adipose tissue, and macrophages. With apolipoprotein CII, LPL mediates the hydrolysis of triacylglycerol component of circulating chylomicrons and very-low-density lipoproteins. It plays a central role in lipid metabolism and transport.29,30 Mutations in the LPL gene are frequently associated with dyslipidemia and atherosclerosis.29,31 In line with our findings on normal cells, other investigators have failed to detect LPL expression in normal purified B and T lymphocytes.32 However, they found that it was expressed and secreted by NK cells, where it was shown to modulate their cytotoxic activity. Reasons for its high expression in UM CLL B cells are unknown. Because UM CLL B cells have been shown to be more responsive to stimulation through their antigen receptor,33,34 we speculate that LPL could play a role in lipid raft formation or stabilization, a biologic process known to be important in B-cell activation.35 The ADAM29 gene encodes a member of the disintegrin and metalloproteinase family of transmembrane proteins, which have been shown to mediate cell-to-cell or cell-to-matrix interactions (or both) as well as the proteolytic shedding of cell surface molecules.36,37 In contrast to other ADAMs that are expressed in various tissues, ADAM29 transcripts are highly restricted to the testis.38,39 Because its precise functions are so far unknown, its role in the biology of CLL remains unexplained. The fact that for these 2 genes: (1) similar results were obtained on total CLL lymphocyte populations and from purified leukemic cells, and (2) their absence of detection or very low level in normal peripheral B cells, indicates that their overexpression could be tumor specific. Alternatively, it might reflect a restricted expression in a minor B-lymphoid subpopulation, which is expanded in CLL. Further experiments on purified B-cell subsets are warranted to prove whether these hypotheses are correct.
The discovery that ZAP70 gene expression could differentiate MT from UM CLLs has also emerged from microarray experiments.18,19 Further studies confirmed its presence at the protein level by flow cytometry and showed that it had prognostic impact.20-23 We evaluated the ability of the L/A ratio as well as ZAP-70 expression and the IGVH mutational status to predict clinical outcome in our cohort of patients with CLL. In univariate analysis, all 3 parameters correlated with EFS in the whole population. In multivariate analysis, however, ZAP-70 was no longer selected. For stage A patients, these 3 biologic parameters were predictive of EFS. When considering the patients with stage B and C disease, the L/A ratio was the only parameter that correlated significantly with survival in univariate analysis. The IGVH UM status became a significant risk factor only after adjustment for sex and age in multivariate analysis. Rassenti et al reported recently that ZAP-70 was a better predictor of time from diagnosis to initial therapy than the IGVH mutational status.23 The lack of information concerning the patients' clinical stage in this cohort makes comparison with our data difficult. In our study, most of patients in stages B and C received early treatment, whereas it was delayed in those with stage A disease until progression was observed. Therefore, only patients in stage A can be compared. As noted, ZAP-70, the L/A ratio, and the IGVH mutational status were all similarly capable of predicting treatment-free survival for this group of patients. In contrast, ZAP-70 expression had no prognostic value in patients in stages B and C. Similar findings have also been reported by Crespo et al.20 It remains to be elucidated why ZAP-70, which is a strong prognostic parameter for patients in stage A, does not predict outcome for patients in stages B and C. The availability of biologic prognostic indicators such as the L/A ratio for stage B and C CLL cases may therefore be of great importance for future risk-adapted treatments. For the cases with discordant results for ZAP-70, L/A ratio, and IGVH genes, it was unclear which of these 3 biologic parameters was the most reliable prognostic indicator. Larger series will be necessary to determine which most accurately predict clinical evolution.
In the present study, we also evaluated the correlations between these 3 biologic prognostic markers. The concordance rate with IGVH mutational status was slightly higher for the L/A ratio (92%) than for ZAP-70 (85%). For this latter parameter, the threshold value that best correlated with the IGVH mutational status was found to be 20%, similar to that previously described by Crespo et al20 and Rassenti et al,23 but higher than that used by Orchard et al22 (10%). The discordance rate with the IGVH status was slightly higher than that observed by Crespo et al20 and Orchard et al22 (respectively, 5.4% and 7.8%) but lower than that of Rassenti et al (23%). In this latter work, the investigators used another anti-ZAP-70 antibody and a different gating strategy.23 These discordant results might be due to technologic reasons, in addition to the type of patients studied. Further studies evaluating the different technologic aspects of its detection are clearly needed before ZAP-70 expression can be used in a clinical perspective. Interestingly, concordant results for ZAP-70, L/A ratio, and IGVH genes were found in about 80% of cases. Provided that these results are confirmed in larger series of patients, combining ZAP-70 and L/A ratio quantification may represent an alternative to sequencing the IGVH genes in about 80% of patients.
We also describe an alternative method to assess simultaneous expression of LPL and ADAM29 by a multiplex competitive RT-PCR technique. Although it was not informative in a minority of cases, in our hands this assay proved to be simple and reproducible. Its extension to a routine test, however, requires further evaluation by independent laboratories. The determination of the IGVH mutational status is an expensive labor-intensive technique not well suited as a routine test in most clinical laboratories. Simpler techniques such as flow cytometric quantification of ZAP-70 or PCR-based expression analysis of LPL and ADAM29 genes represent promising alternatives. Standardization of these techniques, as well as evaluation of their cost, including comparison of RQ-PCR and multiplex RT-PCR for LPL and ADAM29, will be important for future therapeutic protocols. A cost comparison evaluation of the different techniques to assess prognosis in stage A CLL is currently being performed in France.
In summary, our study shows that LPL and ADAM29 constitute new and strong prognosis markers in CLL. Importantly, they provide better prognostic information than ZAP-70 for advanced CLL cases. In addition, combination of the L/A ratio with ZAP-70 expression predicts accurately the IGVH mutational status in 80% of CLL cases, thus rendering sequencing unnecessary in these patients.
Appendix
Members of the advisory board and scientific committee of the French Cooperative Group on CLL are as follows, in alphabetic order: J. L. Binet, B. Cazin, S. Chevret, F. Cymbalista, F. Davi, A. Delmer, G. Dighiero, M. Divine, B. Dreyfus, C. Dumontet, J. P. Fermand, R. Garand, V. Leblond, M. Leporrier, V. Levy, K. Maloum, H. Merle-Béral, M. Michallet, L. Sutton, P. Travade, X. Troussard.
Prepublished online as Blood First Edition Paper, March 31, 2005; DOI 10.1182/blood-2004-08-3344.
Supported by grants from the French Cooperative Group on CLL and the Ministère de la Recherche et de la Technologie. Y.V. was supported by Coordena ão de Aperfei
ão de Aperfei oamento de Passaol de Nível Superior (CAPES) and Funda
oamento de Passaol de Nível Superior (CAPES) and Funda ao de Amparo à Pasquisa do Estado de São Paulo (FAPESP). P.O. was supported by the French Academy of Medicine.
ao de Amparo à Pasquisa do Estado de São Paulo (FAPESP). P.O. was supported by the French Academy of Medicine.
P.O. and Y.V. contributed equally to this work.
A complete list of the members of the French Cooperative Group on CLL appears in the “Appendix.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Rodrigo Proto-Siquera and Dr Marco Antônio Zago for providing patient samples, Ariane Michel for expert technical assistance, and Elizabeth Macintyre for critical reading of the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal