Activating mutations in the FMS-like tyrosine kinase 3 gene (FLT3), including internal tandem duplications (ITDs) in the juxtamembrane (JM) domain or point mutations (PMs) in the activation loop, are the most common genetic aberration in acute myeloid leukemia (AML).1 Recently, Paietta et al2 investigated the presence of FLT3 mutations in 69 adult T-cell acute lymphoblastic leukemia (T-ALL) patients. Three positive cases (2 ITDs and 1 PM) were identified sharing a similar early prothymocytic T-cell developmental state exclusively expressing cKIT/CD117, and a trial to test the efficacy of FLT3 inhibitors for this T-ALL subset was suggested.
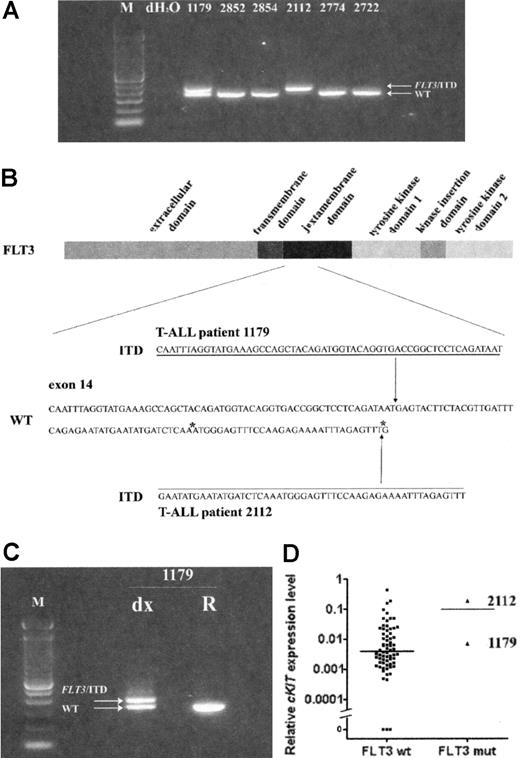
To validate the incidence of FLT3 mutations and to investigate a relation to outcome and other parameters, we screened 72 diagnostic pediatric T-ALL samples for FLT3 mutations, as previously described.3,4 We identified FLT3/ITD mutations in 2 pediatric T-ALLs (Figure 1A), whereas no point mutations in the kinase domain were detected. Sequence analysis confirmed a 51-base pair insertion in patient 2112 and a 57-base pair insertion in patient 1179 (Figure 1B). Moreover, no wild-type FLT3 was identified in patient 2112, suggesting loss of the wild-type allele.1
Immunophenotypic analyses revealed a similar profile for both FLT3-mutated patient samples (ie, TdT+, CD2+, CD5+, CD7+, CD4+/CD8-, cytoplasmic CD3+, surface CD3-, and CD10-). CD34 expression was detected in 24% and 21% of the leukemic blasts in patients 2112 and 1179, respectively. Only patient 2112 weakly expressed CD13 (24%) but not CD33. Although representing early T-cell differentiation stages for both patient samples, the maturation stage seems more advanced compared with the FLT3-mutated adult T-ALL cases (CD34+, CD4-/CD8-).2 Since no additional patient material was left for flow cytometry, cKIT/CD117 expression was determined by real-time quantitative polymerase chain reaction (RQ-PCR) on isolated blasts5 (> 90% leukemic cells) from all pediatric T-ALL samples (Figure 1D). Whereas only the 3 FLT3-mutated adult T-ALL patients highly expressed cKIT,2 most pediatric T-ALL samples expressed cKIT mRNA to some extent. Patient 2112 highly expressed cKIT, whereas patient 1179 showed a weak cKIT expression that was about 26-fold lower. Since various non-FLT3-mutated T-ALL samples highly expressed cKIT/CD117 at levels comparable to patient 2112, we conclude that cKIT/CD117 expression is not exclusively associated with FLT3 mutations. Nevertheless, transcript levels do not necessarily correlate with protein expression levels.2 In line with previous observations,2 leukemic blasts of FLT3-mutated samples highly expressed LYL1 and LMO2. Both pediatric samples carried a HOX11L2 translocation in contrast to the FLT3-mutated adult T-ALL cases.2
Activating FLT3 mutations in pediatric T-ALL. (A) Genomic PCR analysis for the FLT3 gene. PCR results for 6 pediatric T-ALL patient samples are shown. T-ALL patient 1179 shows a heterozygous FLT3/ITD mutation. No wild-type FLT3 is detected in T-ALL patient 2112, probably due to loss of heterozygosity of the wild-type allele. (B) Overview of the functional domains in the FLT3 tyrosine kinase receptor. Genomic position of the FLT3/ITD mutations detected in patients 1179 and 2112 are shown. Mutation position annotation is based on the FLT3 reference sequence NM_004119. (C) FLT3 mutation analysis of diagnosis and relapse material of T-ALL patient 1179. The FLT3/ITD mutation was present at diagnosis but absent at relapse. (D) Relative cKIT/CD117 mRNA expression indicated as percentage of GAPDH expression for the investigated pediatric T-ALL cohort (FLT3 wt vs FLT3 mut). For both groups the median FLT3 mRNA expression is shown. wt indicates wild type; mut, mutated; dx, at diagnosis; R, at relapse; and *, genomic position of the recently identified FLT3/ITD mutations in adult T-ALLs.2
Activating FLT3 mutations in pediatric T-ALL. (A) Genomic PCR analysis for the FLT3 gene. PCR results for 6 pediatric T-ALL patient samples are shown. T-ALL patient 1179 shows a heterozygous FLT3/ITD mutation. No wild-type FLT3 is detected in T-ALL patient 2112, probably due to loss of heterozygosity of the wild-type allele. (B) Overview of the functional domains in the FLT3 tyrosine kinase receptor. Genomic position of the FLT3/ITD mutations detected in patients 1179 and 2112 are shown. Mutation position annotation is based on the FLT3 reference sequence NM_004119. (C) FLT3 mutation analysis of diagnosis and relapse material of T-ALL patient 1179. The FLT3/ITD mutation was present at diagnosis but absent at relapse. (D) Relative cKIT/CD117 mRNA expression indicated as percentage of GAPDH expression for the investigated pediatric T-ALL cohort (FLT3 wt vs FLT3 mut). For both groups the median FLT3 mRNA expression is shown. wt indicates wild type; mut, mutated; dx, at diagnosis; R, at relapse; and *, genomic position of the recently identified FLT3/ITD mutations in adult T-ALLs.2
Patient 1179 relapsed 13 months after initial diagnosis, whereas patient 2112 is in continued complete remission (CCR; 61+). Interestingly, patient 1179 showed no FLT3/ITD mutation at relapse (Figure 1C), possibly due to loss of the mutated allele during therapy, or, alternatively, the FLT3/ITD-positive clone was eliminated during chemotherapy with a subsequent relapse from a non-FLT3-mutated parental clone.
In conclusion, we confirm the presence of FLT3 mutations in pediatric T-ALL (2/72; 2.7%). Although both immature, the immunophenotypes of the FLT3-mutated pediatric and adult2 T-ALL cases differed. In addition, a link between mRNA expression of cKIT/CD117 and FLT3 mutations could not be demonstrated. Since patient 2112 is in CCR and relapse material of patient 1179 did not show evidence for FLT3 mutation, the FLT3-mutated T-ALL subclone seems to be effectively eradicated by current chemotherapy. This suggests that the application of FLT3 inhibitors for FLT3-mutated T-ALL, as suggested by Paietta et al,2 may not further improve treatment outcome in pediatric T-ALL.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal