In classic Hodgkin lymphoma (HL) and posttransplantation lymphoproliferative disease (PTLD), 2 malignancies frequently associated with Epstein-Barr virus (EBV), the tumor cells often appear to derive from B-cell receptor (BCR)–deficient and therefore preapoptotic germinal center (GC) B cells. To test whether EBV can rescue BCR-less GC B cells, we infected human tonsillar CD77+ GC B cells in vitro with EBV. More than 60 monoclonal lymphoblastoid cell lines (LCLs) were established. Among these, 28 cell lines did not express surface immunoglobulin (sIg). Two of the sIg-negative cell lines carry obviously destructive mutations that have been introduced into originally functional VH gene rearrangements during the process of somatic hypermutation. Quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) showed that in most other lines the sIg deficiency was not simply the result of transcriptional down-regulation, but it was rather due to posttranscriptional defects. These findings strongly support the idea that EBV plays a central role in the pathogenesis of classic HL and PTLD by rescuing BCR-deficient, preapoptotic GC B cells from apoptosis, and that EBV infection renders the cells independent from survival signals normally supplied by a BCR. The monoclonal LCLs represent valuable models for early stages of lymphoma development in classic HL and PTLD.
Introduction
Epstein-Barr virus (EBV) is a human γ-herpes virus that infects greater than 90% of humans worldwide and establishes a latent infection in B cells. Although the virus is normally a harmless passenger, EBV is also associated with several malignancies, including B-cell lymphomas.1 EBV is found in the lymphoma cells in nearly all cases of endemic Burkitt lymphoma and posttransplantation lymphoproliferative disease (PTLD), whereas it is associated with Hodgkin lymphoma (HL) and sporadic Burkitt lymphoma in about 40% and 20% to 30% of cases, respectively.1 Gene expression and immunoglobulin (Ig) V gene studies revealed that each of these lymphomas originates from germinal center (GC) B cells, indicating a specific role of EBV-infected GC B cells in the development of these lymphomas.1-6
Normally, B cells are stringently selected for expression of a functional B-cell receptor (BCR) throughout their life, and loss of BCR expression leads to rapid cell death.7 GC B cells are particularly apoptosis prone, and, if such cells acquire disadvantageous mutations (eg, nonsense mutations or mutations decreasing the affinity to the immunizing antigen) through somatic hypermutation, they are very efficiently eliminated.8 Surprisingly, in about a quarter of cases of classic HL as well as in a fraction of PTLD cases somatic V gene mutations were identified that rendered originally functional Ig gene rearrangements nonfunctional.3-6 From these findings and additional features of the 2 types of lymphomas, it has been speculated that the tumor cells in HL generally and in PTLD in a fraction of cases are derived from preapoptotic GC B cells that were rescued from apoptosis by some transforming event.2-6 One of the EBV-encoded genes expressed in these lymphomas, the latent membrane protein 2A (LMP2A), can mimic a BCR and leads to the survival of BCR-deficient immature B cells in transgenic mouse models.9-11 From this, it has been speculated that, in EBV-positive HL and PTLD, EBV might play an important role in lymphoma pathogenesis by rescuing BCR-deficient GC B cells from death.3,5 This idea implies that LMP2A does not only allow the survival of BCR-deficient immature murine B cells, but also that LMP2A may even rescue human BCR-deficient GC B cells on infection by EBV.
Because EBV can transform B cells in vitro, giving rise to lymphoblastoid cell lines (LCLs), we wanted to find out whether EBV can rescue BCR-deficient GC B cells on infection in vitro (first attempts to transform GC B cells with EBV were performed by Gregory et al,12 but these cell lines were studied for only a few weeks, and clonality or Ig gene mutation pattern was not determined). We show here that EBV can indeed rescue not only BCR-positive, but also BCR-deficient GC B cells, supporting a central role of EBV in early stages of the pathogenesis of HL and PTLD and providing an explanation of how BCR-less GC B cells can give rise to lymphoma clones in these malignancies.
Materials and methods
Purification of human GC B cells
GC B cells were isolated from human tonsils from patients undergoing routine tonsillectomy. Approval was obtained from the University of Duisburg-Essen Medical School Institutional Review Board, for these studies. Informed consent was provided according to the Declaration of Helsinki. Mononuclear cells were obtained by Ficoll density centrifugation (Amersham, Freiburg, Germany). GC centroblasts were enriched by magnetic cell separation with anti–CD77–fluorescein isothiocyanate (FITC; Becton Dickinson, BD, Heidelberg, Germany) and anti-FITC microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Magnets, columns, and buffers were precooled at 4°C, and all purification steps were performed on ice to prevent rapid apoptosis of GC B cells.
Transformation of isolated centroblasts and cell culture
EBV infection was performed with 5 mL supernatant of the marmoset cell line B95-8 per 107 B cells. Twenty-four hours after infection the cells were seeded into 96-well plates at 50, 100, 500, and 103 cells per well in either 100% LCL culture supernatant from a cell line that was split 1:2 24 hours in advance or on autologous feeder cells.13 Some additional cell lines were established through limiting dilution cloning of cells proliferating in bulk culture for up to 2 weeks. Uninfected centroblasts were seeded at 103 cells per well into 96-well plates in LCL supernatant or on feeder cells as a control for the outgrowth of in vivo EBV-infected cells. We never obtained lines from these controls. Autologous feeder cells were prepared from tonsillar mononuclear cells that were incubated with 50 μg/mL mitomycin for 30 minutes at 37°C in an atmosphere containing 5% CO2. Cells were washed and resuspended in RPMI 1640 (Gibco, Karlsruhe, Germany) containing cyclosporine A (50 mg/mL). A 96-well plate with feeder cells but without addition of isolated B cells confirmed that the mitomycin- and cyclosporin-treated feeder cells could not proliferate. Outgrowing cell clones were transferred into 24-well plates 2 to 3 weeks after EBV infection.
Cell lines and culture
The marmoset B95-8 cell line was used for EBV production. BL41 is a Burkitt lymphoma cell line. LCL 940410, LCL 031006, LCL 030515, LCL 970402, and IMEBV are LCLs established from peripheral blood B cells of healthy donors. All cell lines and centroblast-derived LCLs (CB)–LCLs were grown in RPMI-1640 with Glutamax-1 (Gibco) supplemented with 10% fetal calf serum and 100 U/mL penicillin/streptomycin at 37°C in an atmosphere containing 5% CO2.
Immunofluorescence staining
Cytospins with 5 × 104 cells were dried overnight and fixed for 7 minutes with cooled acetone (-20°C). Cells were incubated for 1 hour with EBV nuclear antigen 2 (EBNA2; clone PE2; Dako, Glostrup, Denmark), LMP1 (clone CS1-4; Dako), or B-cell lymphoma 6 (BCL6; clone PG-B6; Dako) monoclonal antibodies at a dilution of 1:100, 1:100, or 1:10, respectively. Cells were washed 3 times for 5 minutes with phosphate-buffered saline (PBS). The secondary anti–mouse IgG Alexa 594 antibody (Invitrogen GmbH, Karlsruhe, Germany) was diluted 1:500 in PBS/0.5% bovine serum albumin (BSA). Cells were incubated for 1 hour and washed 3 times for 5 minutes with PBS. Hoechst dye was diluted to 20 μg/mL, and cells were stained for 10 minutes and washed 3 times for 5 minutes with PBS. All incubations and washing steps were performed at room temperature in a wet chamber in the dark. Cytospins were closed with polyvinylalcohol medium with Dabko (Sigma-Aldrich, Steinheim, Germany) and analyzed with a fluorescence microscope. For flow cytometry, 5 × 105 cells were stained with anti–CD77-FITC (clone 5B5; BD) or anti–CD38-FITC (clone HIT2; BD) for 10 minutes on ice. Cells were washed and resuspended in PBS/0.5% BSA for analysis with a fluorescence-activated cell sorter (FACS)–Canto (BD).
Ig gene characterization
VH, Vλ, and Vκ gene family–specific polymerase chain reaction (PCR) and primers were described earlier.5,14 Briefly, VH, and Vκ gene rearrangements were detected by combinations of primers binding to the leader regions of VH or Vκ genes with primer mixes for the respective J gene segments, and Vλ gene rearrangements were amplified by Vλ framework region I family–specific primers together with primers for the Jλ genes. DHJH rearrangements and a fragment indicative for germ line configuration of the IgH locus were amplified as described, using 7 DH gene family–specific primers together with JH gene primers.14
Liposome staining
Anti-Igκ (clone G20-193) and anti-Igλ (clone JDC-12) antibodies were obtained from BD, Heidelberg, Germany. These antibodies were then coupled to streptavidin (Squarix GmbH, Marl, Germany). Cells were preincubated with 1% polyglobulin N in PBS/5% BSA (Bayer AG, Leverkusen, Germany), to prevent unspecific binding of the antibodies and stained with cyanine 5 (Cy5)–filled, biotin-coupled fluorescent liposomes as described.15
Quantitative RT-PCR
Total RNA was isolated according to the spin protocol of the RNeasy Mini kit (Qiagen GmbH, Hilden, Germany), including an additional on-column DNA digestion step. cDNA was synthesized with the Omniscript RT kit (Qiagen GmbH). Reverse transcriptase (RT)–PCR was performed in triplicates. Primers and 6-carboxyfluorescein (FAM)–labeled probes for Cκ (Hs00415042_m1), Cμ (Hs00383067_g1), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Hs99999905_m1) were obtained as assays on demand, and one assay on design each was generated for the 2 Cα genes (CαF, 5′-GCTACAGCGTGTCCAGTGT-3′; CαR, 5′-GGCAGCAGGTGGACCTC-3′; CαFAM, 5′-ATGTGTTTCCGGATTTTG-3′) the 4 Cγ genes (CγF, 5′-CAAGTGCAAGGTCTCCAACAAAG-3′;CγR, 5′-GGCTGACCTGGTTCTTGGT-3′;CγFAM, 5′-CAGCCCCGAGAACCA-3′), and the 4 Cλ genes (CλF, 5′-AGCCAACAAGGCCACACT-3′; CλR, 5′-CCTTCCAGGCCACTGTCA-3′;CλFAM, 5′-TCCCGGGTAGAAGTCACT-3′) (Applera GmbH, Darmstadt, Germany). cDNA was amplified with TaqMan Universal PCR Master Mix, No AmpErase UNG (Applera GmbH). The amplification program consisted of 60 seconds at 95°C, followed by 45 cycles of 15 seconds at 95°C and 60 seconds at 60°C. GAPDH was used as endogenous control. cDNA prepared from magnetic-activated cell sorting (MACS)–isolated CD19+ peripheral blood B cells from 2 donors was pooled and used as a calibrator for Ig expression. To compare the transcript levels in the LCLs with those in polyclonal CD19+ B cells, we determined the frequencies of B cells expressing the various heavy chain isotypes and κ or λ light chains by flow cytometry. The mean values 83.5% IgM+, 9% IgG+, 7.5% IgA+, 57.5% Igκ+, and 42.5% Igλ+, which are similar to published data,16 were used for the calculations. Controls of cDNA synthesis without reverse transcriptase were consistently negative.
Western blotting
Immunoblotting was performed under reducing conditions. Fresh cell lysates were heated at 70°C for 5 minutes and separated by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, blocked with 4% milk (Töpfer, Dietmannsried, Germany) for 1 hour at room temperature. Incubation with rat anti-LMP2A (clone 14B7) was performed at a dilution of 1:10 in milk at 4°C overnight. Donkey antirat antibody labeled with horseradish peroxidase (A9542; Sigma-Aldrich, Schnelldorf, Germany) was diluted 1:8000 in milk, and incubation was performed for 2 hours at room temperature. Visualization was performed with enhanced chemiluminescence (ECL) Plus detection reagent (Amersham).
Results
Establishment and characterization of monoclonal LCLs from GC B cells
To study the role of EBV in the rescue of BCR-deficient GC B cells during early steps of lymphoma pathogenesis, we isolated human CD77+ GC B cells (centroblasts) from tonsils of 3 patients undergoing routine tonsillectomy, infected the cells with EBV, and established stable LCLs. The purity of isolated CD77+ cells was 97% to 99% (data not shown). Time until EBV infection of isolated cells was crucial, because greater than 75% of human centroblasts die within the first 16 hours of in vitro culture.8 Therefore, the infection was performed immediately after cell isolation (a maximum of 4.5 to 5.5 hours after the operation). To obtain monoclonal cell lines the cells were either seeded at 103, 500, 100, or 50 cells/well into 96-well plates 24 hours after infection or at 2, 1, and 0.5 cells/well 2 weeks after infection. Cell lines were expanded from plates with cells proliferating in less than one third of wells, because these should be mostly monoclonal.17
The validation of clonality of CB-LCLs was performed with a VH gene family–specific PCR for VH gene rearrangements and sequence analysis that identified 62 monoclonal CB-LCLs from the 3 donors (Table 1). In 61 of the cell lines, an in-frame VH gene rearrangement was obtained; twelve of these carried in addition an out-of-frame VH gene rearrangement on the second allele, and one line harbored 2 in-frame VHDHJH joints. The 62 cell lines were further analyzed by a PCR that amplifies DHJH joints and a fragment indicative of germ line configuration of the IgH locus, showing that none of the lines had more than 2 distinct IgH alleles, thereby confirming their monoclonality (data not shown). Sixty of the 62 CB-LCLs carried somatically mutated VH gene rearrangements with a mean mutation frequency of 4.2% (range, 0.5%-15.5%). Two monoclonal CB-LCLs were unmutated and may derive from EBV transformation of GC founder cells that are often still unmutated.18 The results of the mutation analysis are hence in line with a derivation of the CB-LCLs from GC B cells. The derivation of the CB-LCLs from in vitro EBV-infected B cells and not from GC B cells that were already EBV infected in vivo was ascertained by a PCR analysis that distinguishes wild-type and the B95-8 strain of EBV (data not shown).19
Ig light chain surface expression of 62 CB-LCLs from three different donors
Donor of GC B cells . | No. of monoclonal cell lines . | No. of slg κ+ cell lines . | No. of slg λ+ cell lines . | No. of slg- cell lines . |
|---|---|---|---|---|
| A | 15 | 7 | 1 | 7 |
| B | 24 | 13 | 1 | 10 |
| C | 23 | 8 | 4 | 11 |
Donor of GC B cells . | No. of monoclonal cell lines . | No. of slg κ+ cell lines . | No. of slg λ+ cell lines . | No. of slg- cell lines . |
|---|---|---|---|---|
| A | 15 | 7 | 1 | 7 |
| B | 24 | 13 | 1 | 10 |
| C | 23 | 8 | 4 | 11 |
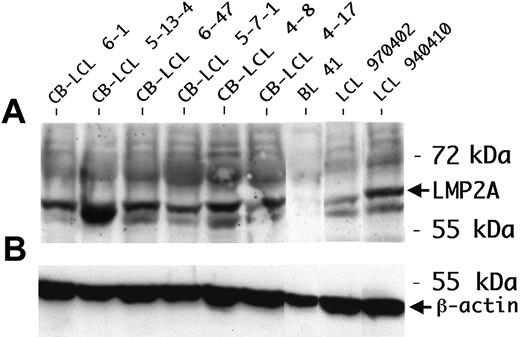
We performed a phenotypic characterization of the CB-LCLs, in comparison to tonsillar centroblasts and LCLs established from peripheral blood B cells (PB-LCLs). Expression of LMP1, LMP2A, and EBNA2 was detected in all CB-LCLs analyzed, demonstrating that the cells show a typical EBV latency III gene expression pattern (Table 2; Figure 1). CD77 expression was still detectable at low levels on 3 of 8 CB-LCLs analyzed, but also 1 of 3 PB-LCLs showed low CD77 expression, indicating that this marker may occasionally be induced in PB-LCLs. The GC B-cell marker CD38 was expressed not only on CB-LCLs, but also on PB-LCLs, in line with previous reports that this activation marker is expressed in LCLs (Table 2).20 The GC B-cell–specific transcription factor Bcl-6 was down-regulated in the CB-LCL, presumably because of LMP1 expression, which, by mimicking an active CD40 receptor, is known to lead to Bcl-6 down-regulation.21 Thus, on EBV infection, centroblasts partially down-regulate expression of typical GC B-cell markers.
Phenotypic characterization of CB-LCLs
Cell type . | CD77 . | CD38 . | Bcl-6 . | LMP1 . | LMP2A . | EBNA2 . |
|---|---|---|---|---|---|---|
| CD77+ B cells | 3/3 | 1/1* | ND* | ND | ND | ND |
| PB-LCLs | 1/3† | 3/3 | 0/3 | 3/3 | 2/2 | 3/3 |
| CB-LCLs | 3/8† | 8/8 | 0/7 | 7/7 | 6/6 | 7/7 |
Cell type . | CD77 . | CD38 . | Bcl-6 . | LMP1 . | LMP2A . | EBNA2 . |
|---|---|---|---|---|---|---|
| CD77+ B cells | 3/3 | 1/1* | ND* | ND | ND | ND |
| PB-LCLs | 1/3† | 3/3 | 0/3 | 3/3 | 2/2 | 3/3 |
| CB-LCLs | 3/8† | 8/8 | 0/7 | 7/7 | 6/6 | 7/7 |
Monoclonal CB-LCLs were analyzed for expression of CD77 and CD38 by flow cytometry; for expression of Bcl-6, LMP1, and EBNA2, by immunofluorescence staining on cytospins; and for expression of LMP2A, by Western blot analysis. As a control, LCLs established from peripheral blood B cells were also analyzed. The number of positive samples per samples analyzed is indicated.
ND indicates not determined.
CD77 is expressed in the positively scored cell lines at low levels
Screening for BCR-deficient CB-LCLs
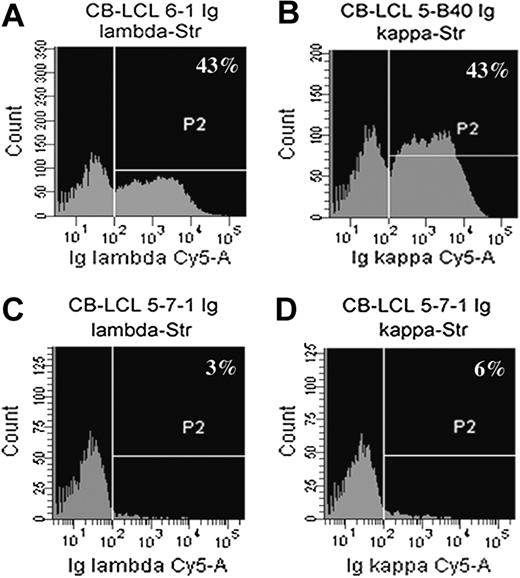
In search for Ig-negative cell lines we determined BCR surface expression of all monoclonal CB-LCLs by flow cytometry with anti-κ and anti-λ light chain antibodies. Because the EBV-encoded transcription factor EBNA2 leads to the down-regulation of Ig heavy chain transcription,22 we used fluorescent liposomes that increase detection sensitivity of surface molecules by 100- to 1000-fold.15 Surprisingly, almost half of the monoclonal CB-LCLs showed no detectable BCR expression (Table 1; Figure 2). We amplified and sequenced the rearranged heavy and light chain genes of the 28 surface BCR-negative cell lines and identified two cell lines that carry somatic mutations rendering the originally functional VH gene rearrangement nonfunctional (Figure 3A,C). In-frame, potentially functional VL gene rearrangements were obtained for all 28 cell lines.
One of the Ig-crippled cell lines (CB-LCL 4-17) harbored a nonsense mutation at the beginning of framework region 3 (FR3), and the other line (CB-LCL 5-7-1) showed deletions of 3 and 51 bp within FR3. Because FR3 is highly conserved and because larger deletions within this region have never been found in sIg+ memory B cells, it is obvious that this deletion of almost half of the FR3 is incompatible with expression of a functional BCR.23,24 The second allele in both crippled cell lines carried a DHJH gene rearrangement (Figure 3B,D), ruling out the presence of a functional VH gene rearrangement on the second IgH allele. Thus, we identified 2 CB-LCLs with destructive VH gene mutations that prevent the expression of a BCR in these lines. LMP2A protein expression as the anticipated survival signal could be validated in these cell lines (Table 2; Figure 1). Analysis of 23 CB-LCLs (including one of the crippled lines) for ongoing hypermutation activity revealed lack of intraclonal V gene diversity in 21 of the lines. Importantly, the crippled line CB-LCL 4-17 also lacked intraclonal diversity, strongly arguing against acquisition of the crippling mutations in vitro (data not shown).
LMP2A expression in CB-LCLs. (A) LMP2A protein expression was determined by Western blot analysis. The peripheral blood B-cell–derived LCL 970402 and LCL 940410 were used as positive controls. The protein lysate of the EBV-negative cell line BL41 was blotted as a negative control. All LCLs analyzed were consistently positive for LMP2A. (B) β-actin was used as a loading control.
LMP2A expression in CB-LCLs. (A) LMP2A protein expression was determined by Western blot analysis. The peripheral blood B-cell–derived LCL 970402 and LCL 940410 were used as positive controls. The protein lysate of the EBV-negative cell line BL41 was blotted as a negative control. All LCLs analyzed were consistently positive for LMP2A. (B) β-actin was used as a loading control.
Flow cytometric analysis of Ig light chain surface expression with fluorescent liposomes. Cells were stained with anti-Igκ– and anti-Igλ–streptavidin antibodies and then incubated with biotin-coupled, Cy5-filled liposomes. (A-D) Shown are examples of a sIgλ+ cell line (A; CB-LCL 6-1), a sIgκ+ cell line (B; CB-LCL 5-B40), and a sIg-negative cell line (C-D; CB-LCL 5-7-1).
Flow cytometric analysis of Ig light chain surface expression with fluorescent liposomes. Cells were stained with anti-Igκ– and anti-Igλ–streptavidin antibodies and then incubated with biotin-coupled, Cy5-filled liposomes. (A-D) Shown are examples of a sIgλ+ cell line (A; CB-LCL 6-1), a sIgκ+ cell line (B; CB-LCL 5-B40), and a sIg-negative cell line (C-D; CB-LCL 5-7-1).
Down-regulation of Ig transcription in CB-LCLs
We wondered whether the lack of sIg expression in the 26 CB-LCLs without detectable crippling V gene mutations was due to down-regulation of Ig heavy and/or light chain gene transcription or could be rather explained by posttranscriptional defects (eg, inability of proper heavy and/or light chain folding or pairing, perhaps because of destructive replacement mutations). Down-regulation of heavy chain (but not light chain) transcription in LCLs has indeed been described.22 We therefore performed quantitative real-time RT-PCR for Ig heavy and light chain constant region transcripts for 8 of the surface BCR- and 3 BCR+ CB-LCLs. The Ig transcription levels in these cell lines were compared with the respective levels in peripheral blood CD19+ B cells. We observed Ig heavy chain transcript levels in the sIg-negative CB-LCLs 1.6- to 100-fold lower than the expression levels of normal peripheral blood B cells. Several of the cell lines expressed 2 Ig isotype transcripts (Table 3). This might be due to intraclonal class-switching in culture, to germ line transcription of CH genes, or to transcription of DHJH joints on the second allele. In 2 of the lines, expression of 2 isotypes might also reflect the fact that each of these contained 2 VH gene rearrangements. CB-LCL 5-5-1 harbored an in-frame and an out-of-frame VH gene rearrangement, and CB-LCL 4-8 carried 2 in-frame VH gene rearrangements. Rare human B cells carrying 2 in-frame VH gene rearrangements have been described.25 Although there is a relatively low level of heavy chain transcription in the sIg- CB-LCLs, a similar level was also observed in the sIg+ CB-LCLs (Table 3). Thus, down-regulation of Ig heavy chain transcription does not easily explain the lack of sIg expression.
Quantitative RT-PCR of the constant regions of Ig heavy and light chains from CB-LCLs and PB-LCLs
LCLs . | IgM, % . | IgA, % . | IgG, % . | Igκ, % . | Igλ, % . | In-frame VL gene rearrangements . |
|---|---|---|---|---|---|---|
| sIg– CB-LCLs | ||||||
| CB6-14 | 16 | < 0.01 | < 0.01 | < 0.01 | 8 | Vλ2-11/Jλ1 |
| CB5-17-3 | < 0.01 | < 0.01 | 9 | 1 | 0.04 | Vκ3-20/Jκ3 |
| CB5-B3 | < 0.01 | < 0.01 | 32 | < 0.01 | 14 | Vλ1-47/Jλ7 |
| CB5-5-1 | 0.8 | < 0.01 | 32 | < 0.01 | 0.1 | Vκ3-15/Jκ5 |
| CB5-B4 | < 0.01 | < 0.01 | 9 | 6* | 3 | Vλ2-11/Jλ1 |
| CB5-13-4 | 0.1 | < 0.01 | 10 | 1* | 20 | Vλ2-14/Jλ2/3 |
| CB4-21 | 1 | < 0.01 | 63 | 0.8 | 128 | Vλ3-1/Jλ2/3 |
| CB4-8 | 9 | < 0.01 | 18 | 2* | 40 | Vλ3-21/Jλ1 |
| sIg+ CB-LCLs | ||||||
| CB6-1 | 0.3 | 3 | < 0.01 | < 0.01 | 6 | Vλ3-19/Jλ2/3 |
| CB6-44 | 0.4 | < 0.01 | 15 | 0.2 | 11 | Vλ1-51/Jλ2/3 |
| CB5-B40 | 30 | 0.02 | < 0.01 | 14 | 0.1 | Vκ1-9/Jκ4 |
| PB-LCLs† | ||||||
| IMEBV | 5 | 0.1 | 5 | 0.7 | 0.03 | ND |
| LCL 031006 | 0.5 | < 0.01 | 0.3 | < 0.01 | 3 | ND |
| LCL 030515 | 8 | 0.02 | < 0.01 | 0.09 | 2 | ND |
| LCL 940410 | < 0.01 | 0.2 | < 0.01 | < 0.01 | 0.7 | ND |
LCLs . | IgM, % . | IgA, % . | IgG, % . | Igκ, % . | Igλ, % . | In-frame VL gene rearrangements . |
|---|---|---|---|---|---|---|
| sIg– CB-LCLs | ||||||
| CB6-14 | 16 | < 0.01 | < 0.01 | < 0.01 | 8 | Vλ2-11/Jλ1 |
| CB5-17-3 | < 0.01 | < 0.01 | 9 | 1 | 0.04 | Vκ3-20/Jκ3 |
| CB5-B3 | < 0.01 | < 0.01 | 32 | < 0.01 | 14 | Vλ1-47/Jλ7 |
| CB5-5-1 | 0.8 | < 0.01 | 32 | < 0.01 | 0.1 | Vκ3-15/Jκ5 |
| CB5-B4 | < 0.01 | < 0.01 | 9 | 6* | 3 | Vλ2-11/Jλ1 |
| CB5-13-4 | 0.1 | < 0.01 | 10 | 1* | 20 | Vλ2-14/Jλ2/3 |
| CB4-21 | 1 | < 0.01 | 63 | 0.8 | 128 | Vλ3-1/Jλ2/3 |
| CB4-8 | 9 | < 0.01 | 18 | 2* | 40 | Vλ3-21/Jλ1 |
| sIg+ CB-LCLs | ||||||
| CB6-1 | 0.3 | 3 | < 0.01 | < 0.01 | 6 | Vλ3-19/Jλ2/3 |
| CB6-44 | 0.4 | < 0.01 | 15 | 0.2 | 11 | Vλ1-51/Jλ2/3 |
| CB5-B40 | 30 | 0.02 | < 0.01 | 14 | 0.1 | Vκ1-9/Jκ4 |
| PB-LCLs† | ||||||
| IMEBV | 5 | 0.1 | 5 | 0.7 | 0.03 | ND |
| LCL 031006 | 0.5 | < 0.01 | 0.3 | < 0.01 | 3 | ND |
| LCL 030515 | 8 | 0.02 | < 0.01 | 0.09 | 2 | ND |
| LCL 940410 | < 0.01 | 0.2 | < 0.01 | < 0.01 | 0.7 | ND |
The transcript levels of Ig heavy and light chains are normalized to the Ig expression levels of CD19+ peripheral blood B cells, set to 100%. ND indicates not determined.
The VκJκ rearrangements of CB-LCL 5-B4, 5-13-4, and 4-8 are out of frame but show somatic mutations as an indication that these rearrangements are not inactivated by rearrangement of the κ deleting element, so that they might still be transcribed
Lines IMEBV and LCL031006 are biclonal. The monoclonal lines LCL 030515 and LCL 940410 lacked detectable sIg expression
Expression levels were also determined for Ig light chains. Two of the sIg- cell lines showed light chain transcript levels comparable to CD19+ peripheral blood B cells (40% and 128%), and 5 cell lines had a 5- to 100-fold lower level. Only one cell line with an in-frame VκJκ rearrangement had very low κ transcript levels (< 0.01%), which likely explains the lack of sIg expression (Table 3). In 2 sIg- and 1 sIg+ CB-LCL with in-frame and mutated VκJκ joints, indication for low-level transcription of a Cλ gene was obtained, although no VλJλ joint could be amplified, perhaps reflecting Cλ germ line transcription.
Taken together, in 7 of the 8 sIg- cell lines analyzed the sIg-deficiency cannot be assigned to a lack of Ig transcription, but it is probably due to posttranscriptional effects. These presumably include destructive somatic replacement mutations that prevent proper folding of Ig heavy or light chains, thereby leading to the inability to express a functional BCR on the cell surface. Western blot analysis of sIg-negative cell lines indeed identified a line with easily detectable heavy chain protein, suggesting an impairment of surface expression of this polypeptide.26 However, 2 of 4 monoclonal PB-LCLs analyzed for sIg expression also lacked detectable BCR expression (data not shown), implying that in some instances posttranscriptional mechanisms other than somatic mutations can lead to sIg down-regulation in EBV-transformed cell lines, because peripheral blood B cells are all sIg positive.
VHDHJH and DHJH gene rearrangements of the 2 Ig-crippled cell lines. (A,C) Shown are the VHDHJH gene sequences of CB-LCL 4-17 and CB-LCL 5-7-1 from the 5′ end of FR3 to the start of the JH primer sequence. (B,D) The DHJH gene sequences are depicted from upstream of the recombination signal sequences (RSS; composed of nonamer, 12 base pair [bp] spacer and heptamer) 5′ of the DH segments to the start of the JH primer. (A,B) CB-LCL 4-17, (C,D) CB-LCL 5-7-1. N represents N nucleotides. (A) The 2 point mutations giving rise to a nonsense codon in CB-LCL 4-17 are located within the first codon of FR3 and are underlined. (C) CB-LCL 5-7-1 contains a 3-bp and a 51-bp deletion in FR3 that are indicated by black boxes. The 51-bp deletion renders FR3 of CB-LCL 5-7-1 nonfunctional. In this sequence a DH segment could not be identified but may be part of the sequence marked by N.
VHDHJH and DHJH gene rearrangements of the 2 Ig-crippled cell lines. (A,C) Shown are the VHDHJH gene sequences of CB-LCL 4-17 and CB-LCL 5-7-1 from the 5′ end of FR3 to the start of the JH primer sequence. (B,D) The DHJH gene sequences are depicted from upstream of the recombination signal sequences (RSS; composed of nonamer, 12 base pair [bp] spacer and heptamer) 5′ of the DH segments to the start of the JH primer. (A,B) CB-LCL 4-17, (C,D) CB-LCL 5-7-1. N represents N nucleotides. (A) The 2 point mutations giving rise to a nonsense codon in CB-LCL 4-17 are located within the first codon of FR3 and are underlined. (C) CB-LCL 5-7-1 contains a 3-bp and a 51-bp deletion in FR3 that are indicated by black boxes. The 51-bp deletion renders FR3 of CB-LCL 5-7-1 nonfunctional. In this sequence a DH segment could not be identified but may be part of the sequence marked by N.
Discussion
We show here that EBV can rescue GC B cells from apoptosis and transform them into stable LCLs even if these cells have lost a functional BCR through somatic mutations in the GC. Thus, as previous work in a transgenic mouse model showed that LMP2A can allow survival of immature B cells, the present study demonstrates that EBV can even rescue apoptosis-prone human GC B cells.10,11
Although we identified 2 clearly crippled cell lines, the quantitative RT-PCR for Ig transcript levels indicates that, among the other sIg-negative lines, there are additional ones that have a nonfunctional BCR, such as because of posttranscriptional defects, such as an impairment of correct protein folding as a result of amino acid exchanges in FRs. Thus, EBV can replace the survival signals normally supplied by the BCR, presumably through expression of LMP2A, which mimics the BCR. Whereas EBV is found in nearly all cases of PTLD, only 40% of cases of HL in the Western world are EBV positive. Therefore, it remains to be determined which pathogenetic mechanisms might replace the function of EBV in the EBV-negative cases of HL. For instance, Hodgkin and Reed-Sternberg (HRS) cells consistently show constitutive nuclear factor κB (NFκB) activity which might be caused not only by EBV but also by other means, such as somatic mutations in the IKBA gene.27
It is controversially discussed whether in vivo EBV can directly infect GC B cells or gains access to this B-cell compartment through activation of EBV-infected naive B cells.2,19,28 The experiments described here (and similar results in the accompanying article by Mancao et al26 in this issue of Blood beginning on page 4339, and also by Chaganti et al29 ) not only show that EBV can transform GC B cells in vitro but also that GC B cells that acquired destructive mutations and therefore activated the apoptosis program can still be rescued by EBV. Thus, in light of indications that EBV infection may be incompatible with a “normal” GC B-cell differentiation program, the present results suggest that EBV infection of (crippled) GC B cells may be able to rescue these cells, and that such cells in rare instances may give rise to HL or PTLD tumor clones.
In EBV-positive PTLD, the tumor cells usually express the latency III program. The expression of the same set of EBV latent proteins in the CB-LCLs make them ideal in vitro models to study early steps in the pathogenesis of BCR-positive as well as BCR-negative PTLD cases. In EBV-associated HL, however, a latency II expression program is observed. Nevertheless, it may well be that EBV-infected HRS-cell precursors in the GC may also show a latency III program and that the program switches to latency II in the course of malignant transformation, similar to the proposed switch of the EBV latency program when EBV-infected naive B cells differentiate into GC B cells.28 Although a latency III profile of EBV-infected GC B cells has not been described yet, the viral gene expression pattern of such cells is indeed still a matter of debate. On the one hand, it has been shown that in healthy virus carriers EBV-positive GC B cells express transcripts of LMP1 and LMP2A, but not EBNA2, consistent with a latency II profile.2,28,30 On the other hand, Araujo et al31 analyzed EBV-positive GC B cells from tonsils of healthy children from Brazil and Germany and did not find detectable LMP1 expression in 9 of 11 tonsils. EBNA2 was absent in all EBV-harboring GC B cells in that study. Moreover, EBV-infected GC B cells in infectious mononucleosis express only EBNA2 without LMP1 and LMP2A.2,19 In conclusion, these studies indicate that, depending on the particular situation, EBV-infected GC B cells may use different latency programs. Alternatively, genetic or epigenetic changes occurring in HRS-cell precursors may influence the EBV latency status, promoting a latency II in the established HRS-cell clones in vivo.
Taken together, the results presented here support the idea that EBV plays a major role in the pathogenesis of HL and PTLD, presumably by rendering the infected cells independent from BCR-derived survival signals. The GC B-cell–derived cell lines may become valuable in vitro models not only to study the influence of EBV on GC B cells in vitro but also to disclose the effect of LMP2A on receptorless human B cells and to reveal pathogenetic mechanisms of EBV in lymphomagenesis.
Prepublished online as Blood First Edition Paper, August 30, 2005; DOI 10.1182/blood-2005-06-2342.
Supported by the Deutsche Forschungsgemeinschaft (through KU 1315/3-1) and by the Interne Forschungsförderung Essen (IFORES) program of the University of Duisburg-Essen, Medical School, Essen, Germany.
D.B. and R.K. designed the research; D.B. and J.K. performed the research; C.U. contributed vital material; D.B., J.K., and R.K. controlled and analyzed the data; D.B. and R.K. wrote the manuscript; and all authors checked the final version of the manuscript.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Michaela Fahrig, Gwen Lorenz, and Kerstin Heise for excellent technical assistance; Mirela Stecki and Sabine Albrecht for sequencing work; and Alexander Scheffold for providing fluorescent liposomes. We thank George Klein and Gerald Niedobitek for providing LCLs established from peripheral blood, Ines Pfeil for helpful advice, and Wolfgang Hammerschmidt and Christoph Mancao for sharing unpublished data and stimulating discussions.
The authors declare that they have no competing financial interest.

![Figure 3. VHDHJH and DHJH gene rearrangements of the 2 Ig-crippled cell lines. (A,C) Shown are the VHDHJH gene sequences of CB-LCL 4-17 and CB-LCL 5-7-1 from the 5′ end of FR3 to the start of the JH primer sequence. (B,D) The DHJH gene sequences are depicted from upstream of the recombination signal sequences (RSS; composed of nonamer, 12 base pair [bp] spacer and heptamer) 5′ of the DH segments to the start of the JH primer. (A,B) CB-LCL 4-17, (C,D) CB-LCL 5-7-1. N represents N nucleotides. (A) The 2 point mutations giving rise to a nonsense codon in CB-LCL 4-17 are located within the first codon of FR3 and are underlined. (C) CB-LCL 5-7-1 contains a 3-bp and a 51-bp deletion in FR3 that are indicated by black boxes. The 51-bp deletion renders FR3 of CB-LCL 5-7-1 nonfunctional. In this sequence a DH segment could not be identified but may be part of the sequence marked by N.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/13/10.1182_blood-2005-06-2342/2/m_zh80240588070003.jpeg?Expires=1765891876&Signature=JSQq0VnWu0GAbVAQFBezZv8vyLco1gp3ryj~kQ0wrzB1oT9jWyKFFuv7MgqpkqTt~x6o09PHD-cSE~Q2GOcaa3CJYVyjJIGqAB~jfHE8xlshxyOwYm7A4InMPc89pLKKQRZfZvt2td~MrgNM1NaowHbrxYAXv~4wp1SAKhB3ph5acgB4fMJliEv6PMjXzaz8E3WvQq5OaTRMiB2wPTgruyyYPLuJwnd-NCvzKSkS1atcbHZe69oJcKTZlJkFkqWvRKAU44zlAYJZtFShLa5SDWqavLJPLwTx1kn23FP0T1SR~XS~~pmeA2MmHl1QefZrHvqKTobMaJZUE3A~LVh2LA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal