Abstract
Vascular endothelial growth factor receptor-3 (VEGFR-3) plays a key role for the remodeling of the primary capillary plexus in the embryo and contributes to angiogenesis and lymphangiogenesis in the adult. However, VEGFR-3 signal transduction pathways remain to be elucidated. Here we investigated VEGFR-3 signaling in primary human umbilical vein endothelial cells (HUVECs) by the systematic mutation of the tyrosine residues potentially involved in VEGFR-3 signaling and identified the tyrosines critical for its function. Y1068 was shown to be essential for the kinase activity of the receptor. Y1063 signals the receptor-mediated survival by recruiting CRKI/II to the activated receptor, inducing a signaling cascade that, via mitogen-activated protein kinase kinase-4 (MKK4), activates c-Jun N-terminal kinase-1/2 (JNK1/2). Inhibition of JNK1/2 function either by specific peptide inhibitor JNKI1 or by RNA interference (RNAi) demonstrated that activation of JNK1/2 is required for a VEGFR-3–dependent prosurvival signaling. Y1230/Y1231 contributes, together with Y1337, to proliferation, migration, and survival of endothelial cells. Phospho-Y1230/Y1231 directly recruits growth factor receptor–bonus protein (GRB2) to the receptor, inducing the activation of both AKT and extracellular signal–related kinase 1/2 (ERK1/2) signaling. Finally, we observed that Y1063 and Y1230/Y1231 signaling converge to induce c-JUN expression, and RNAi experiments demonstrated that c-JUN is required for growth factor–induced prosurvival signaling in primary endothelial cells.
Introduction
Vascular endothelial growth factor receptor-1 (VEGFR-1) (also known as Flt-1), VEGFR-2 (also known as KDR/Flk-1), and VEGFR-3 (also known as Flt-4) play a central role in vasculogenesis and angiogenesis.1,2 They are activated by the family of angiogenic factors that is composed of several members with partially overlapping function due to the promiscuity of receptor recognition. Placental growth factor (PlGF) and vascular endothelial growth factor-B (VEGF-B) recognize VEGFR-1, VEGF is the ligand of both VEGFR-1 and VEGFR-2, while VEGF-C and VEGF-D each recognize and activate VEGFR-2 and VEGFR-3. These receptors are highly related, with an extracellular ligand binding site containing 7 immunoglobulin-homology segments, a single polypeptide chain transmembrane sequence, and an interrupted intracellular tyrosine kinase domain. Gene disruption experiments demonstrated that each of these receptors is essential for vascular development and angiogenesis.2 Both VEGFR-1 and VEGFR-2 act early during endothelial differentiation although with different functions: while Vegfr-1–/– embryos die for excessive endothelial proliferation, Vegfr-2–/– die with no endothelial or hematopoietic cells.3,4
The absence of Vegfr-3 does not impair the differentiation of endothelial cells or the formation of the primary vascular plexus; rather, it affects the remodeling of the primary vascular network.5 In adults, VEGFR-3 is expressed mainly in lymphatic endothelial cells.6,7 VEGFR-3 is also expressed in quiescent endothelial cells of fenestrated capillaries and microvessels of several tissues.8-10 It is transiently induced in vessels undergoing active angiogenesis during wound healing and in tumors.11-14 In freshly isolated human umbilical vein endothelial cells (HUVECs) VEGFR-3 is abundantly expressed and functional.15 Moreover, inactivation of VEGFR-3 during tumor-induced angiogenesis, by means of monoclonal antibody, suggested that VEGFR-3 might play a role in the maintenance of vessel integrity by negatively modulating VEGFR-2.13,16
Although the overall structure of VEGFR-3 split tyrosine kinase is similar to VEGFR-1 and VEGFR-2, VEGFR-3 shows few significant differences with respect to VEGFR-1 and VEGFR-2. Its extracellular domain is cleaved within the fifth immunoglobulin-like domain, and the 2 resulting polypeptides are connected by a disulfide bridge.17 It is selectively associated to α5β1 but not with αvβ3.18,19 The sequence of its C-terminal tail is expressed in 2 splice variants, due to a retroviral insertion, that generate 2 isoforms designated VEGFR-3 long, which is the predominant form in most tissues, and VEGFR-3 short, which lacks the last 65 amino acids of the cytolasmic tail.20,21 In lymphatic cells activation of VEGFR-3 induces proliferation, migration, and survival, and it has been shown to activate p42/p44 mitogen–activated protein kinase (MAPK) (extracellular signal–regulated kinase 1/2 [ERK1/2]) and AKT phosphorylation.22 Ectopically expressed VEGFR-3 in fibroblasts induced transformation by recruiting Src homology domain containing growth factor receptor–bound protein 2 (SHC-GRB2) to Y1337.23,24 Analysis of VEGFR-3 phosphorylation demonstrated that upon receptor dimerization, in addition to Y1337, other 4 tyrosine residues at the VEGFR-3 C-terminal domain are potential transphosphorylated sites,25 thus suggesting that a more complex signaling is originated from this receptor.
Here we investigated, in a systematic way, the role of VEGFR-3 tyrosine residues by site-directed mutagenesis of the putative tyrosines involved in signaling in a chimeric receptor obtained by fusion of the extracellular domain of the epidermal growth factor receptor (EGFR) with the transmembrane and intracellular tail of VEGFR-3. The analysis of the signaling pathways originating from the chimeric EGFR–VEGFR-3 (E-R3) receptor in endothelial cells allowed us to dissect the signaling that, originating from the receptor, induces a phenotypic response in endothelial cells. Besides the identification of the tyrosine residue required for VEGFR-3 kinase activation, we identified 2 additional docking sites important for the receptor signaling. Phosphotyrosine 1063 induces a survival signaling cascade that recruiting CRKI/II to the receptor induces c-JUN expression via c-JUN N-terminal kinase-1/2 (JNK1/2). Phosphorylation of tyrosine residues 1230/1231 by the direct recruiting of GRB2 to the receptor contributes to VEGFR-3–dependent proliferation, migration, and survival, which activate the classic signaling ERK1/2 and AKT. These results define VEGFR-3 signaling and permit us to understand its role in endothelial cells.
Materials and methods
DNA constructs
The full-length cDNA of human VEGFR-3 (residues 1 to 1363), kindly obtained from Dr Daniel Birnbaum (Institut National de la Santé et de la Recherche Médicale [INSERM], Marseille, France), was subcloned in pcDNA3 expression vector (Invitrogen, Carlsbad, CA) at the XbaI restriction site. VEGFR-3 mutants were performed on the VEGFR-3/pcDNA3 construct by using the Quick Change Mutagenesis Site-Directed Mutagenesis Kit (Promega, Madison, WI) according to the manufacturer's instructions. The DNA fragment coding for the N-terminal domain of human EGFR (residues 1 of 652) was amplified with the oligonucleotides (5′-GAGAAGATCTGACCAGCATGGGACCCTCCGGGACGGCCGGG-3′ and 5′-GAGAGGTACCGACAAGGATCACGATGGCGATGGACGGGATCTTAGGCCCATT-3′) from a human muscle cDNA library. The resulting fragment was then subcloned in frame with VEGFR-3 at the KpnI site, and the chimeric construct was inserted in the retrovirus vector PINCO.26 The VEGFR-3 tyrosine mutant constructs were obtained by subcloning the VEGFR-3 cDNA fragment of each site-specific mutant between the FseI-PmeI sites of the wild-type chimeric construct. The resulting mutant chimera were tested by sequence analysis. Expression vectors for CRK–Src homology domain 2 (SH2) and GRB2-SH2 were kindly provided by Dr Pier Giuseppe Pellicci (European Institute of Oncology, Milan, Italy). The cDNA coding for the dominant negatives of MAP kinase kinase-3 (MKK3), MKK4, and MKK7, cloned in the expression retrovirus vector MIGR1, was kindly obtained from Dr Guido Franzoso (University of Chicago, IL).
The retroviral cassette to express small hairpin RNA (shRNA) under the U6 promoter was described by Zippo et al.27 Oligonucleotides coding for JNK1 (oligo sense 5′-TCGAGAATGTCCTACCTTCTCTATTCAAGAGATAGAGAAGGTAGGACATTCTTTTTCTGCA-3′; oligo antisense 5′-GAAAAAGAATGTCCTACCTTCTCTATCTCTTGAATAGAGAAGGTAGGACATTC-3′), JNK2 (oligo sense 5′-TCGAGAATGTCCTACCTTCTTTATTCAAGAGATAAAGAAGGTAGGACATTCTTTTTCTGCA-3′; oligo antisense 5′-GAAAAAGAATGTCCTACCTTCTTTAT CTCTTGAATAAAGAAGGTAGGACATTC-3′), GRB2 (oligo sense 5′-TCGAGAAACGATGTGCAGCACTTAACTTGAGAAAGTGCTGCACATCGTTTCTTTTTCTGCA-3′; oligo antisense 5′-GAAAAAGAAACGATGTGCAGCACTTTCTCAAGTTAAGTGCTGCACATCGTTTC-3′), c-JUN (oligo sense 5′-TCGAGCAGCAGTTGCAAACATTTAACTTGAGAAAATGTTTGCAACTGCTGCTTTTTCTGCA-3′; oligo antisense 5′-GAAAAAGCAGCAGTTGCAAACATTTTCTCAAGTTAAATGTTTGCAACTGCTGC-3′), and green fluorescence protein (GFP) (oligo sense 5′-TCGAGCCACAAGTTCAGCGTGTCTTCAAGAGAGACACGCTGAACTTGTGGCTTTTTCTGCA-3′; oligo antisense 5′-CGGTGTTCAAGTCGCACAGAAGTTCTCTCTGTGCGACTTGCCACAAAACACCGAAAAAG-3′) were annealed and cloned into ClaI-SalI of a lentiviral vector as previously described.27
Virus preparations
A total of 7 × 106 293GPG cells28 were transfected with 24 μg plasmid constructs E-R3, mutants Y812F, Y830F, Y833F, Y1063F, Y1068F, Y1230F, Y1231F, Y1265F, Y1337F, E-R3ΔC, and PINCO vector by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The collected supernatant was ultracentrifuged in a Beckman ultracentrifuge into Ultraclear tubes (Beckman, Hialeah, FL). Infected HUVECs were assayed by flow cytometry for the expression level of the GFP protein expressed by the PINCO vector and the expression level of chimeric receptors on the cell surface. The results were confirmed by Western blot analysis using the whole-cell lysate.
Antibodies, EGF, and inhibitors
Anti-EGFR (sc-528), VEGFR-3 (sc-321), P-tyrosine (sc-7020), GRB2 (sc-255), anti–c-JUN (sc-45X), and anti-SP1 (sc-59) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antiphosphatidylinositol-3 kinase (anti-PI3K) P85 was purchased from Upstate Biotechnology (Lake Placid, NY). Anti–P-ERK1/2, P–stress-activated protein kinase (P-SAPK)/JNK (Thr183/Tyr185), P–c-JUN (Ser63), P-AKT (Ser473), anti-SAPK/JNK, anti-AKT, and anti-p44/42 MAPK were obtained from Cell Signaling Technology (Beverly, MA); anti-JNK1/2 monoclonal antibody was from BD Pharmingen (San Diego, CA). Anti-CRK was purchased from Transduction Laboratories (BD Biosciences, San Jose, CA).
Human epidermal growth factor (EGF) was purchased from Sigma (St Louis, MO). Wortmanin, LY294002, and PD98059 were purchased from Calbiochem (San Diego, CA). U0126 was purchased from Cell Signaling Technology.
Immunoprecipitates, Western blotting, recombinant proteins, and Far Western analysis
Immunoprecipitates were performed as described.15 A total of 2 × 106 subconfluent cells were starved in M199, 1% bovine serum albumin (BSA), 0.1% serum for 20 hours. Cells were stimulated for the indicated time at 37°C with 10 ng/mL EGF. After washing with ice-cold phosphate-buffered saline (PBS) containing 0.1 mM sodium orthovanadate, cells were solubilized in 1 mL lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane] HCI pH 7.4, 150 mM NaCl, 1% Nonidet P-40 [NP-40], 0.25% Nadeoxycholate, 1 mM EGTA [ethylene glycol tetraacetic acid], 1 mM EDTA [ethylenediaminetetraacetic acid], 2.5 mM Na-pyrophophate, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM Na3VO4 pH 10.0, antiprotease cocktail; Sigma), collected, and incubated on ice for 10 minutes. Cell lysate was centrifuged at 10 000g for 15 minutes, and the supernatants were precleared for 1 hour at 4°C by incubation with 15 μL protein A– or protein G–agarose (Roche, Milan, Italy). The precleared lysates were used for immunoprecipitation either with the receptor-specific antibodies (EGFR, VEGFR-3) or antibodies recognizing GRB2 or CRK. After incubation the solution was centrifuged at 200g for 4 minutes and washed 3 times with 0.5 mL lysis buffer and once with ice-cold PBS containing 1 mM Na3VO4. Immunoprecipitates were separated by 6% to 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and immunoblotted with the specified antibodies.
Glutathione S-transferase (GST)–CRK-SH2 and GST-GRB2-SH2 used for Far Western analysis and GST–c-JUN (1-79) used for the kinase assay were expressed and purified following the pGEX vector supplier's instructions (Amersham Pharmacia Biotech, Piscataway, NJ). Far Western blotting was performed as described.29 One milligram of recombinant domain fusion proteins in PBS was biotinylated at room temperature (RT) for 1 hour with 100 μg biotinamidocaproate-N-hydroxysuccinimide ester (Sigma). Biotinylated proteins were purified by gel filtration on disposable columns (BioSpin 6 Columns; Bio-Rad, Hercules, CA) according to manufacturer's instructions. The final biotinylated products were used at a concentration of 2 μg/mL in 5% BSA, 1 mM dithiothreitol (DTT) in Far Western blotting of nitrocellulose membrane. The staining was performed by a chemiluminescence assay using enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech). EGF was obtained from GIBCO-Bethesda Research Laboratories (Bethesda, MD).
DNA synthesis, chemotaxis, and sprouting assays
Mitogenic activity of HUVECs was assayed in 96-well plates. A total of 6 × 103 cells per well were grown for 16 hours in complete M199 medium. The cells were then starved for 24 hours in M199 medium containing 2% fetal bovine serum (FBS) and 0.5% BSA. Cells were then incubated with EGF at 10 ng/mL in the presence of 3H-thymidine (1 μL per well corresponding to 1 μCi [3.7 × 104 Bq] per well) and incubated for 24 hours at 37°C. They were trypsinized, and 3H-thymidine incorporation was measured by using a γ-counter (Hewlett Packard, Palo Alto, CA).
Chemotaxis activity was performed in Boyden chambers as described.15 A total of 1.5 × 106 HUVECs were grown for 16 hours in complete M199 medium. The cells were then starved for 24 hours in M199 medium containing 2% FBS and 0.5% BSA. Polyvinylpyrrolidone-free polycarbonate filters (Nucleopore; Corning Costar, Acton, MA) with a pore size of 8 μm coated with 0.1% gelatin were used. Sprouting assays were performed as described.30 Briefly, HUVECs resuspended in complete M199 culture medium containing 0.12% methylcellulose were seeded in nonadherent round-bottom 96-well plates. Single spheroids per well (800 cells per spheroid) were embedded into rat tail collagen gels (Boehringer Mannheim, Mannheim, Germany). HUVEC spheroids were resuspended in M199 containing 20% FBS and 2% methylcellulose, with or without 10 ng/mL EGF, and mixed with an equal volume of 0.4% collagen solution (rapidly transferred into 48-well plates prewarmed at 37°C and allowed to polymerase) and incubated for 18 to 24 hours. Sprouting was quantified by measuring the length of the capillary sprouts (ocular grid at × 100 magnification) that had grown out of each spheroid (5 to 10 spheroids per experimental group and experiment were measured).
Apoptosis assay
A total of 5000 cells per cm2 were grown in complete M199 medium for 16 hours on a 1% gelatin-coated 8-well Chamber slide. Cells were starved for 24 hours in 0.25% BSA M199 with and without EGF and fixed in 2% paraformaldehyde in PBS; apoptosis was assessed by terminal deoxynucleotidyl transferase deoxyuridine triphosphate (dUTP) nick end labeling (TUNEL) staining (Promega), following the manufacturer's instructions. The percentage of apoptotic cells was calculated from the number of TUNEL-positive cells divided by the total number of counted DAPI (4,6 diamidino-2-phenylindole)–stained cells.
JNK activation and peptide inhibition
JNK activity was measured with a commercially available kit (Cell Signaling Technology) according to the manufacturer's instructions. Equal amounts of proteins were shaken with c-JUN protein beads for 16 hours at 4°C. The kinase reaction was performed at 30°C for 30 minutes. The reaction was stopped by adding 3 × sample buffer followed by boiling for 5 minutes. Samples were loaded on the 10% SDS-polyacrylamide gel, and Western blots were performed with an antibody against phosphospecific P–c-JUN (Ser 63) (Cell Signaling Technology). JNK1/2 inhibition using the peptide JNKI1 was performed as described.31 Either 1 μM JNKI1 or 1 μM TAT peptide was added to cells for 1 hour prior to the EGF cell treatment.
Cell cultures
HUVECs were grown on gelatin-coated plastic Petri dishes as described.32 GPG293 adenovirus 5–transformed human embryonic kidney 293GPG packaging cells were used for the generation of retrovirus vector particles.28 The 293GPG cells were grown at 37°C and 5% CO2 in high-glucose Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated FBS, 50 U/mL penicillin-streptomycin, and 2 mM l-glutamine. Drugs selections of 293GPG cells were performed at 2 μg/mL puromycin (Sigma), 0.3 mg/mL G418 (GIBCO). All growth media were supplemented with 1 μg/mL tetracycline (Sigma).
Results
Functional analysis of VEGFR-3 tyrosine site-specific mutants
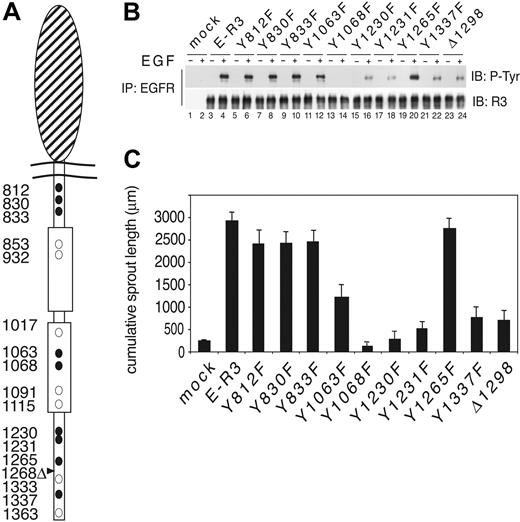
To elucidate VEGFR-3 signaling pathways in HUVECs without the interference from the VEGFR-2 receptor, we generated a VEGFR-3 chimeric receptor by fusion of the EGFR extracellular domain (amino acids 1 to 652) with the transmembrane and cytoplasmic domain of VEGFR-3 (amino acids 779 to 1363) and named it E-R3 (EGFR–VEGFR-3) (Figure 1A). This approach was possible because HUVECs do not express detectable endogenous EGFR by Western blotting or fluorescence-activated cell sorter (FACS) analysis and do not respond to EGF stimulation.33,34 To dissect VEGFR-3 signaling, the tyrosine residues potentially phosphorylated were substituted with phenylalanine residues. In addition, we generated a short form of the receptor by deletion of the C-terminal domain from residue 1298. This construct mimics the naturally occurring VEGFR-3 short, which lacks the tyrosine residues 1333, 1337, and 1363 (Figure 1A). The chimeric receptor wild type and mutants were then transduced in early passages HUVECs using the retroviral expression vector PINCO.26 The efficiency of cell infection was measured by GFP signal coexpressed in the viral vector, and the expression of each chimeric receptor on the cell surface was monitored by FACS analysis (not shown). Transduced cells were stimulated with 10 ng/mL EGF. The whole lysates were immunoprecipitated with an antibody against EGFR N terminus and immunoblotted with rabbit polyclonal antibody to the C terminus of VEGFR-3. EGF treatment induced optimal phosphorylation of the wild-type E-R3 chimeric receptor (Figure 1B, lanes 3 and 4). The mutants Y812F, Y830F, Y833F, and Y1265F showed phosphorylation levels similar to the wild-type E-R3. Mutant Y1068F was defective in receptor phosphorylation, thus suggesting that this tyrosine residue is necessary for the activation of the receptor kinase domain. This result is in agreement with previous observations on VEGFR-2. In fact, Y1068 of VEGFR-3 corresponds in VEGFR-2 to the Y1059 that has been demonstrated to be required for the activation of the kinase domain.34 Receptor mutants Y1063F, Y1230F, Y1231F, Y1337F, and E-R3Δ1298 all showed significant reduction of receptor phosphorylation. Functional analysis of receptor mutants was performed by assessing the in vitro sprouting (ie, the formation of tubelike structures resembling early capillaries) of HUVECs in a 3-dimensional collagen matrix. This assay provides information on endothelial cell differentiation as well as other endothelial cell functions like vitality and cell organization that could not be assayed in 2-dimensional culture. Cells transduced with the wild-type chimeric receptor as well as with the mutants Y812F, Y830F, Y833F, and Y1265F demonstrated efficient sprouting activity in response to EGF, suggesting that these mutations do not affect the chimeric receptor function. On the contrary, HUVECs transduced with mutant Y1063F showed about 50% reduction of sprouting activity, and Y1230F, Y1231F, Y1337F, and E-R3ΔC all showed more than 80% reduction in the formation of capillary sprouts (Figure 1C) (a representative sprouting assay is shown in supplemental Figure S1, available at the Blood website; see the Supplemental Figure link at the top of the online article).
Functional analysis of VEGFR-3 tyrosine mutants. (A) Schematic representation of the EGF–VEGFR-3 chimera. The large oval represents the extracellular EGFR domain. Tyrosine residues are indicated by circles. • represents the tyrosine residues that have been mutated in this study. Positions of the tyrosines are indicated by numbers. Deletion mutant is indicated by an arrowhead. (B) HUVECs expressing the chimeric E-R3 mutant receptors untreated or treated with 10 ng/mL EGF for 10 minutes. Immunoprecipitates with EGFR receptor were revealed with anti–VEGFR-3 C-terminal domain and antiphosphotyrosine-specific antibodies as indicated. (C) Quantitative analysis of 3 independent sprouting experiments measured by digitized image analysis of HUVECs embedded as spheroids in a collagen matrix treated with 10 ng/mL EGF and photographed 18 to 24 hours after induction. Quantification of the cumulative sprout length of 5 spheroids per experimental group. The results are expressed as the mean ± SD of 3 independent experiments.
Functional analysis of VEGFR-3 tyrosine mutants. (A) Schematic representation of the EGF–VEGFR-3 chimera. The large oval represents the extracellular EGFR domain. Tyrosine residues are indicated by circles. • represents the tyrosine residues that have been mutated in this study. Positions of the tyrosines are indicated by numbers. Deletion mutant is indicated by an arrowhead. (B) HUVECs expressing the chimeric E-R3 mutant receptors untreated or treated with 10 ng/mL EGF for 10 minutes. Immunoprecipitates with EGFR receptor were revealed with anti–VEGFR-3 C-terminal domain and antiphosphotyrosine-specific antibodies as indicated. (C) Quantitative analysis of 3 independent sprouting experiments measured by digitized image analysis of HUVECs embedded as spheroids in a collagen matrix treated with 10 ng/mL EGF and photographed 18 to 24 hours after induction. Quantification of the cumulative sprout length of 5 spheroids per experimental group. The results are expressed as the mean ± SD of 3 independent experiments.
Y1063 is the major mediator of the survival signal, while Y1230/Y1231 contributes together with Y1337 to proliferation, migration, and survival
Next we analyzed the contribution of each tyrosine in proliferation, migration, and survival mediated by the E-R3 chimeric receptor. As expected, Y1068F was deficient in all tests. Mutant Y1063F showed no significant reduction in cell proliferation measured as 3H-thymidine incorporation and cell mobility, assayed as migration in a Boyden chamber (Figure 2A-B). However, the analysis of E-R3–dependent survival measured by TUNEL staining revealed that, while the wild-type chimera induced more than 50% protection from apoptosis by serum deprivation, mutant Y1063F showed about 10% to 15% protection from apoptosis (Figure 2C), thus suggesting that Y1063 is the major single site of VEGFR-3 involved in prosurvival signaling. Differently from mutant Y1063, mutants Y1230F, Y1231F, Y1337F, and E-R3Δ1298 each showed about 70% reduction of proliferation and migration. In the survival assay these mutants showed a minor reduction of protection from apoptosis, because each was still able to induce more than 30% protection in this assay. Taken together these experiments identified on VEGFR-3 intracellular domain 3 new tyrosine residues implicated in the receptor signaling in primary endothelial cells: Y1063 required for the survival signaling and Y1230/Y1231 that contributed together with Y1337 to endothelial cell proliferation, migration, and survival. Notably, of the remaining tyrosine residues our assays did not show evidence of biologic activities of the tyrosines 812, 830, and 833 located within the juxtamembrane domain as well as tyrosine 1265 within the C-terminal tail. The deletion mutant E-R3Δ1298 showed biologic activity comparable with the mutant Y1337F in all tests, suggesting that Y1333 and Y1363, contained in this region, do not provide a significant functional contribution in the biologic tests assayed.
Tyrosine residues 1063, 1068, 1230, 1231, and 1337 play a functional role in E-R3–dependent proliferation, migration, and survival. (A) Mitogenic response of HUVECs transduced with E-R3 mutant receptors as indicated. Quantitative analysis of [3H] incorporation of HUVECs treated with EGF. The results are expressed as the mean ± SD of 3 independent experiments. (B) Chemotactic response of HUVECs transduced with E-R3 mutants as indicated. Cells seeded in the upper well of a Boyden chamber at 37°C and the lower well contained 10 ng/mL EGF. Cells migrating through the polycarbonate filter were quantified by staining with Giemsa solution, and counting was performed in a light microscope (magnification, × 40). The results are expressed as the mean ± SD of 3 independent experiments. (C) Apoptosis of HUVECs transduced with E-R3 mutant constructs as indicated. Quantification of DNA fragmentation by TUNEL staining expressed as a percentage over total nuclei stained with DAPI of cells after 24 hours of serum starvation or in the presence of 10 ng/mL EGF. The results are expressed as the mean ± SD of 3 independent experiments.
Tyrosine residues 1063, 1068, 1230, 1231, and 1337 play a functional role in E-R3–dependent proliferation, migration, and survival. (A) Mitogenic response of HUVECs transduced with E-R3 mutant receptors as indicated. Quantitative analysis of [3H] incorporation of HUVECs treated with EGF. The results are expressed as the mean ± SD of 3 independent experiments. (B) Chemotactic response of HUVECs transduced with E-R3 mutants as indicated. Cells seeded in the upper well of a Boyden chamber at 37°C and the lower well contained 10 ng/mL EGF. Cells migrating through the polycarbonate filter were quantified by staining with Giemsa solution, and counting was performed in a light microscope (magnification, × 40). The results are expressed as the mean ± SD of 3 independent experiments. (C) Apoptosis of HUVECs transduced with E-R3 mutant constructs as indicated. Quantification of DNA fragmentation by TUNEL staining expressed as a percentage over total nuclei stained with DAPI of cells after 24 hours of serum starvation or in the presence of 10 ng/mL EGF. The results are expressed as the mean ± SD of 3 independent experiments.
Tyrosine 1063 associates with CRKI/II and activates JNK1/2
Mutation of Y1063F showed strong reduction of the VEGFR-3–mediated survival from serum starvation–induced apoptosis as well as reduced sprouting, which is a test for endothelial cell differentiation and survival ability in the 3-dimensional collagen matrix.35
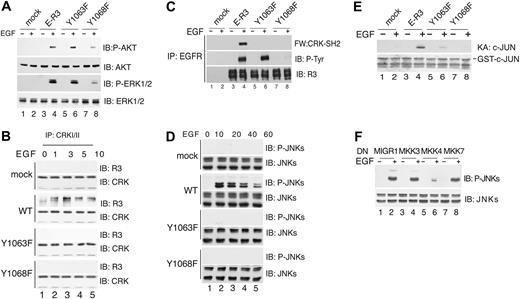
Because the major signal pathway that induces cell survival activates AKT, we analyzed whether Y1063F mutant showed reduced AKT activation. Immunoblotting with phosphospecific antibodies of AKT phosphorylation at serine 473 or ERK1/2 phosphorylation at threonine 202 and tyrosine 204 showed that mutant Y1063F activated AKT and ERK1/2 to levels comparable to the wild-type receptor (Figure 3A), thus allowing us to exclude that Y1063 signals cell survival via AKT or ERK1/2. This result pointed out a difference between Y1063, which was signaling survival by a pathway independent from AKT, and Y1230/Y1231, whose mutation showed inhibition of AKT and ERK1/2 phosphorylation (Figure 5C). We therefore investigated in greater detail the signaling pathway originating from Y1063. This tyrosine residue is contained within the sequence YKDP, which represents a putative consensus motif for CRKI/II (P-YXXP).36 Pulldown assays with GST-CRK-SH2 fusion suggested that CRK could bind to VEGFR-3 via its SH2 domain to phosphotyrosine 1063 of activated receptor (data not shown). Time-course coimmunoprecipitation assay with monoclonal antibody anti-CRKI/II showed that E-R3 was coprecipitated with CRKI/II in a ligand-dependent manner with a peak at 3 minutes, while no binding was observed with cells transduced with the mutant Y1063F as well as with the kinase-inactive mutant Y1068F used as a control (Figure 3B). Direct interaction between CRK and the receptor could be demonstrated by Far Western analysis. Serum-starved HUVECs expressing either E-R3, Y1068F, or Y1063F mutant receptors were induced with EGF. Immunoblotting using biotinylated GST-CRK fusion protein on extracts immunoprecipitated for the chimeric receptor with anti-EGFR showed direct binding of the recombinant GST-CRK-SH2 to the wild-type receptor following EGF treatment, whereas receptor mutants Y1063F and Y1068F were negative (Figure 3C). These results suggested that upon receptor activation CRK is directly recruited to phosphotyrosine 1063.
Phospho-Y1063 recruits CRK and activates JNK1/2 via MKK4. (A) AKT and ERK1/2 activation by wild-type and mutant E-R3 chimeric receptors as indicated. Immunoblot analysis of HUVEC cell extracts untreated or treated with 10 ng/mL EGF for 20 minutes with specific antibodies as indicated. AKT activation was detected using phospho-S473 AKT-specific antibody, and ERK1/2 activation was revealed with phospho-T202/Y204 ERK-specific antibody. (B) Time-course analysis of CRK binding to ER-3 by coimmunoprecipitation. HUVECs transduced with E-R3 mutants as indicated either untreated or treated with 10 ng/mL EGF at the times indicated were immunoprecipitated with anti-CRK–specific antibodies and revealed with the anti–VEGFR-3 C-terminal domain. (C) CRK-SH2 binds directly to E-R3 chimeric receptor. Far Western blotting of extracts from HUVECs transduced with E-R3 mutants as indicated were immunoprecipitated with anti-EGFR N-terminal–specific antibodies and revealed with biotinylated recombinant CRK-SH2. (D) Time-course analysis of JNK activation in HUVECs transduced with wild-type E-R3 receptor and the receptor mutants as indicated. JNK activation was assessed by phosphorylation of threonine 183 and tyrosine 185 using phosphospecific antibodies. (E) HUVEC extracts assayed for JNK-dependent kinase activity toward recombinant GST–c-JUN 1-79 in vitro by phosphorylation of serine 63 and 73 with phosphospecific antibodies. Ponceau staining of Escherichia coli extracts is shown. (F) Inhibition of JNK activation in HUVECs transduced with E-R3 chimera by dominant-negative (DN) MAPKs as indicated transduced with a retroviral vector (MIGR1).
Phospho-Y1063 recruits CRK and activates JNK1/2 via MKK4. (A) AKT and ERK1/2 activation by wild-type and mutant E-R3 chimeric receptors as indicated. Immunoblot analysis of HUVEC cell extracts untreated or treated with 10 ng/mL EGF for 20 minutes with specific antibodies as indicated. AKT activation was detected using phospho-S473 AKT-specific antibody, and ERK1/2 activation was revealed with phospho-T202/Y204 ERK-specific antibody. (B) Time-course analysis of CRK binding to ER-3 by coimmunoprecipitation. HUVECs transduced with E-R3 mutants as indicated either untreated or treated with 10 ng/mL EGF at the times indicated were immunoprecipitated with anti-CRK–specific antibodies and revealed with the anti–VEGFR-3 C-terminal domain. (C) CRK-SH2 binds directly to E-R3 chimeric receptor. Far Western blotting of extracts from HUVECs transduced with E-R3 mutants as indicated were immunoprecipitated with anti-EGFR N-terminal–specific antibodies and revealed with biotinylated recombinant CRK-SH2. (D) Time-course analysis of JNK activation in HUVECs transduced with wild-type E-R3 receptor and the receptor mutants as indicated. JNK activation was assessed by phosphorylation of threonine 183 and tyrosine 185 using phosphospecific antibodies. (E) HUVEC extracts assayed for JNK-dependent kinase activity toward recombinant GST–c-JUN 1-79 in vitro by phosphorylation of serine 63 and 73 with phosphospecific antibodies. Ponceau staining of Escherichia coli extracts is shown. (F) Inhibition of JNK activation in HUVECs transduced with E-R3 chimera by dominant-negative (DN) MAPKs as indicated transduced with a retroviral vector (MIGR1).
Because the adaptor CRK through its SH3 domain binds to several target molecules and activates the JNK pathway without stimulating ERK or p38,37 we tested whether VEGFR-3 signaling activated JNK and this activation was dependent upon Y1063. As shown in Figure 3D, in cells transduced with E-R3, phosphospecific antibodies for JNK threonine 183 and tyrosine 185 showed that JNK1/2 phosphorylated already at 10 minutes after EGF induction, decreasing thereafter. On the contrary, JNK1/2 were not phosphorylated in cells expressing the kinase-inactive receptor Y1068F or with the mutant Y1063F. JNK activity assay, measured by phosphorylation of GST–c-JUN, confirmed JNK activation only in cells expressing the wild-type receptor and not in cells transduced with Y1068F or Y1063F (Figure 3E). These results demonstrated that E-R3 signaling induced JNK1/2 and this activation was dependent on Y1063.
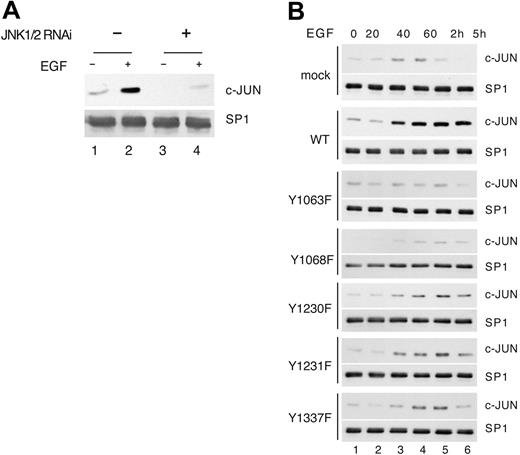
JNK has been shown to be phosphorylated either by MKK4 or MKK7 in independent complexes, which ensure specificity of the signal transduction.38 As shown in Figure 3F, the infection of endothelial cells with a viral vector expressing the dominant-negative MKK4, but not MKK3 or MKK7, inhibited JNK phosphorylation on threonine 183 and tyrosine 185 induced by VEGFR-3 signaling, demonstrating that MKK4 lies upstream of JNK1/2. Taken together these results define the pathway E-R3/CRK/MKK4/JNK.
VEGFR-3 survival signaling is mediated by JNK1/2
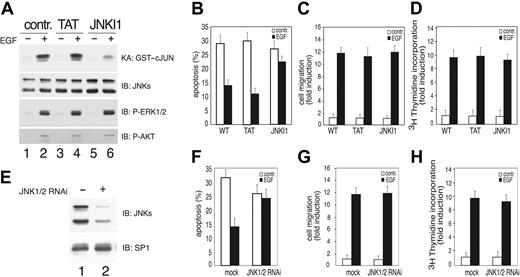
JNK signaling has been associated with apoptosis as well as with cell survival.39,40 To analyze whether in endothelial cells VEGFR-3–dependent JNK activity was inducing a prosurvival signaling, we specifically inhibited JNK by using the peptide inhibitor JNKI1.41 This peptide is quite specific because, differently from SP6000125 (data not shown), it blocked E-R3–dependent activation of JNK without affecting the activation of other MAPKs in primary endothelial cells (Figure 4A). Pretreatment of HUVECs with JNKI1 significantly reduced the survival effect mediated by E-R3. In fact, the addition of JNKI1 prior to the addition of EGF resulted in apoptotic cell death of approximately 25%, whereas treatment of cells with EGF in the absence of JNKI1 resulted in apoptotic cell death of approximately 15% (Figure 4B). The prosurvival activity of JNK was quite specific, as its inhibition did not affect ER-3–dependent migration or DNA synthesis (Figure 4C-D). In an independent set of experiments we inhibited JNK's expression by generating lentiviral vectors expressing shRNA designed to inhibit both JNK1/2 under the control of the polymerase III U6 (Figure 4E). Also in this case we obtained reduction of E-R3–mediated survival (Figure 4F) without affecting migration or proliferation, thus demonstrating that in endothelial cells JNK1/2 is required for VEGFR-3–dependent protection from growth factor deprivation–induced apoptosis.
JNK activity is required for E-R3–dependent survival signal. (A) HUVECs transduced with E-R3 chimera untreated or treated with 10 ng/mL EGF pretreated with 1 μM mock peptide (TAT) or JNK inhibitory peptide (JNKI1) as indicated. The synthetic peptide JNKI1 specifically inhibits JNK1/2 activity measured by phosphorylation of the recombinant GST–c-JUN 1-79 in vitro assessed by phosphospecific antibodies. The same extracts were assayed for ERK and AKT phosphorylation with phosphospecific antibodies. (B) Quantification by TUNEL staining of apoptotic HUVECs pretreated with mock peptide (TAT) or with JNK inhibitory peptide (JNKI1). (C-D) Chemotaxis and mitogenic response of HUVECs treated as in panel A in the presence of TAT or JNKI1 as indicated. (E) Western blot analysis of JNK1/2 silencing obtained by transducing HUVECs with lentiviral vectors expressing JNK1 and JNK2 shRNA. (F) Quantification by TUNEL staining of HUVEC apoptosis of cells expressing a mock lentiviral construct or JNK1/2 shRNAas indicated. (G-H) Chemotaxis and mitogenic response of HUVECs either mock infected or silenced by retroviral infection with a vector expressing JNK1/2 shRNA. The results are expressed as the mean ± SD of 3 independent experiments.
JNK activity is required for E-R3–dependent survival signal. (A) HUVECs transduced with E-R3 chimera untreated or treated with 10 ng/mL EGF pretreated with 1 μM mock peptide (TAT) or JNK inhibitory peptide (JNKI1) as indicated. The synthetic peptide JNKI1 specifically inhibits JNK1/2 activity measured by phosphorylation of the recombinant GST–c-JUN 1-79 in vitro assessed by phosphospecific antibodies. The same extracts were assayed for ERK and AKT phosphorylation with phosphospecific antibodies. (B) Quantification by TUNEL staining of apoptotic HUVECs pretreated with mock peptide (TAT) or with JNK inhibitory peptide (JNKI1). (C-D) Chemotaxis and mitogenic response of HUVECs treated as in panel A in the presence of TAT or JNKI1 as indicated. (E) Western blot analysis of JNK1/2 silencing obtained by transducing HUVECs with lentiviral vectors expressing JNK1 and JNK2 shRNA. (F) Quantification by TUNEL staining of HUVEC apoptosis of cells expressing a mock lentiviral construct or JNK1/2 shRNAas indicated. (G-H) Chemotaxis and mitogenic response of HUVECs either mock infected or silenced by retroviral infection with a vector expressing JNK1/2 shRNA. The results are expressed as the mean ± SD of 3 independent experiments.
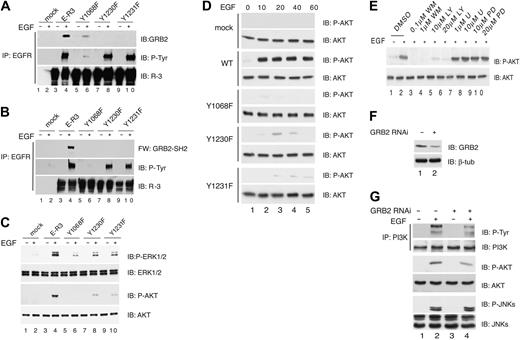
Phospho-Y1230/Y1231 recruits GRB2 and activates ERK1/2 and AKT. (A) Coimmunoprecipitation of chimeric E-R3 with GRB2. HUVECs transduced with wild-type E-R3 or the mutants as indicated either untreated or induced with 10 ng/mL EGF for 10 minutes were immunoprecipitated with anti-EGFR N-terminal–specific antibodies and revealed with anti-GRB2, phosphotyrosine, or VEGFR-3 C-terminal domain as indicated. (B) GBR2-SH2 binds directly to E-R3 chimeric receptor. Far Western blotting of extracts from HUVECs transduced with E-R3 mutants as indicated were immunoprecipitated with anti-EGFR N-terminal–specific antibodies and revealed with biotinylated recombinant GRB2-SH2. (C) Wild-type E-R3, but not mutants Y1230 and Y1231, fully activates AKT and ERK phosphorylation measured with phosphospecific antibodies. (D) Time-course analysis of AKT activation of HUVECs transduced with wild-type ER-3 or with the mutants as indicated. (E)AKT activation is mediated by PI3K. Effect on ER-3–dependent AKT phosphorylation by inhibition of PI3K by wortmanin (WM), or LY294002 (LY), MEK1/2 by U0126 (U), PD98059 (PD). (F) Western blot analysis of GRB2 silencing obtained by transducing HUVECs with lentiviral vectors expressing GRB2 shRNA. (G) PI3K and AKT phosphorylation are dependent upon GRB2 expression. PI3K was immunoprecipitated with antibodies anti-PI3K and P85. The Ip was immunoblotted with anti–P-Tyr. P-AKT and P-JNKs were revealed with phosphospecific antibodies. IP indicates immunoprecipitated.
Phospho-Y1230/Y1231 recruits GRB2 and activates ERK1/2 and AKT. (A) Coimmunoprecipitation of chimeric E-R3 with GRB2. HUVECs transduced with wild-type E-R3 or the mutants as indicated either untreated or induced with 10 ng/mL EGF for 10 minutes were immunoprecipitated with anti-EGFR N-terminal–specific antibodies and revealed with anti-GRB2, phosphotyrosine, or VEGFR-3 C-terminal domain as indicated. (B) GBR2-SH2 binds directly to E-R3 chimeric receptor. Far Western blotting of extracts from HUVECs transduced with E-R3 mutants as indicated were immunoprecipitated with anti-EGFR N-terminal–specific antibodies and revealed with biotinylated recombinant GRB2-SH2. (C) Wild-type E-R3, but not mutants Y1230 and Y1231, fully activates AKT and ERK phosphorylation measured with phosphospecific antibodies. (D) Time-course analysis of AKT activation of HUVECs transduced with wild-type ER-3 or with the mutants as indicated. (E)AKT activation is mediated by PI3K. Effect on ER-3–dependent AKT phosphorylation by inhibition of PI3K by wortmanin (WM), or LY294002 (LY), MEK1/2 by U0126 (U), PD98059 (PD). (F) Western blot analysis of GRB2 silencing obtained by transducing HUVECs with lentiviral vectors expressing GRB2 shRNA. (G) PI3K and AKT phosphorylation are dependent upon GRB2 expression. PI3K was immunoprecipitated with antibodies anti-PI3K and P85. The Ip was immunoblotted with anti–P-Tyr. P-AKT and P-JNKs were revealed with phosphospecific antibodies. IP indicates immunoprecipitated.
Y1230/Y1231 associates with GRB2 and activates ERK1/2 and AKT pathways
Mutations of the tyrosine residues 1230/1231 reduced VEGFR-3–mediated proliferation, migration, and survival signaling. These tyrosine residues are contained within the motif YYNW, which upon phosphorylation might generate a consensus for GRB2 (P-YXXW).36 To assess whether VEGFR-3 interacts with GRB2 cells, extracts were immunoprecipitated with anti-EGFR N-terminus and the filter was immunoblotted with anti-GRB2 antibodies. GRB2 was coprecipitated with wild-type E-R3 in a ligand-dependent manner while no binding was observed for the Y1230F and Y1231F mutants (Figure 5A). GRB2 direct binding to the receptor was demonstrated by Far Western analysis. Serum-starved HUVECs expressing either E-R3 wild-type, Y1068F, Y1230F, or Y1231F mutant receptors were induced with EGF. Cell extracts were immunoprecipitated with anti-EGFR antibody and subsequently immunoblotted with biotinylated GST-GRB2-SH2 fusion protein. Following EGF stimulation GST-GRB2-SH2 interacted with E-R3, whereas Y1230F and Y1231F mutants did not show detectable binding (Figure 5B). These results demonstrated the direct recruitment of GRB2 to the receptor at phosphorylated tyrosine residues 1230/1231. Next we analyzed the activation of MAPK and AKT by assessing their phosphorylation. Using phosphospecific antibodies we could observe both ERK1/2 and AKT phosphorylation following EGF stimulation while mutants Y1230F and Y1231F showed a significantly reduced phosphorylation of both ERK1/2 and AKT (Figure 5C). Time-course analysis demonstrated that AKT phosphorylation was high at 10 minutes after EGF treatment and remained sustained within 1 hour while both mutants Y1230 and Y1231 showed impaired AKT phosphorylation at all time points considered (Figure 5D). The PI3K inhibitors wortmanin (0.1 and 1 μM) and LY 294002 (10 and 20 μM) abolished AKT phosphorylation in response to EGF treatment (Figure 5E). Moreover, GRB2 knockdown by shRNA (Figure 5F) reduced about 50% of P-PI3K and P-AKT levels without affecting the levels of P-JNK1/2 (Figure 5G), demonstrating that GRB2 is required for E-R3–dependent activation of AKT via PI3K.
c-JUN protects endothelial cells from serum-deprivation–induced apoptosis
Because c-JUN stability and transcriptional activity are increased by JNK phosphorylation,42 we analyzed the c-JUN levels in endothelial cells silenced for JNK1/2. Western blot analysis showed that the expression of c-JUN was strongly reduced in the absence of JNK both in unstimulated and EGF-treated cells (Figure 6A). Next we analyzed c-JUN induction in HUVECs transduced with E-R3 wild type and mutant upon EGF treatment. As expected, EGF treatment induced c-JUN, whose expression increased from 40 minutes and reached a peak of expression at about 2 hours, decreasing thereafter (Figure 6B). A strong inhibition of c-JUN induction could be observed in cells expressing mutant Y1063F, which we previously demonstrated was impaired for JNK activation, therefore demonstrating that the signaling originated from Y1063 is upstream of c-JUN. Reduced levels of c-JUN, although less dramatic, were also observed with the mutants Y1230F, Y1231F, and Y1337F impaired for ERK1/2 activation. Taken together the above results suggest that ERK1/2 and JNK independent pathways contribute to c-JUN induction.
c-JUN induction by E-R3 depends on JNK and ERK signaling. (A) c-JUN expression was detected by Western blot analysis in mock-infected cells or in cells infected with lentiviral vectors expressing JNK1/2 shRNA. (B) Western blot analysis of c-JUN expression in HUVECs transduced with E-R3 mutant chimeric receptor as indicated at various times after treatment with 10 ng/mL EGF.
c-JUN induction by E-R3 depends on JNK and ERK signaling. (A) c-JUN expression was detected by Western blot analysis in mock-infected cells or in cells infected with lentiviral vectors expressing JNK1/2 shRNA. (B) Western blot analysis of c-JUN expression in HUVECs transduced with E-R3 mutant chimeric receptor as indicated at various times after treatment with 10 ng/mL EGF.
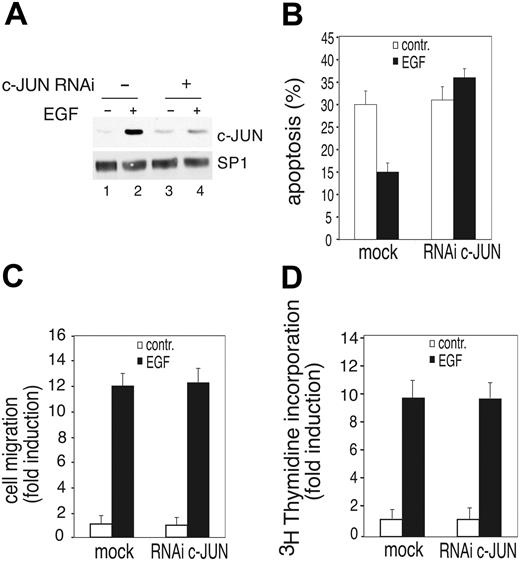
c-JUN expression is required for E-R3–dependent survival signal. (A) Western blot analysis of c-JUN from mock-infected cells or cells silenced for c-JUN obtained by transducing HUVECs with a lentiviral vector expressing c-JUN shRNA before or after 2 hours of treatment with 10 ng/mL EGF. (B) Quantification by TUNEL staining of HUVEC apoptosis expressing a mock lentiviral construct or c-JUN shRNA. (C-D) Chemotaxis and mitogenic response of HUVECs either mock infected or silenced by retroviral infection with a vector expressing c-JUN shRNA. The results are expressed as the mean ± SD of 3 independent experiments.
c-JUN expression is required for E-R3–dependent survival signal. (A) Western blot analysis of c-JUN from mock-infected cells or cells silenced for c-JUN obtained by transducing HUVECs with a lentiviral vector expressing c-JUN shRNA before or after 2 hours of treatment with 10 ng/mL EGF. (B) Quantification by TUNEL staining of HUVEC apoptosis expressing a mock lentiviral construct or c-JUN shRNA. (C-D) Chemotaxis and mitogenic response of HUVECs either mock infected or silenced by retroviral infection with a vector expressing c-JUN shRNA. The results are expressed as the mean ± SD of 3 independent experiments.
c-JUN has been linked to cell proliferation, survival, or apoptosis in different cell types or growth conditions.43,44 We therefore analyzed whether the increased expression of c-JUN in response to VEGFR-3 signaling exerts prosurvival activity in primary endothelial cells silencing its expression by c-JUN shRNA (Figure 7A). Apoptotic cell death measured by TUNEL staining revealed that HUVECs silenced for c-JUN were impaired in their response to E-R3–mediated survival signaling while no significant effect was observed in EGF-dependent migration or DNA synthesis (Figure 7B-D). Taken together these results suggest that in endothelial cells c-JUN plays a pivotal role in growth factor–mediated cell survival.
Discussion
VEGFR-3 is essential for proper vascular development and contributes to adult angiogenesis and lymphangionenesis; however, its signal transduction mechanisms are still incompletely understood. Previous analysis identified the potential VEGFR-3 phosphorylation sites.25 In this study we have analyzed the functional role of VEGFR-3 in endothelial cells by generating site-directed mutagenesis of the tyrosine residues potentially involved in the receptor signaling. The mutants were analyzed by viral transduction into HUVECs of EGFR–VEGFR-3 chimeric molecules to avoid the interference of VEGFR-2. This analysis demonstrated that tyrosine residues 1063, 1068, 1230, 1231, and 1337 play a functional role in VEGFR-3 signaling. We show that tyrosine 1068, located within the second kinase domain of VEGFR-3, is essential for the receptor autophosphorylation, suggesting that this tyrosine may act as an essential residue for the stabilization of receptor in its activated-loop conformation upon ligand stimulation. This residue corresponds to Y1059, previously described as required for positive regulation of VEGFR-2 kinase activity.34 Although also located within the activation loop, mutation of tyrosine 1063 showed only a minor reduction of the receptor phosphorylation compared with the wild type, thus demonstrating that phosphorylation of tyrosine 1063 is not essential for receptor activation. The phenotypic analysis of endothelial cells transduced with this mutant showed a significant reduction of sprouting activity and impaired survival signaling, demonstrating that this tyrosine residue is the single major site for the receptor-mediated survival signaling from serum-deprivation apoptosis. Taken together our results unveiled a novel survival signaling pathway that originated from Y1063 of VEGFR-3 that, via CRK/MKK4/JNK, reaches the nuclear factor c-JUN. Functional experiments of JNK1/2 inhibition clearly demonstrated that induced JNK1/2 mediated by E-R3 receptor activates a prosurvival signaling in endothelial cells. Although activation of JNK is known to be implicated in the induction of the apoptotic response to stress stimuli, few studies demonstrated JNK implication in survival signaling.39,40 In this respect it is notable that the prosurvival signaling originated by phospho-Y1063 activates JNK via MKK4, but not MKK7 or MKK3, thus suggesting that different phenotypic outcomes might be mediated by independent complexes. The sustained activation of JNK by stress or resulting from the inhibition of its feedback signaling leading to apoptosis has been extensively studied, and the genes involved are known, whereas the molecular targets of JNK involved in prosurvival activity are yet unknown.45 Two recent reports addressed this issue. In hematopoietic pro-B FL5.12 cells, JNK-dependent phosphorylation of BAD at threonine 201 has been shown to be required for interleukin-3 (IL-3)–mediated survival,46 and in fibroblasts, JNK-dependent JunD activation was shown to collaborate with nuclear factor–κB (NF-κB) to mediate a survival signaling.47
We here found that c-JUN, induced by VEGFR-3 signaling, is a downstream prosurvival JNK effector in endothelial cells. c-JUN was strongly induced in endothelial cells following E-R3 activation. This induction was abolished by mutation at Y1063 and reduced by the mutations of Y1230, Y1231, and Y1337. Thus, its down-regulation correlated with the reduced survival phenotype of these mutants, and it is in agreement with previous studies in fibroblasts showing that c-Jun is induced by MAPK and stabilized by JNK-mediated phosphorylation of serine 63 and 73.42,48,49 Importantly, we observed that c-JUN silencing significantly reduced VEGFR-3–mediated survival, demonstrating its survival role in primary endothelial cells. Elevated c-Jun levels have been correlated to apoptosis in cells exposed to genotoxic stress in response to UV irradiations, or by its ectopic overexpression.43 However, knock-out experiments also demonstrated that c-Jun can be antiapoptotic depending on the cellular type and status.43 Although, after E-R3 induction, we observed strong c-JUN induction, while BAD phosphorylation and JUND induction were barely detectable in primary endothelial cells (data not shown), our results cannot exclude a contribution of JNK-dependent BAD phosphorylation and JUND activation for cell survival mediated by VEGFR-3. In this respect, it is relevant to remember that c-JUN is necessary for JNK-dependent JUND phosphorylation, because JUND lacks a JNK docking site and most likely is phosphorylated through heterodimerization with c-JUN.50
Functional and biochemical analysis of the E-R3 chimera also identified the contribution of tyrosines 1230/1231 to VEGFR-3–dependent proliferation, migration, and survival. Our data show that these tyrosine residues activate the classic PI3K/AKT and ERK pathways upon GRB2 direct recruitment to the receptor. This site must collaborate with Y1337, which was previously shown to recruit SHC and indirectly GRB2.23,51 Analysis of VEGFR-3 phosphorylation demonstrated that upon VEGFR-2/VEGFR-3 heterodimerization Y1063 and Y1230/Y1231 were still phosphorylated while Y1337 could not be any longer phosphorylated.25 Taken together, these results suggest that during the angiogenic process, VEGFR-2– and VEGFR-3–positive endothelial cells respond to VEGF-C or VEGF-D by forming functional heterodimers that would maintain elevated levels of survival signaling through Y1063, but attenuated endothelial-cell proliferation and spreading as the contribution of Y1337 would be missing. Moreover, Y1063 and Y1230/Y1231 are both located upstream of the truncation in the receptor short form,20,21 suggesting that differential expression of these two forms might contribute to modulation of VEGFR-3 signaling pathways as well.
Prepublished online as Blood First Edition Paper, August 2, 2005; DOI 10.1182/blood-2005-04-1388.
Supported by grants from Associazione Italiana Ricerca sul Cancro (AIRC), Ministero Italiano dell'Istruzione, dell'Unversità e della Ricerca (MIUR), and Fondazione Monte dei Paschi di Siena (MPS).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Guido Franzoso, Pier Giuseppe Pelicci, and Daniel Birnbaum for DNA constructs, Andrea Graziani for the critical reading of the manuscript, Beatrice Grandi for technical help and all members Chiron vaccines for DNA sequence and FACS facilities.

![Figure 2. Tyrosine residues 1063, 1068, 1230, 1231, and 1337 play a functional role in E-R3–dependent proliferation, migration, and survival. (A) Mitogenic response of HUVECs transduced with E-R3 mutant receptors as indicated. Quantitative analysis of [3H] incorporation of HUVECs treated with EGF. The results are expressed as the mean ± SD of 3 independent experiments. (B) Chemotactic response of HUVECs transduced with E-R3 mutants as indicated. Cells seeded in the upper well of a Boyden chamber at 37°C and the lower well contained 10 ng/mL EGF. Cells migrating through the polycarbonate filter were quantified by staining with Giemsa solution, and counting was performed in a light microscope (magnification, × 40). The results are expressed as the mean ± SD of 3 independent experiments. (C) Apoptosis of HUVECs transduced with E-R3 mutant constructs as indicated. Quantification of DNA fragmentation by TUNEL staining expressed as a percentage over total nuclei stained with DAPI of cells after 24 hours of serum starvation or in the presence of 10 ng/mL EGF. The results are expressed as the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/10/10.1182_blood-2005-04-1388/6/m_zh80220586940002.jpeg?Expires=1769085880&Signature=ahzeb5gZarkkItS6uWp4k193C8PjDCxm7Kj3ckMFcDvOpbNjWJ91wPRxHZmgrHdzL0eHPN-CKqr6otRVyJ6jh0GK9x0fBcuTgNAS4SSq~WIZA4FqBV1s6~8B67jYow8McpQyauhKPv7kdZG-1Rr8VjPoi5HTJjZ0T3bOH7jsLQIInaPfvxJukm8jZn47YuAlggg9nsAoAq5-C1K2Vlb0nwTM4xy~9gABFs8iLLkX01fSuj7J4EvBOLLp1tJtwWfCxXQgH9F6TQyoPstNGft~WAFRoByGt41HHGVOq5xY9CEIlejx4pNKscUt3TbZHDH3-o820G4FOa2Uzr3XUTgV4g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal