Comment on Bergsagel et al, page 296
Early oncogenic events in multiple myeloma include IgH translocations and numeric chromosomal changes. Using gene expression profiling, Bergsagel and colleagues have identified several new groups that share dysregulation of cyclin D.
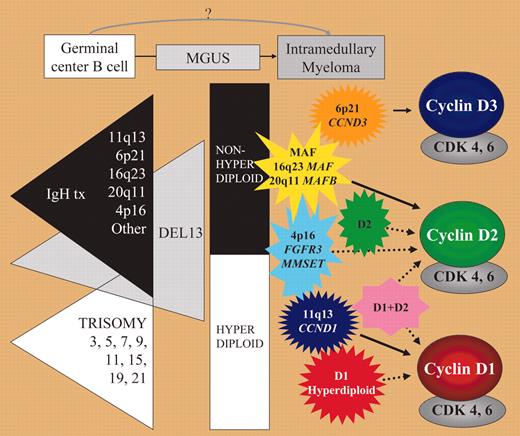
In contrast to acute leukemia, identification of early oncogenic events in multiple myeloma (MM) has remained somewhat elusive. Low proliferative capacities of MM cells and difficulties in isolating premalignant cells and establishing valid animal models explain this lack of a unifying model of oncogenesis in MM. Cytogenetic studies have identified recurrent translocations involving immunoglobulin heavy-chain (IgH) and numeric changes including hyperdiploidy characterized by trisomy of nonrandom chromosomes, and deletions of chromosome 13.1 Patients with MM seem to be divided into 2 subgroups: one with hyperdiploid karyotypes with a low incidence of chromosome 13 abnormalities and one nonhyperdiploid group characterized by recurrent IgH translocations and a higher incidence of chromosome 13 deletions, suggesting that these 2 subgroups represent 2 oncogenic pathways. Mutations of N- and K-RAS, FGFR3, inactivation of p53, and dys-regulation of c-myc seem to be involved later in the course of the disease. These findings draw a complex and only partial picture which until now has not led to precise identification of common mechanisms involved in MM ontogeny. Gene expression profiling techniques offer the opportunity to identify common oncogenic pathways and to define new subgroups in an unbiased approach.FIG1
Early oncogenic events in MM. See the complete figure in the article beginning on page 296.
Early oncogenic events in MM. See the complete figure in the article beginning on page 296.
In this issue of Blood, Bergsagel and colleagues have defined 8 groups of translocation/cyclin D (TC) MM, 4 based on recurrent translocations (4p16, maf, 6p21, 11q13) and 4 based solely on CCND1 and/or CCND2 expression (D1, D1+D2, D2, and none for absence of cyclin D overexpression). All these TC groups have distinct gene expression profiles and clinical evolution. However, they share overexpression of at least one of the 3 cyclins D (cyclins D1-D3). CCND1 appears biallelically dysregulated in almost 40% of MM. Interestingly, monoclonal gammopathy of undetermined significance (MGUS) samples could be assigned into 5 of these TC groups. Bergsagel and colleagues propose a model of early oncogenesis in MM where the TC groups fit into the hyperdiploid or nonhyperdiploid pathway. An intriguing finding of this study and others exploring the role of cyclin D in MM is the contrast between the low proliferative index of MM cells and the apparently consistent finding of deregulated proteins involved in the G1 phase. Methylation of p16INK4a and p15INK4b has been observed in MGUS and MM and this probably contributes together with cyclin D overexpression to disruption of the Rb/pathway.2 However, at least one other pathogenic event is necessary for malignant transformation. Gene profiling data have not identified obvious candidates, though several recent published works suggest that Notch signaling is involved in the interaction between neoplastic plasma cells and their bone marrow microenvironment.3-5 More specifically, the Notch ligand JAG2 was found overexpressed in MGUS as a consequence of hypomethylation of the JAG2 promoter.3 It will be interesting to investigate the role of Notch signaling in the different TC groups defined by Bergsagel and colleagues. Another important finding of their study is that TC groups have different clinical behaviors in spite of the early occurrence of these pathogenic events in the course of the disease. This might suggest that pathogenic events that define these TC groups contribute not only to the early stage of the disease but also to further progression, providing clues for new therapeutic targets. Further validation of the gene profiles defined by the TC groups on other large independent cohorts of patients will be necessary to explore the questions raised by this study. ▪

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal