Abstract
The Ras-like guanine-nucleotide–binding protein Rap1 controls integrin αIIbβ3 activity and platelet aggregation. Recently, we have found that Rap1 activation can be blocked by the nitric oxide/cyclic guanosine monophosphate (NO/cGMP) signaling pathway by type 1 cGMP-dependent protein kinase (cGKI). In search of possible targets of NO/cGMP/cGKI, we studied the expression of Rap1-specific GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs) in platelets. We could detect mRNAs for a new protein most closely related to Rap1GAP and for postsynaptic density-95 discs-large and zona occludens protein 1 (PDZ)–GEF1 and CalDAG-GEFs I and III. Using 5′–rapid amplification of cDNA ends (RACE), we isolated the complete cDNA of the new GAP encoding a 715-amino acid protein, which we have termed Rap1GAP2. Rap1GAP2 is expressed in at least 3 splice variants, 2 of which are detectable in platelets. Endogenous Rap1GAP2 protein partially colocalizes with Rap1 in human platelets. In transfected cells, we show that Rap1GAP2 exhibits strong GTPase-stimulating activity toward Rap1. Rap1GAP2 is highly phosphorylated, and we have identified cGKI as a Rap1GAP2 kinase. cGKI phosphorylates Rap1GAP2 exclusively on serine 7, a residue present only in the platelet splice variants of Rap1GAP2. Phosphorylation of Rap1GAP2 by cGKI might mediate inhibitory effects of NO/cGMP on Rap1. Rap1GAP2 is the first GTPase-activating protein of Rap1 found in platelets and is likely to have an important regulatory role in platelet aggregation.
Introduction
Platelets are of great physiologic importance as regulators of clot formation and inflammation in the vasculature, and they have been established as major therapeutic targets in cardiovascular disease.1 Platelets contain high levels of Rap1, a Ras-like guanine-nucleotide–binding protein. Recently, Rap1 was identified as a potent regulator of integrin function.2-5 For example, the regulation of lymphocyte and macrophage adhesion by integrins αLβ2 and αBM2, respectively, is controlled by Rap1.6,7 Rap1 also facilitates the activation of platelet integrin αIIbβ3, which is required for fibrinogen binding and aggregation.8,9 Recently, Rap1 was shown to control the aggregation of mouse platelets.10
Many different platelet agonists, including thrombin, adenosine diphosphate (ADP), collagen, and thromboxane, induce Rap1 activation.11,12 However, the exact pathways involved in Rap1 activation in platelets are unknown. Data from other cell types suggest that receptor-mediated formation of second messengers can lead to the activation of Rap1-specific guanine nucleotide exchange factors (GEFs). Calcium and diacylglycerol activate CalDAG-GEFI (also termed RasGRP2), a protein detected in mouse megakaryocytes and platelets,10,13 and CalDAG-GEFIII (also termed RasGRP3), involved in neuronal differentiation and B-cell development.14,15 Cyclic adenosine monophosphate (cAMP) activates cAMP-dependent Epacs,16,17 and C3G is regulated by tyrosine phosphorylation18 and by binding to adaptor proteins.19,20 Postsynaptic density-95 discs-large and zona occludens protein 1 (PDZ)–GEF1 is a ubiquitously expressed GEF of Rap1 and contains a negative regulatory domain, though the physiologic activator is unknown.21,22 Finally, dedicator of cytokinesis protein 4 (DOCK4) has been described as activating Rap1.23 Thus far, platelet expression has been reported only for mouse CalDAG-GEFI.10
Inactivation of Rap1 requires specific GTPase-activating proteins (GAPs), and 2 major groups of Rap1-specific GAPs have been identified. The first consists of Rap1GAP24 and its splice variant, Rap1GAPII.25 Rap1GAP is expressed in brain,24,26 thyroid cells,27 and some other tissues.28 The activity of Rap1GAP can be regulated by direct interaction with Gα proteins.25,29-31 The second group of Rap1-specific GAPs encompasses the structurally related proteins signal-induced proliferation-associated protein-1 (SPA-1), found in lymphoid tissues,28 and E6TP1α (SPAR/SPAL), expressed in hippocampal neurons and other tissues.32-34 SPA-1 activity is controlled by the cytoskeleton-anchoring protein afadin-6 (AF-6).35
Platelet activation and aggregation are negatively regulated by the nitric oxide/cyclic guanosine monophosphate (NO/cGMP) pathway.36-39 Many antiplatelet effects of NO/cGMP are mediated by type 1 cGMP–dependent protein kinase (cGKI),40,41 and we have recently observed that cGKI potently inhibits agonist-induced activation of Rap1 in human platelets.42 Therefore, we were interested in identifying possible substrates of cGKI mediating this effect on Rap1 activity.
In this study, we provide evidence that human platelets express a distinct subgroup of Rap1-GEF proteins composed of PDZ-GEF1 and CalDAG-GEFs I and III. Furthermore, only one GAP of Rap1 appears to be present in platelets. We have characterized this new protein, termed Rap1GAP2, as a potent Rap1-specific GAP. We show colocalization of endogenous Rap1GAP2 and Rap1 in platelets. Additionally, we show that Rap1GAP2 is phosphorylated by cGKI, and we have mapped the single phosphorylation site to serine 7 of the Rap1GAP2a and Rap1GAP2b splice variants.
Materials and methods
Materials
Eukaryotic expression vectors for HA-tagged Rap1B, Rap1GAP1, Ras and PDZ-GEF1 and for GST-RalGDS-RBD and GST-Raf1-RBD for bacterial expression were kindly provided by J. Bos (Department of Physiological Chemistry, Utrecht, The Netherlands). KIAA1039 partial cDNA clone and CalDAG-GEFIII (KIAA0846) full-length cDNA clone were obtained from Kazusa DNAResearch Institute (Japan). The eukaryotic expression vector pSG8, containing N-terminal His6- and VSV-epitope tags, was a kind gift from S. Gross (Institute for Biochemistry II, Frankfurt, Germany).
Preparation of platelet RNA
Leukocyte-reduced platelet concentrate (a gift from the local blood bank) was subjected to 3 low-speed spins (150g) to remove aggregated platelets and residual contaminating red and white blood cells. The resultant platelets were pelleted at 1500g and washed 3 times in Tyrode buffer/ethylenediaminetetraacetic acid (EDTA), selecting only the top 66% of the pellet with each cycle. Total RNA was extracted using Tri-Reagent, as described by the manufacturer (Molecular Research Center, Cincinnati, OH). Total RNA from human brain and spleen was obtained from BD Biosciences.
RT-PCR analysis
First-strand synthesis was performed with 0.5 to 1.0 μg total RNA and Superscript II (Invitrogen, Carlsbad, CA) at 42°C for 1 hour, using specific primers for the target mRNAs and oligo(d)T primers in parallel. Polymerase chain reaction (PCR) with specific primers was performed for 30 cycles at different annealing temperatures (50°C, 55°C, and 60°C). PCR-downstream primers were selected with a minimum distance of 200 bp to the respective first-strand primers. In addition, control PCR was performed with upstream and first-strand primers in oligo(d)T-primed first-strand reactions. Table 1 summarizes the primer sequences used.
Primer sequences
Name . | Accession no. . | First-strand primer . | PCR primer downstream . | PCR primer upstream . |
|---|---|---|---|---|
| Rap1GAP1a/Rap1GAP | M64788 | AGCCTGCACCACGACGTA | GCACAGGATAGAACCGATC | CATTCCATACCCGAGCGT |
| Rap1GAPIb/Rap1GAPII | AB003930 | AGCCTGCACCACGACGTA | GCACAGGATAGAACCGATC | CTGGCAGAACACAGATCTAT |
| SPA-1 | AB005666 | GCAATTGAACACTACGCG | GTAGTGGTCACCTCGTTG | CTAGACACCAAAACGGATT |
| E6TP1α | AF090989 | TCTCTGTCTCCAAAGTCCGAT | GAACCAGTATCACTGGATGAA | CCTCAAAATCTTCTATGAACG |
| Tuberin | X75621 | GAAGTTGGAGAAGACGTATCG | CATGTAGTCGCAGACGCAGTA | TGGTGTCCTATGAGATCGTCC |
| KIAA1039 | AB028962 | GCGGAATTTCAGGTTCGA | CATGGAGTGGCTGCGTAC | AGCTGCCATTTACCGACG |
| KIAA1389 | AB037810 | TACTCCTGCCTCGTTCGA | TTGTCTAGCTGAGCTCGATA | TGTCCCTCAATACCGTGC |
| KIAA0545 | AB011117 | TGGGTGCTTATTGCGTAC | TGGGTGCTTATTGCGTAC | GCTGCTCTTTCGAATCGT |
| p761J1523 (DKFZ) | AL136573 | CATCAAATTGGTGTCGATAA | CATATCCAGGGTACGCCG | TGTCCCTCAATACCGTGC |
| CD-GEFI | AF081194 | TGGAAGTTGTGTACGAAGC | TCCACTCCTCCAATACCG | CAGGACTATCACAGTTTCGT |
| CD-GEFIII | AB020653 | CCTCACAGAGATCTTGCG | CAAAGGCCTTGCGGTAATTG | CCTTGGAAAGATCGATTGCT |
| DOCK4 | AY233380 | TTGAAGAGCTCGCGGTAT | GTCTATCTGTCATTTGTCGTAAT | ATGGTACGTGGACCGGAT |
| Epac | AF103905 | GAGGGCCAGTCGGTATAC | CTCCAGTCGTGGTCCGTC | GAGGCAGCAGATCTTGCG |
| Epac2 | U78516 | TGAAATGTCTATGGTCGACG | AAACTTTCAAACTCCGCATAG | ATGTTTCAGTATTTACGACGC |
| Repac | D87467 | GGTCTGCGACAGTCGACT | CCAGGTCTTTCCGGTAGA | TCAGATGTTCCGCGTAGG |
| PDZ-GEF1 | AB002311 | AGAAAGGAACTACGACGTAC | TGGAATGGAGTAGCGACT | AGGCTGTTGAATATCGCG |
| PDZ-GEF2 | AF478567 | TCCAATACTCTCCGTTTAAC | CAGTGACCTCTGTCGAAAC | AAATTGCACTTCATTGTCGAGA |
| C3G | D21239 | CATTGAAGAAGTTTATAATGTCG | ATGGCTGTGAAAGTCGTGC | ATTCAAGAAGCGCGTCAGC |
| smg GDS | X63465 | GCGAAGTTTGGATACGAC | TTTTGATTTACTGTGTCGTATAAG | TGTTACGATAGCCATTCGCT |
Name . | Accession no. . | First-strand primer . | PCR primer downstream . | PCR primer upstream . |
|---|---|---|---|---|
| Rap1GAP1a/Rap1GAP | M64788 | AGCCTGCACCACGACGTA | GCACAGGATAGAACCGATC | CATTCCATACCCGAGCGT |
| Rap1GAPIb/Rap1GAPII | AB003930 | AGCCTGCACCACGACGTA | GCACAGGATAGAACCGATC | CTGGCAGAACACAGATCTAT |
| SPA-1 | AB005666 | GCAATTGAACACTACGCG | GTAGTGGTCACCTCGTTG | CTAGACACCAAAACGGATT |
| E6TP1α | AF090989 | TCTCTGTCTCCAAAGTCCGAT | GAACCAGTATCACTGGATGAA | CCTCAAAATCTTCTATGAACG |
| Tuberin | X75621 | GAAGTTGGAGAAGACGTATCG | CATGTAGTCGCAGACGCAGTA | TGGTGTCCTATGAGATCGTCC |
| KIAA1039 | AB028962 | GCGGAATTTCAGGTTCGA | CATGGAGTGGCTGCGTAC | AGCTGCCATTTACCGACG |
| KIAA1389 | AB037810 | TACTCCTGCCTCGTTCGA | TTGTCTAGCTGAGCTCGATA | TGTCCCTCAATACCGTGC |
| KIAA0545 | AB011117 | TGGGTGCTTATTGCGTAC | TGGGTGCTTATTGCGTAC | GCTGCTCTTTCGAATCGT |
| p761J1523 (DKFZ) | AL136573 | CATCAAATTGGTGTCGATAA | CATATCCAGGGTACGCCG | TGTCCCTCAATACCGTGC |
| CD-GEFI | AF081194 | TGGAAGTTGTGTACGAAGC | TCCACTCCTCCAATACCG | CAGGACTATCACAGTTTCGT |
| CD-GEFIII | AB020653 | CCTCACAGAGATCTTGCG | CAAAGGCCTTGCGGTAATTG | CCTTGGAAAGATCGATTGCT |
| DOCK4 | AY233380 | TTGAAGAGCTCGCGGTAT | GTCTATCTGTCATTTGTCGTAAT | ATGGTACGTGGACCGGAT |
| Epac | AF103905 | GAGGGCCAGTCGGTATAC | CTCCAGTCGTGGTCCGTC | GAGGCAGCAGATCTTGCG |
| Epac2 | U78516 | TGAAATGTCTATGGTCGACG | AAACTTTCAAACTCCGCATAG | ATGTTTCAGTATTTACGACGC |
| Repac | D87467 | GGTCTGCGACAGTCGACT | CCAGGTCTTTCCGGTAGA | TCAGATGTTCCGCGTAGG |
| PDZ-GEF1 | AB002311 | AGAAAGGAACTACGACGTAC | TGGAATGGAGTAGCGACT | AGGCTGTTGAATATCGCG |
| PDZ-GEF2 | AF478567 | TCCAATACTCTCCGTTTAAC | CAGTGACCTCTGTCGAAAC | AAATTGCACTTCATTGTCGAGA |
| C3G | D21239 | CATTGAAGAAGTTTATAATGTCG | ATGGCTGTGAAAGTCGTGC | ATTCAAGAAGCGCGTCAGC |
| smg GDS | X63465 | GCGAAGTTTGGATACGAC | TTTTGATTTACTGTGTCGTATAAG | TGTTACGATAGCCATTCGCT |
All primers are in 5′ to 3′ orientation.
Cloning of Rap1GAP2
The complete 5′ end of the mRNA coding for Rap1GAP2a was obtained with the GeneRacer Kit from Invitrogen. The following gene-specific primers (gsp) were selected from the sequence of KIAA1039: gsp1 5′-GCGGAATTTCAGGTTCGA-3′, gsp2 5′-CTCCATGGAGTGGCTGCGTACGCGGAT-3′, and gsp3 5′-CACGTCCAGGCCTCCTCGGAAAC-3′. 5′–rapid amplification of cDNA ends (RACE) produced a 981-bp fragment, which was cloned into the pCR2.1 vector (Invitrogen). A full-length sequence construct of Rap1GAP2a was obtained by modified asymmetric PCR from the 5′-RACE product and the KIAA1039 clone, as described,43 using 5′-TTAAGCGGCCGCCATGTTTGGCCGGAAGCGCAGTGT-3′ and gsp3 for the 5′-RACE product and 5′-GTTTCCGAGGAGGCCTGGACGTG-3′ and 5′-CCCGTCGACTTAGTGACCCGCACCAGAGCTGG-3′ for the KIAA1039 clone. Full-length Rap1GAP2a and CalDAGGEFIII were cloned into the pSG8 expression vector. The products of 5′-RACE, full-length asymmetric PCR, and pSG8 constructs were verified by sequencing of both strands.
For analysis of splice variants of Rap1GAP2 in platelets, the following primers were used: 5′-CAGACAGACATCGGCACGTA-3′ (exon 1), 5′-ATCATGTTTGGCCGGAAGCGCAGTGTCTCC-3′ (exon 2), 5′-CCGGCTTCTGCTCTTCAA-3′,5′-GGACCCCAGAAGAACAAGGA-3′ (exon 6), 5′-GAAAATGCAGGACGACTATAT-3′, and 5′-ATATAGTCGTCCTGCATTTTC-3′ (exon 6 spliced out).
Sequence comparisons
Multiple alignments of the deduced amino acid sequences were constructed by ClustalX version 1.81 (http://www-igbmc.u-strasbg.fr/BioInfo/).44 The program package Phylip version 3.6a (obtained from J. Felsenstein, Seattle, WA) was used for phylogenetic analyses. Distances between pairs of protein sequences were calculated according to the Jones Taylor Thornton substitution model.45 Tree construction was performed by the neighbor joining method.46 The reliability of the trees was tested by bootstrap analysis,47 with 100 replications (Seqboot program from the Phylip package). Trees were drawn using the Treeview software program (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).48
Northern blotting
Northern blots of multiple human tissues (Clontech, Palo Alto, CA) were carried out by overnight hybridization using modified Church buffer (0.5 M NaB2HPO4, 1 mM EDTA, 7% sodium dodecyl sulfate [SDS], pH 7.2). Probes coding for Rap1GAP1 and Rap1GAP2 were generated from the expression vectors by EcoRI/XhoI digestion, gel purification, and α-32[P]CTP-labeling.
Antibody preparation
Female New Zealand White rabbits were immunized with a peptide corresponding to amino acids 1 to 32 of human Rap1GAP2a/b (MFGRKRSVSFGGFGWIDKTMLASLKVKKQELA-NH2, obtained from Schafer-N [Copenhagen, Denmark]). Four injections at 30-day intervals were performed using 100 μg peptide in 0.5 mL phosphate-buffered saline (PBS) mixed with 0.5 mL Freund adjuvant (Sigma, St Louis, MO; complete and incomplete adjuvant at a ratio of 1:2) per injection. After 149 days, the animals were killed and serum was prepared. Antiserum 644.4 (diluted 1:500 in blocking solution containing 4% nonfat dry milk) was used to detect Rap1GAP2a/b in various cells by Western blotting. For preabsorption, the antigenic peptide was incubated with serum (2 mg peptide/mL serum) for 4 hours at room temperature (RT). Antiserum against human CalDAG-GEFI was prepared similarly using the peptide MAGTLDLDKGCTVEELLRGCIEAFDDSG from the N-terminus of the protein. To detect Rap1GAP1 and SPA-1, commercially available antibodies were used (Santa Cruz Biotechnology, Santa Cruz, CA; BD Transduction Laboratories, Lexington, KY).
Immunofluorescence
Washed platelets were diluted 1:100 in resuspension buffer and allowed to attach to glass coverslips for 30 minutes at RT. Alternatively, COS-1 cells were grown on glass coverslips and transfected with plasmids containing Rap1GAP2 cDNA. Cells were fixed with 3.7% paraformaldehyde in PBS for 15 minutes on ice, and then they were permeabilized with 0.2% Triton X-100 in PBS. To detect Rap1GAP2, the antiserum 644.4 was affinity purified using the antigenic peptide, and the purified serum was used at a concentration of 2.5 μg/mL followed by Cy3-antirabbit (COS-1 cells) or Cy5-antirabbit (platelets) secondary antibodies. In platelets, Rap1 was labeled in parallel using a monoclonal antibody (Transduction Laboratories) diluted 1:50, followed by Cy3-antimouse secondary antibody. COS-1 cell staining was observed with a Zeiss-Axiovert 200 microscope equipped with an LD-Achroplan 40×/0.60 Korr objective lens (Carl Zeiss, Oberkochen, Germany), and images were obtained with a digital camera (Hamamatsu C4742-95) using Openlab software (Improvision, Coventry, UK). Platelets were analyzed using a Zeiss LSM 510 confocal laser scanning microscope equipped with a Plan-Apochromat 63×/1.4 oil DIC objective lens and LSM 510 META software (Carl Zeiss).
Rap1-GTP assay
COS-1 cells were seeded onto 10-cm dishes (1 × 106 cells per plate) and were transfected the next day with cDNA for Rap1B and various combinations of GEFs and GAPs using up to 6 μg DNA, 300 μg diethylaminoethanol (DEAE)–dextran, and 200 μM chloroquine in 6 mL serum-free medium for 3 hours. Two days later cells were washed twice with ice-cold PBS, lysed by the addition of 1 mL lysis buffer (200 mM NaCl; 2.5 mM MgCl2; 50 mM Tris-HCl, pH 7.4; 1% NP-40; 1% glycerol; 1 mM phenylmethylsulfonyl fluoride (PMSF); 1 μM leupeptin; and 2 μM aprotinin) to a 10-cm plate, and scraped off with a rubber policeman. After incubation for 15 minutes at 4°C, lysates were clarified by centrifugation at maximum speed for 15 minutes. Then a sample of 50 μL was removed for analysis of the total amounts of transfected proteins. Seventy-five microliters 10% (vol/vol) glutathione Sepharose bead suspension (Amersham Bioscience, Freiburg, Germany) saturated with GST-RalGDS-RBD was added to the supernatant and incubated at 4°C for 45 minutes rotating. Beads were washed 4 times in 1 × lysis buffer and boiled in 3 × sodium dodecyl sulfate (SDS) sample buffer. Samples were analyzed by Western blot using nitrocellulose membranes (Schleicher & Schuell, Keene, NH), and monoclonal antibodies directed against the HA-tag (HA.11; Covance, Princeton, NJ) were used to detect transfected Rap1B, PDZ-GEF, and Rap1GAP1. An antibody against the VSV-tag (P5D4; Sigma) was used to detect Rap1GAP2 and CalDAG-GEFIII followed by horseradish peroxidase (HRP)–coupled secondary antibodies and enhanced chemiluminescence (ECL) detection (Amersham). All experiments shown were performed at least 3 times and produced similar results.
Phosphorylation of Rap1GAP2
COS-1 cells were transfected with cDNA for wild-type Rap1GAP2, N-terminally tagged with VSV, and various mutants of Rap1GAP2 generated with the transformer site-directed mutagenesis kit (Stratagene, La Jolla, CA). After 2 days, cells were washed twice with ice-cold PBS and were lysed in MIPP-buffer (1% Na-deoxycholate, 20 mM Tris/HCl [pH 7.4], 75 mM NaCl, 0.1% SDS, 1% Triton X-100, 10 mM EDTA, 10 mM Na-pyrophosphate, 50 mM NaF, 5 mM p-nitro-phenyl-phosphate, 2 μg/mL aprotinin, 1 μg/mL leupeptin). Rap1GAP2 was precipitated using anti-VSV antibody P5D4 (Sigma) and protein A/G Sepharose (Santa Cruz Biotechnology) and was phosphorylated in vitro with purified cGK I and a catalytic subunit of cAMP-dependent protein kinase, as described.49 Samples were subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and then by blotting on nitrocellulose. Blots were exposed first to film to detect 32P incorporation. Then Rap1GAP2 expression was analyzed using anti-VSV antibodies and ECL.
Results
GEFs and GAPs of Rap1 expressed in human platelets
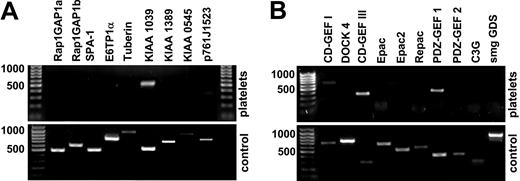
In our analysis of Rap1 inhibition by NO/cGMP/cGKI, we set out to identify the relevant proteins controlling Rap1 activity in human platelets. We performed systematic reverse transcription–polymerase chain reaction (RT-PCR) analysis of known GEFs and GAPs of Rap1 expressed in platelets. For RNA isolation, platelets were purified extensively to avoid contamination by lymphocyte RNA. To verify that the primer sets were able to detect the mRNA for the protein of interest, we performed control experiments using RNA from tissues in which the particular proteins are known to be expressed (Figure 1A-B, lower panels). Platelets contain considerable amounts of RNA, including PolyA+ mRNA, and active protein synthesis is carried out in these cells.50,51 A recent comparison of transcriptome and proteome information from human platelets revealed a high degree of correlation between both types of data.52
Expression of GAPs and GEFs of Rap1 in human platelets. RT-PCR analysis of GAP (A) and GEF (B) expression in washed human platelets was performed using transcript-specific primers for reverse transcription and PCR. As positive control, human brain RNA was used in most reactions except SPA-1, KIAA1039, KIAA0545, and CalDAG-GEFI (CD-GEF I), which were detected in spleen (bottom row of each panel). Results shown were confirmed using oligo-(d)T priming for first-strand synthesis. None of the known GAPs could be detected in platelets. Database searches disclosed a number of cDNAs encoding new potential Rap1GAPs; of these, KIAA1039 was expressed in platelets. The only GEFs of Rap1 detectable in platelets were CalDAG-GEFI (CD-GEF I)/RasGRP2, CalDAG-GEFIII (CD-GEF III)/RasGRP3, and PDZ-GEF1. Shown are representative results from at least 3 (platelets) or 2 (control) similar experiments.
Expression of GAPs and GEFs of Rap1 in human platelets. RT-PCR analysis of GAP (A) and GEF (B) expression in washed human platelets was performed using transcript-specific primers for reverse transcription and PCR. As positive control, human brain RNA was used in most reactions except SPA-1, KIAA1039, KIAA0545, and CalDAG-GEFI (CD-GEF I), which were detected in spleen (bottom row of each panel). Results shown were confirmed using oligo-(d)T priming for first-strand synthesis. None of the known GAPs could be detected in platelets. Database searches disclosed a number of cDNAs encoding new potential Rap1GAPs; of these, KIAA1039 was expressed in platelets. The only GEFs of Rap1 detectable in platelets were CalDAG-GEFI (CD-GEF I)/RasGRP2, CalDAG-GEFIII (CD-GEF III)/RasGRP3, and PDZ-GEF1. Shown are representative results from at least 3 (platelets) or 2 (control) similar experiments.
Surprisingly, none of the established Rap1-GAPs, including Rap1GAP, SPA-1, and E6TP1α, were detectable in platelets (Figure 1A). Therefore, we searched human EST and cDNA databases for sequences homologous to the GAP domain of Rap1GAP. Four types of cDNA were identified, one of which, KIAA1039 (GenBank accession number AB02896253 ), was found to be strongly expressed in human platelets (Figure 1A). We detected mRNAs for PDZ-GEF1 and CalDAG-GEFIII, weakly detected it for CalDAG-GEFI (Figure 1B), but did not detect it for DOCK4, C3G, smgGDS, or any of the Epacs. Recently, CalDAG-GEFI was shown to be expressed in mouse megakaryocytes and platelets,10,13 and we confirmed CalDAG-GEFI protein expression in human platelets by Western blot (not shown). As observed on the mRNA level, the Western blot signal for CalDAG-GEFI was weak, indicating low expression levels of CalDAG-GEFI in human platelets.
Cloning of a new Rap1GAP
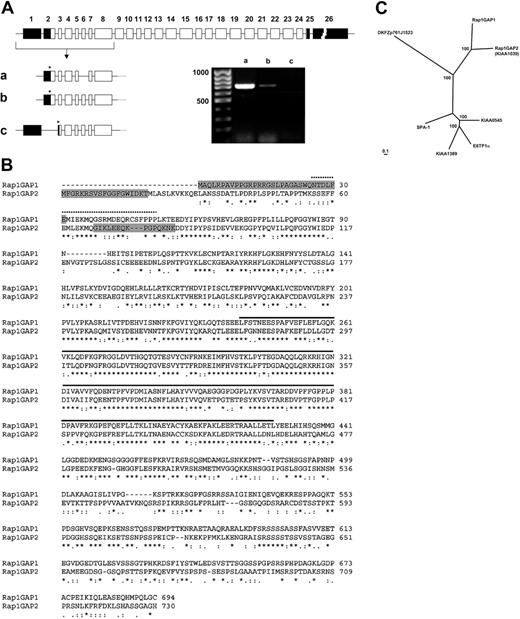
The KIAA1039 sequence represented only a partial cDNA sequence lacking the 5′ end. To obtain the complete cDNA, we performed a 5′-RACE experiment using purified platelet mRNA as template. Complete cDNA was isolated encoding a protein of 715 residues with a predicted molecular weight of 78.4 kDa. Comparison with the Pfam protein motif database showed the presence of a conserved Rap_GAP domain (residues 262-449). The closest relative of the new GAP is Rap1GAP, exhibiting 52% identity (Figure 2B). Two splice variants of Rap1GAP have been described and named Rap1GAP and Rap1GAPII; the latter contains 31 additional amino acids at the N-terminus.25 To comply with standard nomenclature, we have named the new protein Rap1GAP2. Accordingly, we would like to suggest that the splice isoforms Rap1GAP and Rap1GAPII be renamed Rap1GAP1a and Rap1GAP1b. During the course of this study, a new full-length cDNA derived from cerebellum (AK124640) was published containing the KIAA1039 sequence and confirming part of our RACE product. However, the N-terminus of the encoded protein differed from our sequence. Database searches revealed that the gene for Rap1GAP2 is localized on chromosome 17 (17p13.3), spanning 261 kilobases, and that the coding sequence is distributed over 26 exons (Figure 2A). Comparison of genomic and cDNA sequences showed that our platelet sequence and the cerebellar sequence were products of alternative splicing (splice variants a and c in Figure 2A, respectively). Using exon-specific primers, we confirmed variant a as the predominant form in human platelets. In addition, we discovered a third splice variant present in human platelets (designated b in Figure 2A). In summary, 3 splice variants of the new Rap1GAP2, encoding alternative N-termini, have been identified. Variant a lacks exons 1 and 6, which are prevalent in platelets, variant b includes exon 6, and variant c corresponds to the cerebellar clone lacking exon 2 (Figure 2A). Interestingly, a putative GoLoco domain found in Rap1GAP1b/Rap1GAPII25 appears to be conserved in Rap1GAP2b and Rap1GAP2c (Figure 2B). Orthologs of Rap1GAP2 are present in mouse (AK122424) and rat (XP_220692). Sequences of Rap1GAP2a and Rap1GAP2b have been submitted to the GenBank/EMBL/DDBJ database under the accession numbers AJ628447 and AJ628446.
Gene structure and protein sequence of Rap1GAP2. A complete cDNA corresponding to KIAA1039 was cloned from platelet RNA encoding a protein most closely related to Rap1GAP. Consequently, the new protein was named Rap1GAP2. (A) Rap1GAP2 is expressed in 3 different splice variants. The originally cloned variant, a, is the predominant form in platelets. Rap1GAP2a lacks exons 1 and 6, Rap1GAP2b lacks only exon 1, and Rap1GAP2c (AK124640, cloned from cerebellum) is generated by the splicing of exon 2, resulting in the appearance of a new start codon within exon 3 (asterisk indicates start codons, including Kozak initiation sequences). ▪ indicates noncoding regions. Exon 26 comprises 4351 bp. To the right, the results of RT-PCR analysis of variant expression in platelets are shown. Only the splice variants Rap1GAP2a and Rap1GAP2b are detectable in human platelets. This experiment was performed twice with similar results. (B) Sequence comparison between Rap1GAP1 and Rap1GAP2 reveals a conserved Rap1GAP domain (solid line) and a putative GoLoco domain (dotted line). Differences between variants are marked in gray: the box in Rap1GAP1 marks additional sequences found only in the Rap1GAP1b/Rap1GAPII splice variant, the first N-terminal box in the Rap1GAP2 sequence is missing in variant c, the second box in Rap1GAP2 indicates additional sequences derived from exon 6, present only in Rap1GAP2b and Rap1GAP2c. Asterisks, double dots, and single dots indicate different degrees of amino acid conservation. (C) A phylogenetic tree of all presently known Rap1GAPs, including related uncharacterized cDNAs, shows Rap1GAP1 and Rap1GAP2 as distinct subgroups. Accession numbers of used amino acid sequences are presented in “Materials and methods.” Numbers at the branches represent the confidence limits computed by the bootstrap procedure. Bar indicates 0.1 substitutions per site.
Gene structure and protein sequence of Rap1GAP2. A complete cDNA corresponding to KIAA1039 was cloned from platelet RNA encoding a protein most closely related to Rap1GAP. Consequently, the new protein was named Rap1GAP2. (A) Rap1GAP2 is expressed in 3 different splice variants. The originally cloned variant, a, is the predominant form in platelets. Rap1GAP2a lacks exons 1 and 6, Rap1GAP2b lacks only exon 1, and Rap1GAP2c (AK124640, cloned from cerebellum) is generated by the splicing of exon 2, resulting in the appearance of a new start codon within exon 3 (asterisk indicates start codons, including Kozak initiation sequences). ▪ indicates noncoding regions. Exon 26 comprises 4351 bp. To the right, the results of RT-PCR analysis of variant expression in platelets are shown. Only the splice variants Rap1GAP2a and Rap1GAP2b are detectable in human platelets. This experiment was performed twice with similar results. (B) Sequence comparison between Rap1GAP1 and Rap1GAP2 reveals a conserved Rap1GAP domain (solid line) and a putative GoLoco domain (dotted line). Differences between variants are marked in gray: the box in Rap1GAP1 marks additional sequences found only in the Rap1GAP1b/Rap1GAPII splice variant, the first N-terminal box in the Rap1GAP2 sequence is missing in variant c, the second box in Rap1GAP2 indicates additional sequences derived from exon 6, present only in Rap1GAP2b and Rap1GAP2c. Asterisks, double dots, and single dots indicate different degrees of amino acid conservation. (C) A phylogenetic tree of all presently known Rap1GAPs, including related uncharacterized cDNAs, shows Rap1GAP1 and Rap1GAP2 as distinct subgroups. Accession numbers of used amino acid sequences are presented in “Materials and methods.” Numbers at the branches represent the confidence limits computed by the bootstrap procedure. Bar indicates 0.1 substitutions per site.
RNA expression of Rap1GAP2 and Rap1GAP1 in human tissues
Single transcripts of 6.7 kb for Rap1GAP2 and 3.4 kb for Rap1GAP1 were detectable by Northern blot analysis of polyA+ RNA from different human tissues. Expression levels varied between tissues, and both genes tended to be expressed in different locations. For example, Rap1GAP2 is specific for heart, testis, and blood leukocytes, whereas Rap1GAP1 is strongly expressed in brain, kidney, and prostate (Figure 3). Overlapping expression is observed in the pancreas and the gastrointestinal tract, with prominent signals in the stomach. The expression pattern observed for Rap1GAP1 is consistent with previous data.26,28
Tissue distribution of Rap1GAP2 and Rap1GAP1. Northern blots containing PolyA+ RNA from multiple tissues were probed with 32P-labeled probes specific for Rap1GAP2 (top row) or Rap1GAP1 (bottom row). Size of the detected mRNAs is indicated. Shown are representative results from 2 experiments.
Tissue distribution of Rap1GAP2 and Rap1GAP1. Northern blots containing PolyA+ RNA from multiple tissues were probed with 32P-labeled probes specific for Rap1GAP2 (top row) or Rap1GAP1 (bottom row). Size of the detected mRNAs is indicated. Shown are representative results from 2 experiments.
Rap1GAP2 protein is expressed in human platelets
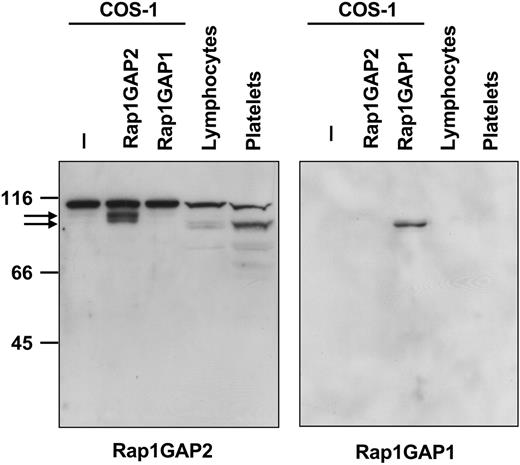
A polyclonal antiserum was raised against a peptide derived from the N-terminus of Rap1GAP2a/b. Western blotting with the obtained antiserum showed the expression of a 95-kDa protein in COS-1 cells transiently transfected with an expression vector for epitope-tagged Rap1GAP2a but not in the cells transfected with Rap1GAP1 (Figure 4). In platelets, and to a lesser degree in lymphocytes, a protein of approximately 90 kDa was detected (Figure 4), corresponding to endogenous Rap1GAP2a/b. The epitope tag included in the transfected protein resulted in an additional 4.4 kDa of calculated molecular weight, which matches almost exactly the difference observed between endogenous and transfected proteins. Preabsorption of the antiserum with the antigenic peptide abrogated labeling of the 90- to 95-kDa Rap1GAP2 band (data not shown). To further verify mRNA data, we analyzed Rap1GAP1 and SPA-1 protein expression in platelets and lymphocytes. As expected, Rap1GAP1 was not detectable (Figure 4), whereas SPA-1 protein was expressed in lymphocytes but not in platelets (data not shown).
Expression of Rap1GAP2 protein. COS-1 cells transiently transfected without or with epitope-tagged Rap1GAP2 or Rap1GAP1, isolated peripheral blood lymphocytes, and isolated washed human platelets were lysed in SDS-containing stop solution. Total protein was separated by SDS-PAGE and immunoblotted with rabbit antiserum against an N-terminal peptide derived from Rap1GAP2 (left) or with an antibody specific for Rap1GAP1 (right). Arrows indicate tagged overexpressed (95 kDa) and endogenous Rap1GAP2 (90 kDa). The antiserum cross-reacts with an unknown protein of 110 kDa. Platelets, and to a lesser extent lymphocytes, contain endogenous Rap1GAP2 but not Rap1GAP1 protein. Shown are examples of experiments performed at least 4 times.
Expression of Rap1GAP2 protein. COS-1 cells transiently transfected without or with epitope-tagged Rap1GAP2 or Rap1GAP1, isolated peripheral blood lymphocytes, and isolated washed human platelets were lysed in SDS-containing stop solution. Total protein was separated by SDS-PAGE and immunoblotted with rabbit antiserum against an N-terminal peptide derived from Rap1GAP2 (left) or with an antibody specific for Rap1GAP1 (right). Arrows indicate tagged overexpressed (95 kDa) and endogenous Rap1GAP2 (90 kDa). The antiserum cross-reacts with an unknown protein of 110 kDa. Platelets, and to a lesser extent lymphocytes, contain endogenous Rap1GAP2 but not Rap1GAP1 protein. Shown are examples of experiments performed at least 4 times.
Rap1GAP2 and Rap1 partially colocalize in human platelets
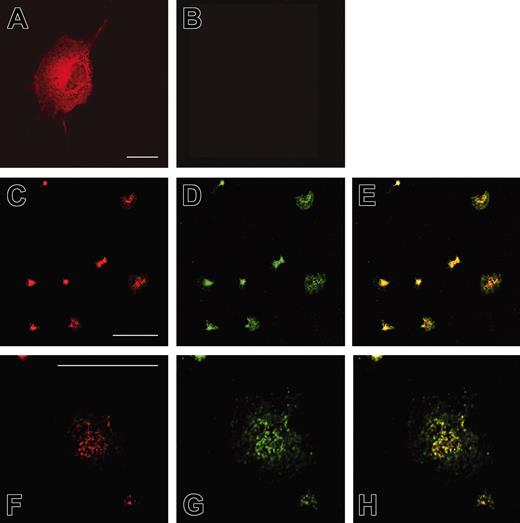
To analyze the subcellular localization of Rap1GAP2, we transfected the cDNA of Rap1GAP2 into COS-1 cells. Staining with the antiserum against Rap1GAP2 revealed a cytosolic and perinuclear localization of overexpressed Rap1GAP2 (Figure 5A). To confirm specificity of the signal, we stained nontransfected cells with the Rap1GAP2 antibody. The signal was absent in nontransfected cells, indicating that the unspecific background protein detected in Western blot (Figure 4) was not labeled in immunofluorescence (Figure 5B). We next investigated the localization of endogenous Rap1GAP2 in human platelets spread on glass coverslips. Rap1GAP2 staining was observed in central, granular structures (Figure 5C, F), and this staining overlapped with the localization of endogenous Rap1 (Figure 5D, G, and merged images E, H). Rap1 tended to exhibit localization that was spread out, whereas Rap1GAP2 was confined to the central structures.
Localization of Rap1GAP2 in transfected COS-1 cells and in platelets. COS-1 cells transiently transfected with Rap1GAP2 (A) and nontransfected control cells (B) were stained with an antibody specific for Rap1GAP2. Rap1GAP2 localized predominantly to cytosolic and perinuclear structures in transfected cells. Nontransfected control cells were not labeled, indicating specificity of the antibody. (C-H) Human platelets spread on glass were double labeled with antibodies specific for Rap1GAP2 (C, F) and Rap1 (D, G, and merged images E, H). (F-H) In the center of each panel, a single large, spread platelet is shown. Both Rap1GAP2 and Rap1 proteins partially colocalized in central vesicular structures. Shown are examples of experiments performed at least 5 times. Bar = 10 μM.
Localization of Rap1GAP2 in transfected COS-1 cells and in platelets. COS-1 cells transiently transfected with Rap1GAP2 (A) and nontransfected control cells (B) were stained with an antibody specific for Rap1GAP2. Rap1GAP2 localized predominantly to cytosolic and perinuclear structures in transfected cells. Nontransfected control cells were not labeled, indicating specificity of the antibody. (C-H) Human platelets spread on glass were double labeled with antibodies specific for Rap1GAP2 (C, F) and Rap1 (D, G, and merged images E, H). (F-H) In the center of each panel, a single large, spread platelet is shown. Both Rap1GAP2 and Rap1 proteins partially colocalized in central vesicular structures. Shown are examples of experiments performed at least 5 times. Bar = 10 μM.
Activation of Rap1 GTPase activity by Rap1GAP2
Given that no physiologic activator of endogenous Rap1GAP2 is known and given that platelets are not amenable to transfection studies, we decided to analyze GTPase activity of Rap1GAP2 in cell lines. We transiently expressed the Rap1GAP2a variant in COS-1 cells and measured Rap1 GTPase activity with the pull-down assay developed by Franke et al.11 Coexpression of Rap1B, together with the platelet GEF CalDAG-GEFIII (Figure 6A) or PDZ-GEF1 (not shown), strongly increased amounts of Rap1B-guanosine triphosphate (GTP). Additional expression of Rap1GAP2 or Rap1GAP1 almost completely blocked the formation of Rap1B-GTP (Figure 6A). Using increasing concentrations of both GAPs, we show that Rap1GAP2 and Rap1GAP1 have similar Rap1-GTPase–activating potential (Figure 6A-B). Recently, the crystal structure of the catalytic domain of Rap1GAP1 was solved, indicating an important role of an asparagine residue for catalytic activity.54 This asparagine residue is conserved in Rap1GAP2 (N342 of Rap1GAP2a), and mutation of this site to alanine completely abolished GAP activity, as expected (data not shown). To verify that the Rap1GAP domain conveys Rap1-specific GAP activity, we studied effects of Rap1GAP2 on Ras-GTP levels using the Ras-binding domain of Raf-kinase as an activation-specific probe. No influence of Rap1GAP2 overexpression on CalDAG-GEFIII–stimulated Ras activity could be observed (data not shown). These data confirm that Rap1GAP2 is indeed a functional and Rap1-specific GTPase-activating protein, as predicted from the sequence homology with known Rap1GAPs.
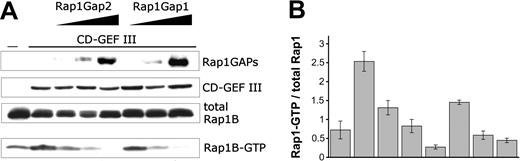
GTPase activity of Rap1GAP2. COS-1 cells were transiently transfected with HA-tagged Rap1B and VSV-tagged CalDAG-GEFIII (CD-GEF III). In addition, increasing amounts of VSV-tagged Rap1GAP2a or HA-tagged Rap1GAP1 were expressed as indicated. Two days after transfection, cells were lysed, and pull-down assays with an activation-specific probe were performed to determine the amounts of Rap1B-GTP (A). Levels of total Rap1B, Rap1GAP2, Rap1GAP1, and CalDAG-GEFIII were determined with tag-specific antibodies. Blots from 4 independent pull-down experiments were scanned and quantified (B). To compensate for differences in total Rap1 expression levels, ratios of Rap1-GTP to total Rap1 signals were calculated. Shown are mean ± SEM. Rap1GAP2 and Rap1GAP1 blocked CalDAG-GEFIII-induced activation of Rap1B with similar potency.
GTPase activity of Rap1GAP2. COS-1 cells were transiently transfected with HA-tagged Rap1B and VSV-tagged CalDAG-GEFIII (CD-GEF III). In addition, increasing amounts of VSV-tagged Rap1GAP2a or HA-tagged Rap1GAP1 were expressed as indicated. Two days after transfection, cells were lysed, and pull-down assays with an activation-specific probe were performed to determine the amounts of Rap1B-GTP (A). Levels of total Rap1B, Rap1GAP2, Rap1GAP1, and CalDAG-GEFIII were determined with tag-specific antibodies. Blots from 4 independent pull-down experiments were scanned and quantified (B). To compensate for differences in total Rap1 expression levels, ratios of Rap1-GTP to total Rap1 signals were calculated. Shown are mean ± SEM. Rap1GAP2 and Rap1GAP1 blocked CalDAG-GEFIII-induced activation of Rap1B with similar potency.
Rap1GAP2 is phosphorylated by cGKI on serine 7
Rap1GAP2 contains a great number of serine residues, which could be substrates for protein phosphorylation. In vivo labeling of transfected COS1-cells with 32P-ortho-phosphate, followed by precipitation of expressed Rap1GAP2, revealed a very strong basal phosphorylation of Rap1GAP2 (data not shown). Rap1GAP2a and Rap1GAP2b contain 2 serine residues that correspond to the consensus sequence -(R/K)2-X-S/T- for phosphorylation by cyclic nucleotide–regulated protein kinases: serine 7 (-RKRS-) and serine 549 (564 in Rap1GAP2b) (-KRRS-). To evaluate whether Rap1GAP2a/b was a substrate of cyclic nucleotide–regulated protein kinases, we performed in vitro kinase assays with purified cGKI and the catalytic subunit of cAMP-dependent protein kinase (cAK). Both kinases strongly phosphorylated Rap1GAP2a (Figure 7A). To identify the exact phosphorylation sites, candidate serine residues were mutated to alanine. Mutation of serine 7 completely abolished Rap1GAP2a phosphorylation by cGKI, whereas phosphorylation by cAK was only marginally reduced (Figure 7A). Mutation of serine 549 did not change cGKI- or cAK-mediated Rap1GAP2a phosphorylation. From these results, we conclude that cGKI phosphorylates Rap1GAP2a/b on a single serine residue. To identify the functional consequences of phosphorylation, we compared GTPase activities of wild-type and serine 7 to alanine, and phosphomimetic serine 7 to aspartate or glutamate, mutants of Rap1GAP2a. However, no significant differences in GTPase activity could be detected in transfected COS-1 cells (Figure 7B). These data might suggest that serine 7 phosphorylation does not directly regulate Rap1GAP2a activity. However, considering the strong activity of Rap1GAP2a in our COS cell assay (Figure 6), subtle changes in catalytic activity cannot be excluded.
Phosphorylation of Rap1GAP2 by cyclic nucleotide–regulated kinases. (A) COS-1 cells were transiently transfected with VSV-tagged wild-type (wt) Rap1GAP2a and mutants of Rap1GAP2a containing serine-to-alanine mutations of serine 7 and serine 549, either singly (S7A and S549A) or in combination (S7A/S549A). Expressed proteins were precipitated with anti-VSV antibody, and in vitro kinase assays were performed using γ-32P]ATP and purified cGKI or a catalytic subunit of cAK. To detect 32P incorporation, samples were separated by SDS-PAGE, blotted onto nitrocellulose, and exposed to film (32P). Then protein amounts were determined using anti-VSV antibodies and ECL detection (WB). Each kinase strongly phosphorylated Rap1GAP2a, and the mutation of serine 7 to alanine abolished Rap1GAP2a phosphorylation by cGKI. (B) Rap1-GTP levels were analyzed in COS-1 cells transiently transfected with Rap1, CalDAG-GEFIII (CD-GEF III), wild-type Rap1GAP2a, and mutants of Rap1GAP2a containing mutations of serine 7 to alanine (S7A), aspartate (S7D) or glutamate (S7E), as described in the legend to Figure 6. Mutation of serine 7 did not change Rap1GAP2 activity in COS-1 cells. Shown are representative results from at least 3 independent experiments.
Phosphorylation of Rap1GAP2 by cyclic nucleotide–regulated kinases. (A) COS-1 cells were transiently transfected with VSV-tagged wild-type (wt) Rap1GAP2a and mutants of Rap1GAP2a containing serine-to-alanine mutations of serine 7 and serine 549, either singly (S7A and S549A) or in combination (S7A/S549A). Expressed proteins were precipitated with anti-VSV antibody, and in vitro kinase assays were performed using γ-32P]ATP and purified cGKI or a catalytic subunit of cAK. To detect 32P incorporation, samples were separated by SDS-PAGE, blotted onto nitrocellulose, and exposed to film (32P). Then protein amounts were determined using anti-VSV antibodies and ECL detection (WB). Each kinase strongly phosphorylated Rap1GAP2a, and the mutation of serine 7 to alanine abolished Rap1GAP2a phosphorylation by cGKI. (B) Rap1-GTP levels were analyzed in COS-1 cells transiently transfected with Rap1, CalDAG-GEFIII (CD-GEF III), wild-type Rap1GAP2a, and mutants of Rap1GAP2a containing mutations of serine 7 to alanine (S7A), aspartate (S7D) or glutamate (S7E), as described in the legend to Figure 6. Mutation of serine 7 did not change Rap1GAP2 activity in COS-1 cells. Shown are representative results from at least 3 independent experiments.
Discussion
Identification of a new Rap1GAP in human platelets
In the present work, we have identified and characterized Rap1GAP2, a new GTPase-activating protein of Rap1. Rap1GAP2 is the first GTPase-activating protein of Rap1 to be described in platelets, and our data suggest that it might be the only one. We show that Rap1GAP2 colocalizes with Rap1 in platelets and can potently inhibit Rap1-GTP formation in COS-cells. Thus, Rap1GAP2 most likely also inhibits endogenous Rap1 in platelets.
Expression patterns of Rap1GAPs and GEFs
The Rap1GAP family of proteins can be grouped into at least 2 subfamilies, Rap1GAP and SPA-1. Expression patterns of the various Rap1-GAPs are tissue specific, whereas Rap1 itself is ubiquitously expressed.55 Rap1GAP1 is prominent in kidney, prostate, and brain, Rap1GAP2 is found in platelets and lymphocytes, and SPA-1 is strongly expressed in lymphoid tissue28 but not in platelets. Rap1GAP2 and SPA-1 are also present together in heart and testis. Rap1GAP1 and Rap1GAP2 are found in the pancreas. In the gastrointestinal tract, these 3 GAPs appear to be coexpressed. Of course, tissue distribution must be further analyzed on the cellular level. Variations in Rap1GAP expression patterns suggest that Rap1 signaling is differentially regulated, depending on the cell type. These patterns might also reflect cell type–specific functions of Rap1.
We have identified 3 Rap1-activating proteins in platelets, namely PDZ-GEF1, CalDAG-GEFI/RasGRP2, and CalDAG-GEFIII/RasGRP3. PDZ-GEF1 had been known to be ubiquitously expressed, though platelet expression had not been analyzed.21 CalDAG-GEFI has been studied in mouse platelets,10 but CalDAG-GEFIII expression was observed only in glial cells of brain, in kidney mesangial cells, and in B cells.14,15 We conclude that Rap1 is activated in a cell type–specific way by different sets of GEFs. Little is known about the regulation of PDZ-GEF1 and CalDAG-GEFs. PDZ-GEF1 function is restricted to the activation of Rap1 and Rap2,22 whereas CalDAG-GEFIII can activate a broad range of Ras family GTPases, including Rap1, Ha-Ras, and R-Ras.14 CalDAG-GEFIII is regulated mainly by diacylglycerol (DAG),15 and CalDAG-GEFI is considered Ca2+-dependent.2,10 CalDAG-GEFIII can also be phosphorylated by protein kinase C (PKC), and phosphorylation is thought to enhance its GEF activity in lymphocytes.15,56
Phosphorylation of Rap1GAP2
In the present study, we show that Rap1GAP2 is phosphorylated by cyclic nucleotide–regulated kinases cAK and cGKI. Cyclic nucleotide–dependent protein kinases often exhibit similar substrate specificities, at least in vitro, and Polakis et al57 provided indirect evidence that Rap1GAP1 could be phosphorylated by cAK on serine residues 490 and 499.58 Serine 490, but not serine 499, is conserved in Rap1GAP2; however, mutation of the corresponding serine 549 of Rap1GAP2a does not affect phosphorylation by cAK or cGKI. Instead we have identified serine 7 as the phosphorylation site of Rap1GAP2a, which is preferred by cGKI over cAK. Interestingly, serine 7 is present only in the 2 platelet splice variants Rap1GAP2a and Rap1GAP2b; it is not present in the Rap1GAP2c isoform found in cerebellum. This might suggest important regulatory functions of the additional N-terminal sequences derived from exon 2 (Figure 2A). cGKI can inhibit platelet aggregation,40,41 and, under certain conditions, activating effects of cGKI on platelet function have been described.59,60 However, none of the hitherto identified cGKI substrates in platelets have been conclusively shown to mediate platelet inhibition/activation by NO/cGMP/cGKI. For example, the deletion of vasodilator-stimulated phosphoprotein (VASP), an established substrate of cGKI involved in platelet adhesion,61,62 only marginally affects NO/cGMP function in platelets.63,64 The new protein Rap1-GTP-interacting adapter molecule (RIAM) might be a link between VASP and Rap1, though platelet functions of RIAM have not been studied.65 Early studies also showed direct phosphorylation of Rap1 by cGKI, but neither GTP-binding nor GTPase activity were influenced by phosphorylation,66,67 and no correlation between Rap1 phosphorylation and inhibition of platelet activation could be detected.68 Rap1GAP2a/b is a new effector of the NO/cGMP pathway, and we speculate that Rap1GAP2 phosphorylation by cGKI could enhance its GAP activity, thereby leading to Rap1 inactivation and ultimately to the inactivation of integrin αIIbβ3. However, serine 7 phosphorylation does not appear to change the GAP activity of Rap1GAP2a/b directly. Instead, serine 7 phosphorylation could regulate the association of Rap1GAP2a/b with binding proteins. The identities of such proteins will have to be determined in future studies. Rap1GAP2 contains a high number of serine and threonine residues that might represent additional phosphorylation sites for different kinases. In vivo labeling of transfected COS-1 cells revealed a strong basal phosphorylation of Rap1GAP2. Because COS-1 cells do not express cGKI, phosphorylation could be mediated in part by cAK. GSK3 was recently shown to phosphorylate Rap1GAP1 on serine 525, resulting in enhanced proteasomal degradation.27 However, this residue is not conserved in Rap1GAP2.
The present work suggests that PDZ-GEFI, CalDAG-GEFs I and III, and Rap1GAP2 control Rap1 activity in human platelets. Additionally, we have identified Rap1GAP2 as new target of the NO/cGMP/cGKI signaling pathway. The mechanisms of Rap1GAP2 regulation deserve further study considering the important role of Rap1 in platelet activation and aggregation.
Supported by the Deutsche Forschungsgemeinschaft (SFB 553).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Prepublished online as Blood First Edition Paper, January 4, 2005; DOI 10.1182/blood-2004-09-3605.
We thank J. Bos for constructs and E. Butt for purified kinases. We also thank all our colleagues at the Institute for Biochemistry II for continuous support and discussions and Barbara Pfeiffer for expert technical assistance, particularly in the generation of antibodies.


![Figure 7. Phosphorylation of Rap1GAP2 by cyclic nucleotide–regulated kinases. (A) COS-1 cells were transiently transfected with VSV-tagged wild-type (wt) Rap1GAP2a and mutants of Rap1GAP2a containing serine-to-alanine mutations of serine 7 and serine 549, either singly (S7A and S549A) or in combination (S7A/S549A). Expressed proteins were precipitated with anti-VSV antibody, and in vitro kinase assays were performed using γ-32P]ATP and purified cGKI or a catalytic subunit of cAK. To detect 32P incorporation, samples were separated by SDS-PAGE, blotted onto nitrocellulose, and exposed to film (32P). Then protein amounts were determined using anti-VSV antibodies and ECL detection (WB). Each kinase strongly phosphorylated Rap1GAP2a, and the mutation of serine 7 to alanine abolished Rap1GAP2a phosphorylation by cGKI. (B) Rap1-GTP levels were analyzed in COS-1 cells transiently transfected with Rap1, CalDAG-GEFIII (CD-GEF III), wild-type Rap1GAP2a, and mutants of Rap1GAP2a containing mutations of serine 7 to alanine (S7A), aspartate (S7D) or glutamate (S7E), as described in the legend to Figure 6. Mutation of serine 7 did not change Rap1GAP2 activity in COS-1 cells. Shown are representative results from at least 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/8/10.1182_blood-2004-09-3605/6/m_zh80080577100007.jpeg?Expires=1763656184&Signature=KP4X593RvNgNi5Ci6yGxi0vESeys5asszF-VXgCuo6655njHdR2OPKdpQDP5MGX9clAEANSdwUM5n5G2kpJ2ZaS3l4YX5cY-S4cQ~zEEXcatwsJyPR7a4nA~XdwQlKr9c9EytybaWep-ITt2oRkL4sPEdebRGzjCADL3I-8mux-fGaTOoBafU3PjQFv7sandAEnLnQTXvc9SADY8FPi0jyYTPSzeMdv515XLti6ONZPbsgdxlc-sxPoFlsg89mGObBTTrRm4Vwy3CcfG1iDJPxwizJYm1ZPFmc~0bkXjqiFj8EXnnZcUrLBW4ucmTT0z-ctapRpM8WeE1rVLF54ZqA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal