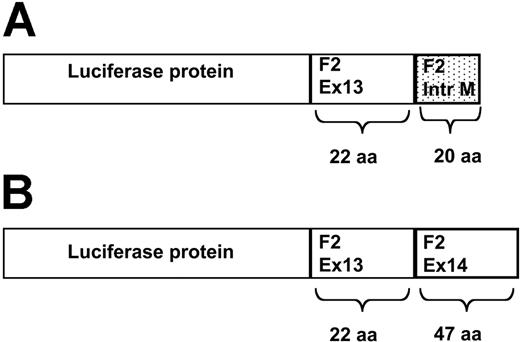
A noncoding polymorphism in the last intron of the prothrombin gene 19911A>G is associated with slightly elevated plasma prothrombin levels. When present in combination with the 20210A mutation on the other allele, it seems to contribute to the risk of thrombosis.1,2 Recently, von Ahsen and Oellerich3 described that splicing efficiency was increased in constructs carrying the 19911G allele, supporting a functional role for the 19911A>G variant. However, they also presented data that contradict these findings. Using constructs in which the genomic fragment containing prothrombin exons 13 and 14 separated by intron 13 was cloned immediately downstream from the luciferase cDNA, protein expression was lower for the 19911G constructs than for the corresponding 19911A constructs.3(Fig2) As a possible explanation for these contrasting findings, the authors suggest that more distant single-nucleotide polymorphisms (SNPs) on the 19911G-20210G allele, which are absent in the constructs, may be responsible for the increased expression of this allele. Although at present this cannot be excluded, we feel that there might be another explanation for their findings. One of the assumptions made by the authors is that the luciferase activity of the different fusion proteins is identical. More specifically, the luciferase protein translated from the nonspliced mRNA has been elongated by 22 amino acids encoded by exon 13 of the prothrombin gene and 20 additional amino acids translated from the 5′ region of intron 13 (Figure 1A), whereas the luciferase protein translated from the spliced mRNA has been elongated with the amino acids from prothrombin exon 13 and 14 (Figure 1B). This results in 2 luciferase molecules with different prothrombin-derived tails, depending on whether or not they have been spliced (Figure 1). The authors do not provide any evidence that these different tails do not affect luciferase activity differently. If the longer exon 13-exon 14 tail affects luciferase activity more negatively than the exon 13-intron 13 tail, increased splicing may be “punished” in this model system by a reduced specific activity. In other words, the 19911G variant may result in increased splicing, and increased protein expression, but the latter is masked by the differences in specific activity of the 2 luciferase fusion proteins. In this context it should be reminded that it has been shown that the C-terminal region of luciferase is important for its activity.4 Similar considerations as presented apply to the β-galactosidase activity of the double reporter constructs.3(Fig6) Correct splicing results in a β-galactosidase-luciferase fusion protein, whereas absence of splicing results in a β-galactosidase protein alone. Because β-galactosidase activity is used for the normalization of luciferase activity, differences in β-galactosidase activity between the constructs influence the luc-gal ratio, possibly leading to a misinterpretation of results.
Schematic representation of the luciferase-prothrombin fusion proteins. (A) Luciferase-prothrombin fusion protein translated from a nonspliced mRNA. Absence of splicing leads to the production of a luciferase protein with a tail derived from exon 13 and part of intron M of prothrombin. (B) Luciferase-prothrombin fusion protein translated from a spliced mRNA. In the event of correct splicing, a luciferase protein connected to a protrombin-derived tail is formed. The prothrombin tail is produced from the translation of exon 13 and 14. aa indicates amino acids.
Schematic representation of the luciferase-prothrombin fusion proteins. (A) Luciferase-prothrombin fusion protein translated from a nonspliced mRNA. Absence of splicing leads to the production of a luciferase protein with a tail derived from exon 13 and part of intron M of prothrombin. (B) Luciferase-prothrombin fusion protein translated from a spliced mRNA. In the event of correct splicing, a luciferase protein connected to a protrombin-derived tail is formed. The prothrombin tail is produced from the translation of exon 13 and 14. aa indicates amino acids.
In conclusion, we believe that the authors should have included experiments with control constructs expressing the relevant fusion proteins for normalization of their luciferase activity assays. Furthermore, analysis of the mRNA species derived from the various constructs would have been helpful to further support the hypothesis that 19911G results in more efficient splicing.
Prothrombin 19911A>G as a functional noncoding variant: the evidence remains
Van der Putten and colleagues criticize a lack of control experiments in our study that addressed functional effects of the prothrombin 19911A>G polymorphism on the splicing efficiency.1 We believe that their concerns are based on a misunderstanding of the products resulting from the various pF2 reporter gene plasmid constructs used. We are happy to clarify this point.
To investigate the effects of the 3′ untranslated region on mRNA expression we introduced the prothrombin gene's last 2 exons and intervening sequence into a construct downstream from the luciferase ORF stop codon. The presence of this stop codon is depicted in Figure 1 in von Ahsen and Oellerich.1 A fusion protein as shown in van der Putten et al's Figure 1 cannot be generated. Changing the stop codon position from the (in terms of the prothrombin gene) ultimate to the penultimate position may render mRNAs from this construct prone to nonsense-mediated decay although many natural genes also terminate in the penultimate exon.2 To control for these effects we removed the stop codons by site-directed mutagenesis so that a fusion protein will be generated in this case. van der Putten and colleagues cite evidence that the C-terminal amino acids of firefly luciferase are functional.3 In fact, this region codes for a peroxisome signal peptide of importance for the native enzyme's function.4 The pGL3 reporter vectors used in our study do not carry this peroxisomal targeting sequence and express a cytosolic form of luciferase termed luc+.5 However, we found comparable results for the pF2 stopmut-type plasmids using the prothrombin stop codon and those using the luciferase stop codon.1(Fig 2) This argues against an appreciable effect of the fusion on protein function.
The parental plasmid for the double-reporter assays with pF2-GALU is pTN23.6 A good correlation between splicing efficiency by reverse transcription-polymerase chain reaction (RT-PCR) and the resulting luciferase-β-galactosidase ratio was shown for this plasmid including the faithful reproduction of effects of known mutations on splicing.6 The pTN23 plasmid is derived from pBgalluc. For the latter plasmid it was shown that the enzymatic activity of the fusion protein is the same as for the nonfused luciferase and β-galactosidase.7 This is in line with a different study using lacZ-luc fusion proteins that also found no evidence for an altered specific activity8 as well as another study using renilla and firefly luciferase fusion proteins.9
In conclusion, we believe that our experiments substantiate that the prothrombin 19911 polymorphism is functional. The 19911G mutation creates an intronic splice enhancer, which is the best explanation for the prothrombotic phenotype observed in case control studies so far. Further studies are needed to prove or disprove whether a further stratification according to additional 19911 carrier status can help to distinguish a higher risk from a lower risk group of 20210G>A heterozygotes.
Correspondence: Nicolas von Ahsen, Georg-August University, Department of Clinical Chemistry, 37099 Göttingen, Germany; e-mail: nahsen@gwdg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal