Abstract
The common prothrombin gene cleavage site mutation 20210G>A is associated with elevated prothrombin levels and thrombosis. The pathomechanism of the 20210G>A mutation was explained by increased mRNA formation and/or more efficient translation. Human studies also showed an influence of the intronic 19911A>G polymorphism on prothrombin activity. We established HepG2 cell lines stably transfected with prothrombin mini-genes containing the last 2 prothrombin exons, the last intron, 3′ untranslated region (UTR), and flanking sequence. The highest mRNA expression and protein activity resulted from the mutant haplotype 19911A-20210A. Haplotypes with wild-type cleavage site (19911A-20210G, 19911G-20210G) also differed significantly as a consequence of the intronic 19911 mutation; the 19911G-20210G haplotype showed lower expression than the 19911A-20210G haplotype, whereas previous clinical studies have reported elevated prothrombin activity with the 19911G-20210G haplotype. The cleavage site pattern was homogeneous with 20210A, which may cause a favorable intracellular processing, and heterogeneous with 20210G. In an independent assay for splicing efficiency, 19911G showed about 30% higher efficiency than 19911A. We conclude that the intronic 19911A>G single nucleotide polymorphism is itself functional and changes splicing efficiency by altering a known functional pentamer motif. Further studies are needed to define the value of additional prothrombin 19911 genotyping for thrombophilia screening, especially in cases heterozygous for 20210G>A.
Introduction
A noncoding mutation in the 3′ untranslated region (UTR) of the prothrombin gene (20210G>A) has been associated with thrombophilia.1 Several studies have since confirmed the association of this common mutation with arterial2 or venous1,3-7 thrombosis. The 20210G>A mutation confers a 3- to 7-fold increased risk for venous thrombosis.8,9 The determination of the prothrombin 20210G>A genotype is recommended as a high-priority test in the investigation of genetic thrombophilia.10 The activity of prothrombin may be partially regulated at a posttranslational level11 and is controlled by a strong genetic background.12 Linkage analysis demonstrated a cosegregation of the mutant allele with elevated prothrombin activity and suggested that this mutation in the 3′ UTR is in itself functional instead of in linkage disequilibrium with another polymorphism.13
Recent reviews have emphasized the role of the mRNA UTR in the pathomechanisms of some human diseases.14,15 Gehring et al demonstrated increased prothrombin mRNA end formation efficiency as a cause of elevated prothrombin protein levels in carriers of the 20210G>A mutation.16 In this study, a β-globin gene vector supplemented with the UTR of the prothrombin gene was used to investigate the polyadenylation reaction. A different study provided no evidence in vivo for altered prothrombin mRNA stability by the 20210A mutation.17 In contrast, the in vitro study by Carter et al18 found an effect of the 20210A genotype on both the efficiency of mRNA processing and mRNA stability.
In addition to these contradictory findings, there is evidence19,20 that a common single nucleotide polymorphism (SNP) (19911A>G)21 in intron M, the prothrombin gene's last intron, may have a functional role. A gene dosage effect of the 19911G allele on prothrombin activity was shown in vivo.19 The allele frequency of this marker is 0.45 for A and 0.55 for G.21 The 19911A allele is in full linkage with the 20210A allele.21 We note that the presence of this SNP completes a G triplet, a common intronic motif known to enhance splicing efficiency.22 However, no functional tests to support a specific function of the prothrombin 19911 base position have been published yet.
This makes a further investigation of functional consequences of haplotypes defined by the prothrombin 19911A>G and 20210G>A polymorphisms worthwhile. To improve and extend the findings of previous in vitro studies16,18 we included additional variables in our own experimental set-up. These are the inclusion of the last 2 exons, the last intron, and the prothrombin 3′ flanking region as a mini-gene in the reporter gene assay system. This should closely simulate the effects of the genomic vicinity including the natural splicing reaction to investigate the effects of the intronic 19911 and 3′ UTR 20210 polymorphisms. Additionally, we directly tested effects of the isolated 19911 SNP with its intron and adjacent splice sites on splicing efficiency.
Materials and methods
The pGL3 control, pGL3 promoter, pCI-neo, and pGEM-T easy vectors were purchased from Promega (Mannheim, Germany). Restriction enzymes were purchased from NEB (Frankfurt, Germany) or Roche (Mannheim, Germany). T4 DNA ligase was from MBI-Fermentas (St Leon-Rot, Germany). Polymerase chain reactions (PCRs) with subsequent cloning steps were done using the Expand-HighFidelity system (Roche); all other PCRs were done using native Taq polymerase (Invitrogen, Karlsruhe, Germany).
Reporter gene vector construction for transient expression
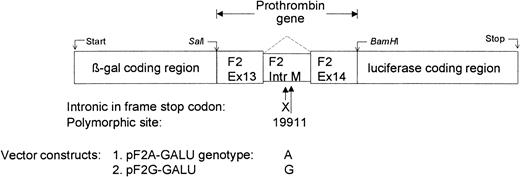
We initially extended the known sequence23 by genome walking (data not shown). While this study was in progress, the identical sequence was published by the ongoing Human Genome Project (GenBank accession number AC115088). The numbers in prothrombin primers refer to base positions in this GenBank entry. Primers F2-99573-99593-for, 5′-CTA GCA CCTAGG ACA AGC CTG ATG AAG GGA AAC, containing an AvrII site (underlined), and F2-100956-100977-rev, 5′-CAC TCGGATCCC CTC CCA CGT AGC TGG GAC TAC, containing a BamHI site, were used for PCR amplification. A 1428-bp fragment is amplified containing 1405 bp prothrombin gene sequence, namely part of exon 13, intron M, exon 14, the prothrombin poly-A signal, 3′ flanking sequence, and attached restriction sites. The DNA samples stemmed from a previous study and were obtained in accordance with the Georg-August-University ethics guidelines.11 Care was taken that the ligation kept the reading frames of the luciferase and prothrombin (class 0 splice sites) gene in phase. PCR products were screened for the UTR 20210G>A genotype with real-time PCR24 and for the intronic 19911A>G genotype by EcoNI digestion.21 Alternatively, a real-time PCR method for the detection of the 19911 polymorphism is presented in “Cleavage site assay and intron M splicing.” PCR products containing desired haplotypes were cloned into the pGEM-T vector and sequenced (thermo sequenase cycle sequencing kit; Amersham Biosciences, Freiburg, Germany) on an automated DNA sequencer (Licor 4200; Licor, Bad Homburg, Germany). Plasmid DNA was then cut with BamHI and AvrII and the resulting fragment ligated into the pGL3 control vector. This vector contains the luciferase gene from Photinus pyralis under control of an SV40 promoter. The vector was prepared by BamHI and XbaI digestion, which completely removed the vectors poly-A signal and viral enhancer sequence from immediately behind the luciferase stop codon. These functional elements were then replaced by the respective elements from the prothrombin gene (Figure 1).
Scheme of vector layouts representing different prothrombin haplotypes. The original SV40 poly-A site cassette was removed from the vector and replaced in its entirety by the prothrombin gene polyadenylation signal, cleavage site, and 3′ flanking region. Vectors of the “short” type are identical to their parent plasmids except for a shortened 3′ flanking region. SV40 Prom. indicates SV40 promoter.
Scheme of vector layouts representing different prothrombin haplotypes. The original SV40 poly-A site cassette was removed from the vector and replaced in its entirety by the prothrombin gene polyadenylation signal, cleavage site, and 3′ flanking region. Vectors of the “short” type are identical to their parent plasmids except for a shortened 3′ flanking region. SV40 Prom. indicates SV40 promoter.
A natural XbaI site in prothrombin intron M prevented cloning with XbaI; we therefore included an XbaI-compatible AvrII site into the primer. Inserts were then ligated into the prepared pGL3 vector and correct vector assembly confirmed by restriction fragment length polymorphism and sequencing. The resulting vector is 6159 bp. In our nomenclature “wt” and “mut” refer to plasmids with either wild-type or mutant cleavage site, respectively, known as the prothrombin 20210G>A mutation. The following letter code denotes the base at position 19911 (intronic SNP) followed by the base at position 20210. Accordingly, vectors were named pF2-wt-AG, pF2-mut-AA, and pF2-wt-GG (Figure 1).
An alternative set of vectors used a shorter 3′ flanking genomic element. This part of the genomic sequence was used for all comparable studies to date and corresponds to the last bases of the first published prothrombin gene sequence.23 These vectors were constructed as explained above by using an alternative reverse primer (F2-100153-100171-rev, 5′-GGATCC ATC CTC CCA CCT CAG CCT C) to amplify a 617-bp fragment containing 599-bp prothrombin gene sequence and attached restriction sites. Resulting plasmids were 5374 bp and designated pF2-wt-AG short, pF2-mut-AA short, and pF2-wt-GG short. All plasmids were prepared using midi or maxi prep kits from Qiagen (Hilden, Germany).
Site-directed mutagenesis
Stop codons were removed from the luciferase coding sequence in the vector constructs pF2-wt-AG, pF2-mut-AA, and pF2-wt-GG by a modified QuikChange method25 to change TAA TTC TAG to CAA TTC CAG. As the primer phosphorylation step25 was unnecessary, it was omitted. Resulting vectors were named pF2-wt-AG stopmut, pF2-mut-AA stopmut, and pF2-wt-GG stopmut.
Cell transfection
HepG2 cells (DGZMS, Braunschweig, Germany) were chosen for the transfection experiments because they express prothrombin (data not shown). For transient assays HepG2 cells were seeded at 2.0 × 106 cells in 6-well dishes and grown in Dulbecco modified Eagle medium (PAA Laboratories, Pasching, Austria). After 24 hours, cells were transfected with 5 μg pSVβGal plasmid DNA (Promega) and 5 μg DNA of appropriate luciferase vector complexed with Lipofectamine 2000 (Invitrogen). At 24 hours after transfection, cells were harvested in reporter lysis buffer (Promega) and lysed by a freeze-thaw cycle. Experimental repeats for each plasmid in Figure 2 were as follows (number of individual experiments/repeats): pF2-wt-AG (6/3), pF2-mut-AA (6/3), pF2-wt-GG (6/3), pF2-wt-AG short (6/1), pF2-mut-AA short (5/1), pF2-wt-GG short (3/1), pF2-wt-AG stopmut (5/2), pF2-mut-AA stopmut (5/2), and pF2-wt-GG stopmut (5/2).
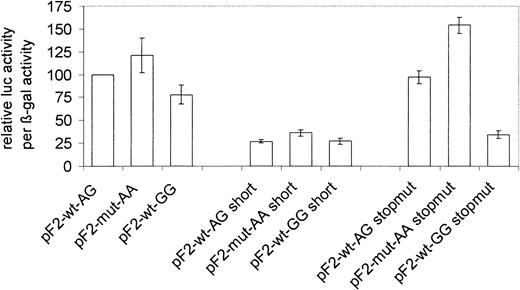
Transient transfection assays of different prothrombin haplotypes. The normalized β-galactosidase (β-gal) corrected luciferase (luc) reporter gene activity is reported. Underlying vector constructs are outlined in Figure 1. Stopmut vectors are derived from respective standard vectors (nos. 1-3 in Figure 1) by mutagenesis of the luciferase stop codon (see “Materials and methods”). For the number of experimental repeats, see “Materials and methods.” Error bars represent the standard error of mean. The leftmost column is used for normalization and has no error bar.
Transient transfection assays of different prothrombin haplotypes. The normalized β-galactosidase (β-gal) corrected luciferase (luc) reporter gene activity is reported. Underlying vector constructs are outlined in Figure 1. Stopmut vectors are derived from respective standard vectors (nos. 1-3 in Figure 1) by mutagenesis of the luciferase stop codon (see “Materials and methods”). For the number of experimental repeats, see “Materials and methods.” Error bars represent the standard error of mean. The leftmost column is used for normalization and has no error bar.
Stably transfected and polyclonal HepG2 cells were seeded at 2.0 × 106 cells in 6-well dishes. Medium was changed after 24 hours. Cells were harvested after a further 24 hours as described.
Reporter gene vector construction for stable expression
pGL3 plasmid–based reporter gene vectors were modified for stable expression in mammalian cells by introduction of the neomycin resistance gene from the pCI-neo plasmid. The resistance cassette (1367 bp) was amplified with the primers 5′-GCGGCCGCG CAG CAC CAT GGC CTG AAA TAA C and 5′-GCGGCCGCA CAC AAA AAA CCA ACA CAC AGA TGT A. The resulting PCR product (1383 bp) was introduced via the unique NotI site into the pF2-wt-AG, pF2-mut-AA, and pF2-wt-GG plasmids. The resulting plasmids were pF2-neo-wt-AG, pF2-neo-mut-AA, and pF2-neo-wt-GG and 7534 bp in size.
Stable cell lines were selected with G418 (PAA Laboratories) and experiments were done with pooled stable clones.
Double reporter assay to detect different splicing in 19911 G or A alleles
The pTN23 plasmid was a kind gift from M. T. Nasim and I. C. Eperon, University of Leicester, Leicester, United Kingdom.26 This plasmid contains a β-galactosidase (β-gal) open reading frame (ORF) without a stop codon followed by a cloning site and a luciferase ORF. Transfection with the plasmid results in transcription and translation of a fusion protein with β-gal and luciferase activity. Provided that an intron with complete splicing signals is cloned between β-gal and luciferase, the ratio of luciferase to β-gal activity will indicate the efficiency of splicing as a function of intronic and surrounding exonic sequences.26 If mRNA splicing is inefficient an in-frame stop codon, within the retained intron, prevents translation of the luciferase ORF.
A SalI site between β-gal and luciferase ORFs was lost during construction of the pTN23 plasmid. This site was reconstructed by mutagenesis and the resulting plasmid was cut with SalI and BamHI. Inserts contained the polymorphic intron 19911 position and were amplified by PCR with primers F2-99620-99640-for, 5′-CGG TCG ACG GGG GAC CCT TTG TCA TGA AG, containing a SalI site, and F2-99796-99816-rev, 5′-CGG GAT CCC ATT TGA TAC CAG CGG TTG TT, containing a BamHI site, cloned by standard techniques, and confirmed by sequencing. Resulting plasmids were designated pF2A-GALU and pF2G-GALU depending on the base at position 19911 (Figure 3).
Scheme of the dual-reporter vector for assaying splicing efficiency. Vectors contained prothrombin intron M with the polymorphic 19911A>G site and adjacent exon sequence fused in-frame to β-galactosidase (β-gal) and luciferase reporter genes. In the case of inefficient splicing, a natural in-frame stop codon (X) is present within the intron. The ratio of luciferase activity to β-gal activity reflects the ratio of expression from spliced mRNA to the total expression of spliced and unspliced RNA. The parental plasmid is pTN23 (see “Materials and methods”).26
Scheme of the dual-reporter vector for assaying splicing efficiency. Vectors contained prothrombin intron M with the polymorphic 19911A>G site and adjacent exon sequence fused in-frame to β-galactosidase (β-gal) and luciferase reporter genes. In the case of inefficient splicing, a natural in-frame stop codon (X) is present within the intron. The ratio of luciferase activity to β-gal activity reflects the ratio of expression from spliced mRNA to the total expression of spliced and unspliced RNA. The parental plasmid is pTN23 (see “Materials and methods”).26
The cloned 213-bp fragment contained 197 bp prothrombin gene sequence from terminal exon 13 (21 bp), complete intron M (146 bp), beginning of exon 14 (30 bp), and attached restriction sites. The reading frame was kept in phase between β-gal, exon 13, exon 14, and luciferase sequence. In the case of intron retention a naturally occurring stop codon (TAG) is present in frame 61 bp in the intron.
HepG2 cells were seeded as described above and transfected with 5 μg plasmid. Cells were harvested after 24 hours and tested for β-gal and luciferase activity as described below. Experiments were repeated 6 times in duplicate.
Luciferase and β-gal assays, neomycin phosphotransferase (NPT) enzyme-linked immunosorbent assay (ELISA)
Luciferase activity was measured in cell lysates according to the manufacturer's instructions (Luciferase assay system; Promega) on a tube luminometer (model 1250; Bio-Orbit, Turku, Finland). β-gal activity was measured with a colorimetric assay (570 nm) using chlorophenol red-β-D-galactopyranoside (CPRG; Roche) as substrate. Lysate (50 μL) was incubated at 37°C with 5 μL of a 50 mM CPRG solution and 200 μL assay buffer (50 mM potassium phosphate buffer, 1 mM MgCl2, pH 7.5) for 1 to 3 hours until color developed. Reactions were stopped by addition of 400 μL 1 M Na2CO3. Resulting luciferase activity was corrected by calculating the luciferase activity to β-gal activity ratio to account for effects caused by different transfection efficiencies.27 19911A-20210G wild-type plasmid activity was set to 100%.
In experiments with stably transfected cell lines we used normalization to the NPT concentration to account for effects due to different genomic integration sites. NPT is the gene product of the neo-R cassette that confers G418 resistance. It is expressed from the same vector as the luciferase/prothrombin product and was measured with the NPT II PathoScreen ELISA Kit (Linaris, Wertheim, Germany).
Reverse transcription (RT)–PCR analysis
Total RNA was isolated from stably transfected cells with the SV total RNA isolation system (Promega). RNA (1 μg) was reverse transcribed with M-MLV RT enzyme (Invitrogen) using oligonucleotide-dT priming. First-strand product (1 μL) was used for quantitative PCR reactions on a LightCycler (Roche) in SYBR-green format with FastStart Taq polymerase (Roche). PCR was performed using a luciferase-specific forward primer (Luc_for, 5′-TGT GGA CGA AGT ACC GAA AGG) and reverse primer (Luc_rev, 5′-TCG TCC ACA AAC ACA ACT CC). Controls with no RT were carried out and showed absence of amplification. mRNA expression was normalized to neo-R as a housekeeping gene. The neo-R PCR was carried out using the primers neo-R_for GGA TGA TCT GGA CGA AGA GC and neo-R_rev AAT ATC ACG GGT AGC CAA CG.
Cleavage site assay and intron M splicing
Position of the cleavage site (eg, the position where the pre-mRNA is cleaved and the poly-A tail attached) was determined by a PCR-based assay. We have devised a method based on the rapid amplification of cDNA ends poly(A) test (RACE-PAT)28 followed by a modified 3′ RACE reaction.29 Total RNA from stably transfected cell lines was extracted with phenol-chloroform.30 RNA (2 μg) was reverse transcribed with SuperScript II RT enzyme (Invitrogen) using a tailed oligonucleotide primer (5′-AAG CAG TGG TAT CAA CGC AGA GTA CT20).29 First-strand product (1 μL) was used in a PCR reaction with the forward primer Luc_for1 5′-ATC CAT CTT GCT CCA ACA CC and a tail-specific universal primer mix comprising the adapter oligonucleotide 5′-GTA ATA CGA CTC ACT ATA GGG CAA GCA GTG GTA TCA ACG CAG AGT and primer 5′-GTA ATA CGA CTC ACT ATA GGG C.29 A nested PCR reaction was performed with the forward primer Luc_for and adapter-specific reverse primer 5′-AAG CAG TGG TAT CAA CGC AGA GT. PCR products were cloned into a pGEM-T vector and positive clones picked. The region including the cleavage site was reamplified on the LightCycler with a prothrombin-specific primer F2-99882-99901-for 5′-CCG CCT GAA GAA GTG GAT AC and the adapter reverse primer 5′-AAG CAG TGG TAT CAA CGC AGA GT together with 0.2 μM F2-99983-100013-pA-anchor 5′-Cy5.5-TTT ATT GGG AAC CATAGT TTTAGAAAC ACAA-PHO and F2-100015-pA-WT-probe 5′-TTC TCG CTG AGA GTC AC-FLU or F2-100015-pA-(WT+C)-probe 5′-TTG CTC GCT GAG AGT CAC-FLU. Probes were synthesized by MWG Biotech (Ebersberg, Germany) including the modifications by Cy5.5, phosphorylation (PHO), and fluoresceine (FLU) as indicated. Detection probes have no end point on genomic DNA because they also hybridize with the mRNA poly-A tail. PCR product length depended on the position where the tailed RT primer bound to the cDNA poly-A tail. Samples were subjected to an analytic melting cycle on the LightCycler from 30°C to 70°C at 0.1°C/min ramp rate. Melting points of the probes allowed an unambiguous assignment of the cleavage site. Index samples for different cleavage sites were sequenced and included in the run.
About 50 positive clones were checked if they contained correctly spliced products by PCR amplification over the splice sites. Intron presence or absence was then determined by hybridization probe binding in a real-time PCR assay. Primers F2-99620-99640-for, 5′-GGG GGA CCC TTT GTC ATG AAG and F2-99796-99816-rev, 5′-CAT TTG ATA CCA GCG GTT GTT and 0.2 μM of F2-99735-99763-IntM-anchor 5′-Cy5.5-CTG ATG TGA CCT TGA ACT TGA CTC TAT TG-PHO and F2-99717-99732-IntM-WT-probe 5′-TTG CCT GGC AGA GGA A-FLU amplified a 197-bp fragment from prothrombin containing intron M. Samples were subjected to an analytic melting cycle on the LightCycler from 30°C to 70°C at 0.1°C/min ramp rate. Melting points of probes allowed an unambiguous assignment of 19911A or G alleles. Clones with correctly spliced inserts had the intron M removed and have no binding site for hybridization probes. Index samples for different alleles were sequenced and included in the run.
Statistical analysis
Between-group comparisons were done with the Wilcoxon signed ranks test using SPSS for Windows (Chicago, IL). Other calculations were performed using Microsoft Excel for Windows (Seattle, WA).
Results
Vectors with the extended 3′ flanking genomic region are more efficiently processed
A transient reporter gene assay was set up for the investigation of functional consequences of the prothrombin gene mutations 19911A>G and 20210G>A. Our cloning strategy included the last 2 exons, the last intron and flanking genomic DNA downstream of the polyadenylation and cleavage site (Figure 1).
In a first series of transfection experiments we determined the vectors that resulted in optimal luciferase activity signal. We compared vector constructs harboring extended or short prothrombin 3′ flanking regions (Figure 1). Luciferase activity was always higher in vectors containing the extended flanking region (Figure 2).
In all experiments the luciferase activity observed from the mutant genotype (pF2-mut-AA plasmids) was higher than that of the others (pF2-wt-AG, pF2-wt-GG plasmids). The resulting absolute luciferase activity of the vectors of the short type was only 27% to 35% that of standard vectors. The novel 3′ flanking sequence obviously contains positional elements that promote efficient processing of vector constructs only by introducing the naturally occurring genomic surrounding.
Nonsense-mediated decay (NMD) effects on mRNA derived from the vector construct
The transcription machinery first encounters the stop codon of the luciferase gene during intracellular processing of the vector-derived mRNA. The stop codon from the last exon of the prothrombin gene is present further downstream (Figure 1). This could target mRNA transcribed from the vector to NMD, a quality control pathway that causes degradation of mRNAs that cannot be fully transcribed.31 Such mRNAs contain premature stop codons with a larger than 50 to 55 bp distance between a stop codon and a downstream exon-exon junction.31 The vectors investigated here have a distance of 75 bp between the luciferase stop codon and the next exon-exon junction and may therefore be prone to NMD, which could influence our results. To test this possibility the luciferase stop codon was removed by mutagenesis. mRNA transcribed from these plasmids uses the natural prothrombin stop codon in the last exon. We again observed that luciferase activity constantly decreased from the mutant genotype (pF2-mut-AA stopmut plasmid) to pF2-wt-AG stopmut (P < .05) and pF2-wt-GG stopmut (P < .05, Figure 2). Direct comparison of plasmids containing a premature stop codon with those without shows no effect in the case of the 19911A-20210G haplotype (100% vs 97% activity), slightly more pronounced activity for the 19911A-20210A haplotype (121% vs 154%), and less activity for the 19911G-20210G (35% vs 78%).
The prothrombin 20210G>A and intronic 19911A>G polymorphisms differentially influence reporter gene activity and mRNA amount
Our initial experiments with RT-PCR from transiently transfected cells were unsatisfactory. RNA isolated with the phenol-chloroform method or silica-spin columns including DNAse digest showed repeatedly positive no-RT controls, likely due to remaining plasmid contamination after RNA isolation. Therefore, stable HepG2 cell lines were established using plasmids expressing mRNA from all 3 investigated prothrombin haplotypes. These plasmids are identical with those used for transient transfection studies except for an additional neo-r gene. The pooled stably transfected cells allowed for reliable RT-PCR analysis.
Our results show that effects from the 20210 cleavage site mutation are generally larger than those observed by the 19911 base variation in otherwise identical plasmids.
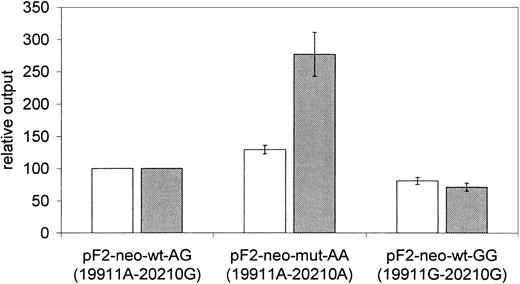
We observed on average 29% higher normalized luciferase activity in experiments with the plasmid pF2-neo-mut-AA (P < .05, Figure 4) compared with pF2-neo-wt-AG. The luciferase activity of plasmid pF2-neo-wt-GG was reduced to 81% of the wild-type pF2-neo-wt-AG (P < .05, Figure 4).
Prothrombin haplotypes differentially influence reporter gene activity and mRNA expression in stably transfected cells. Comparison of normalized corrected luciferase reporter gene activity (NPT corrected, □) and luciferase mRNA expression (neo-R expression corrected, ▤) relative output for plasmids representing different prothrombin haplotypes. Cells are stably transfected and polyclonal; 6 experiments were performed in duplicate. Error bars represent the standard error of mean. Results for the pF2-wt-neo-AG plasmid are used for normalization and have no error bars.
Prothrombin haplotypes differentially influence reporter gene activity and mRNA expression in stably transfected cells. Comparison of normalized corrected luciferase reporter gene activity (NPT corrected, □) and luciferase mRNA expression (neo-R expression corrected, ▤) relative output for plasmids representing different prothrombin haplotypes. Cells are stably transfected and polyclonal; 6 experiments were performed in duplicate. Error bars represent the standard error of mean. Results for the pF2-wt-neo-AG plasmid are used for normalization and have no error bars.
The effect of the 20210A cleavage site mutation was transcriptional as evident from a pronounced increase (277%) of mRNA expression. The order of mRNA expression followed that of the protein activity in the different plasmids. The pF2-neo-wt-AG plasmid showed higher mRNA expression than the pF2-neo-wt-GG plasmid carrying the intronic 19911G mutation (P < .05) but lower expression than pF2-mut-AA plasmids (P < .05, Figure 4).
Variation in the cleavage site position
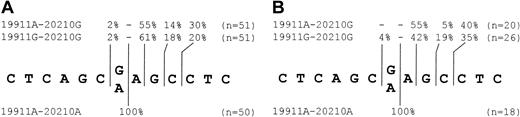
We determined the mRNA cleavage site in about 50 prothrombin-positive clones from each haplotype: 49% (19911G-20210G), 61% (19911A-20210G), and 64% (19911A-20210A) of the mRNAs still contained the intron. However, mRNA containing the intron was also polyadenylated and we observed no specifically different pattern if only correctly spliced clones were included in the analysis (Figure 5). There is a trend toward higher percentage of unspliced products in clones with the 19911A base compared with those with the 19911G base.
Cleavage site choice by different prothrombin haplotypes. The percentage of cleavage site choice and the number of analyzed clones are shown for the different prothrombin haplotypes. The mRNA cleavage sites are projected on the genomic DNA sequence with the polymorphic 20210G>A site indicated. See “Results” and “Discussion” for assignment of cleavage sites. mRNA was isolated from pooled stably transfected HepG2 cells. (A) All clones analyzed. Sum is not always 100% due to rounding. (B) Subgroup analysis of clones containing correctly spliced inserts.
Cleavage site choice by different prothrombin haplotypes. The percentage of cleavage site choice and the number of analyzed clones are shown for the different prothrombin haplotypes. The mRNA cleavage sites are projected on the genomic DNA sequence with the polymorphic 20210G>A site indicated. See “Results” and “Discussion” for assignment of cleavage sites. mRNA was isolated from pooled stably transfected HepG2 cells. (A) All clones analyzed. Sum is not always 100% due to rounding. (B) Subgroup analysis of clones containing correctly spliced inserts.
The cleavage site use is strongly influenced by the 20210 base. In the case of 20210A, polyadenylation always starts at 20210. We note that use of the 20211 or even 20209 sites cannot be excluded since all clones polyadenylated from these positions would score for 20210. Given the preference toward CA dinucleotides for poly-A tail addition,32 cleavage at position 20210 is most likely. The poly-A is usually added after transcription to an A present in the precursor.32
In contrast, the cleavage site is markedly heterogeneous if the 20210G wild-type base is present. We found predominant use of the 20211 cleavage site, followed by use of the 20213, 20212, and very rarely the 20209 cleavage site. Again the use of the 20210 site cannot be excluded.
In a preliminary experiment we determined the cleavage sites but not the splicing status. The resulting data (supplemental material; see the Supplemental Document link at the top of the online article on the Blood website) indicate that significant variation in the determination of cleavage sites is possible.
The 19911 base influences splicing efficiency
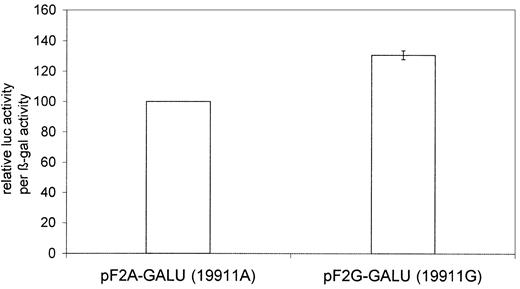
The dual reporter assay with the pF2A-GALU and pF2G-GALU plasmids gave on average 31% higher luciferase per β-gal activity from the intronic G base at position 19911 (Figure 6). This indicates a more efficient splicing and is in line with a trend seen from the number of clones containing unspliced mRNA in the cleavage site assay.
The intronic 19911A>G polymorphism affects splicing. Clones containing the 19911G base have an on average 31% higher ratio of spliced mRNA compared with those containing the 19911A base (P < .05) as judged by their relative luciferase (luc) activity per β-galactosidase (β-gal) activity output. There were 6 experiments performed in duplicate. Error bars represent the standard error of mean. The left column is used for normalization and has no error bar.
The intronic 19911A>G polymorphism affects splicing. Clones containing the 19911G base have an on average 31% higher ratio of spliced mRNA compared with those containing the 19911A base (P < .05) as judged by their relative luciferase (luc) activity per β-galactosidase (β-gal) activity output. There were 6 experiments performed in duplicate. Error bars represent the standard error of mean. The left column is used for normalization and has no error bar.
Discussion
Prothrombin haplotype effects in vivo and in vitro
The second most common monogenetic risk factor for thrombophilia in whites was discovered in 1996 by Poort et al through systematic screening of the prothrombin gene.1 A 20210G>A polymorphism was found at the naturally occurring cleavage site where nascent mRNA is cleaved and polyadenylated. The pathomechanism remained unexplained for several years,8,13 but a direct functional role for this polymorphism was proposed.13 The prothrombin 20210G>A mutation is a gain-of-function polymorphism causing elevated thrombin generation10,33 and elevated prothrombin concentration and/or activity.1,11,33 The accumulation of functional data from different prothrombin haplotypes warrants their further detailed investigation. There are 2 studies that have examined effects of different haplotypes on plasma prothrombin levels.19,20 These studies included genotyping of an A to G transition in the last intron of the prothrombin gene at position 19911. The 20210A allele carries the 19911A marker; this haplotype was found on 459 of 460 alleles.19-21 Only one allele carried a 19911G-20210A haplotype representing a recent recombination event.19 After exclusion of all 20210A carriers, prothrombin levels were 4 U/dL higher for each 19911G allele compared with the 19911A allele.19 This moderate increase was not significantly associated with the risk of venous thrombosis after exclusion of 20210A carriers. However, in heterozygous 20210A carriers the risk of venous thrombosis increased from 1.6 in subjects having 19911A as the other allele to 4.7 in those having 19911G as the other allele,19 a perhaps unexpected consequence of this intronic SNP. Another study by Pérez-Ceballos et al confirmed these findings.20 There was a trend to increasing prothrombin activity with an increasing number of 19911G alleles. The odds ratio for venous thrombosis was higher (5.86 vs 3.34) when carriers of the 19911A-20210A allele had 19911G-20210G as the other allele compared with 19911A-20210G.20
Previous in vitro studies on prothrombin have investigated only the effects of the cleavage site polymorphism and disregarded possible effects from the polymorphism in the last intron.16,18 These approaches did not consider the interdependence between the splicing of the last intron, polyadenylation, and mRNA end formation.34 The study of Gehring et al used transient HeLa-cell transfection of a β-globin fusion gene supplemented with the prothrombin 3′ UTR, poly-A signal, and 140-bp 3′ flanking region.16 Results showed that the mutated cleavage site (20210A) was processed more efficiently, while the amount of pre-mRNA and the poly-A tail length remained unchanged. The study by Carter et al confirmed the increased formation of mRNA and protein from vectors expressing the 20210A variant.18 However, the latter study also did not include the natural splicing reaction and used a viral SV40 polyadenylation signal instead of the natural one to end the message. Our mini-gene study simulates both, the prothrombin genes' natural splicing and polyadenylation reaction. Our data validate the previous finding of a higher protein production from plasmids with the 20210A mutant genotype. The underlying mechanism is transcriptional since the mRNA expression and protein activity are always correlated (Figure 4).
Effects from the prothrombin gene 3′ flanking region
The extended prothrombin gene 3′ flanking region in our vector constructs was beneficial in terms of the resulting luciferase signal compared with vectors having only the previously published short flanking sequence (Figure 2). The flanking sequence is not present on the transcribed mRNA, and therefore its effect is not that of a conventional enhancer element. Translational termination is well defined by a stop codon, but there is no such clear-cut signal for the termination of transcriptional elongation by RNA polymerase II. In vitro transfection studies of the human β-globin locus showed that transcription terminated within a region of 1.5-kb downstream of the poly-A site.35 It was demonstrated that read-through transcription beyond the region of the normally occurring termination is dependent on the presence of a functional poly-A site and a terminal splice acceptor.35 These observations illustrate the orchestrated interaction between splicing, polyadenylation, and transcription termination.36-38 We interpret our findings in terms of an improved transcription termination process leading to better RNA polymerase II recycling.39
NMD effects
Our results could be influenced by NMD. The distance between the luciferase stop codon and the next exon-exon border in our vector constructs is 75 bp. Such constructs are prone to NMD, an mRNA surveillance mechanism that causes degradation of mRNA species with premature stop codons, defined by a distance of more than 55 bp to the next exon-exon junction.31 These messages are degraded to ensure that no truncated proteins are made.40 However, NMD may not necessarily occur (Asselta et al41 and references therein). If NMD affects our constructs we would expect a positive effect from the removal of the first apparently “premature” stop codon on expression as was shown in another study.42 In contrast, we did not find a consistent increase in luciferase activity from such plasmids. The pF2-wt-AG stopmut plasmid showed unchanged activity, the pF2-mut-AA stopmut plasmid slightly higher activity, and the pF2-wt-GG stopmut plasmid lower activity than their stop codon–containing counterparts (Figure 2). As for the plasmids containing the luciferase stop codon, the pF2-mut-AA stopmut showed the highest, pF2-wt-AG stopmut showed intermediate, and pF2-wt-GG stopmut showed the lowest activity. We conclude that NMD cannot explain these results.
Cleavage site heterogeneity depending on base 20210
Both plasmids with the wild-type cleavage site (20210G) produced significantly less protein than those with the mutant cleavage site (20210A) (Figure 4). Pollak et al described that in the case of the 20210A mutation 100% of transcripts were polyadenylated at position 20210, while in the case of the wild-type 20210G cleavage site with unknown 19911 genotype most (74%) of the transcripts were polyadenylated at position 20212 and only 26% at position 20210.17 It is noteworthy that for 20210G, polyadenylation has most likely occurred at position 20211. If the start of the poly-A tail corresponds with an A in the genomic sequence, this is the base to which the tail is added after transcription.32 Accordingly, we score the poyladenylation behind the 20210G position as starting after cleavage at 20211 (Figure 5). Our own experimental data confirm that the 19911A-20210A haplotype polyadenylates only at 20210. This homogeneity may cause a favorable intracellular processing of these mRNAs. An A base is generally preferred for the poly-A addition but there is no specific dinucleotide motif for the cleavage reaction.43 The 19911A-20210G and 19911G-20210G haplotypes displayed a comparable pattern with predominant cleavage at 20211 and 20213, followed by 20212 and 20209. Pollak et al reported a less frequent cleavage at 20210 (ie, 20211) and more frequent cleavage at 20212 for the 20210G genotype.17 These differences might be caused by the fact that we used a hepatic tumor cell line opposed to human liver. Furthermore, significant sampling effects must be considered. We cannot exclude a minor effect of the 19911 base on the cleavage site choice but a significant effect would have been detected under our experimental conditions. Our finding that at least 4 cleavage sites can be used in the case of a weak signal, such as 20210G, further adds to the already observed cleavage site heterogeneity of prothrombin. This heterogeneity is also a common feature in other genes,44 although its causes and consequences remain unknown so far.
Splicing efficiency depending on 19911
Prothrombin intron M is only 146 bp, which is small compared with the median human intron size of 1044 bp.45 The correct processing of vertebrate U2-type introns requires more information than is coded for by splice sites and branch point.46 This information is distributed within the gene46 and can be attributed to auxiliary cis-elements known as splicing enhancers and silencers or splicing control elements that can reside in both intronic or exonic positions.47 A known intronic splicing enhancer is the GGG triplet.22 It was initially thought to be typical for small introns but genome-wide analysis discovered overrepresentation of the GGG motif also in the first and last 200 bp of long introns.48 More specifically, sequence analysis of short introns revealed a subset of 10 pentamers that significantly contribute to the prediction of short introns, including the CAGGG motif.49 The 19911A>G SNP toggles between CAGAG and CAGGG, completing not only a GGG triplet but also a significant pentamer motif.
Our double reporter assay indicates a 31% higher splicing efficiency resulting from the prothrombin intron M 19911G genotype (Figure 6) when only the intron and its splice sites are analyzed. This finding is not unexpected considering the higher amount of spliced mRNA resulting from the 19911G compared with 19911A genotypes because the concentration of an unspliced intron is inversely proportional to its splicing rate.50 This different splicing efficiency is expected to alter gene expression. Effects during the transcription of a nuclear pre-mRNA and the composition of ribonucleoproteins (RNPs) on the mature mRNA determine the translational activity of the cytoplasmic RNA.51 Position and sequence of introns can alter the structure and composition of mRNAs and modulate their translational functionality.52
Interactions between splicing (prothrombin 19911A>G) and end formation (20210G>A) in prothrombin
Splicing and the polyadenylation reaction are tightly coupled.36-38 Human prothrombin has at least 2 common polymorphisms that affect splicing (19911) and polyadenylation via the cleavage site choice (20210). There is a large database supporting the prothrombotic role of prothrombin 20210G>A. Less is presently known about effects caused by 19911A>G. An increasing number of 19911G alleles was associated with increasing prothrombin activity, and carriers of the 19911G allele were overrepresented in controls with prothrombin activity higher than 130% after exclusion of 20210A carriers.19 There are 2 studies that show an increased risk of thrombosis in 20210A carriers with 19911G on the other allele compared with those 20210A carriers with 19911A on the other allele. Alone, 19911A>G was not associated with thrombosis after exclusion of 20210A carriers.19,20
Our prothrombin mini-gene assays confirmed the significantly higher mRNA and protein output of constructs having the 19911A-20210A haplotype (Figure 4). We found a lower protein activity from the 19911G-20210G haplotype, which is contrary to in vivo data.19,20 This may be caused by more distant SNPs that are linked with the 19911G-20210G haplotype and not included in our mini-gene. Known intronic SNPs19,21 that were not included in the mini-gene constructs are possible candidates. The 19911A>G polymorphism itself promotes splicing efficiency (Figure 6), and we expect higher transcript levels from 19911G-20210G alleles in vivo. The allelic variation in gene expression is presently difficult to measure because conventional methods are suitable only for coding SNPs.53 New mass-spectrometry–based methods overcome this limitation but are still very laborious.54 The generated cell lines are homozygous for the inserted gene, and trans effects from 19911G-20210G versus 19911A-20210G wild-type alleles on 19911A-20210A mutation carriers cannot be investigated. Improved expression from 19911G-20210G alleles has the potential to be a crucial trigger toward thrombosis manifestation in 20210A carriers. SNPs at 19911 and 20210 explain part of the genetic background of variable prothrombin activity. More variation can be attributed to other prothrombin mutations in certain populations.55,56 In fact the prothrombin gene's last intron, last exon, and 3′ UTR are equipped with several noncoding regulatory SNPs with the potential function to fine-tune the activity of this central player in coagulation. This fine tuning opposed to possible coding gain-of-function mutations seems to be beneficial for this gene with a plethora of biologic functions.
In conclusion, we established a mini-gene system for the study of prothrombin mRNA splicing and end formation. The extended genomic 3′ flanking region causes higher expression when included in the mini-gene. We provide evidence for a differential influence of prothrombin gene haplotypes on mRNA formation. We confirm previous findings of increased mRNA formation linked to the 20210A mutation. The cleavage site displays marked heterogeneity for 19911A-20210G and 19911G-20210G alleles and is homogenous for 19911A-20210A alleles. In addition, we show that the intronic 19911A>G mutation is itself functional and affects splicing efficiency by altering a functional pentamer motif. Further studies are needed to define the value of additional prothrombin 19911 genotyping for thrombophilia screening especially in cases heterozygous for 20210G>A.
Prepublished online as Blood First Edition Paper, September 22, 2003; DOI 10.1182/blood-2003-02-0419.
Supported by an in-house research grant from the Medical Faculty, Georg-August-University, Göttingen, Germany, to N. von A.
Data from this work were presented in abstract form at the 18th International Congress of Clinical Chemistry and Laboratory Medicine, Kyoto, Japan, October 20-25, 2002; and the 47th annual meeting of the Society of Thrombosis and Haemostasis Research, Innsbruck, Austria, February 15-18, 2003.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Victor W. Armstrong for his valuable help and discussion. Reiner Andag and Sandra Hartung are gratefully acknowledged for their excellent technical assistance.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal