Abstract
Primary mediastinal B-cell lymphoma (PMBL) is a well-defined subtype of diffuse large B-cell lymphoma. Molecular cytogenetics revealed frequent gains of 9p24. JAK2, mapping in this region, is presently regarded as a candidate oncogene because expression profiling showed high Janus kinase-2 (JAK2) transcript levels and JAK2 was found to be constitutively phosphorylated in mediastinal B-cell lymphomas. We confirm that in the MedB-1 mediastinal B-cell line, harboring a trisomy 9, JAK2 transcription is elevated and the product is highly phosphorylated. However, JAK2 is not overexpressed at the protein level. On top, JAK2 protein turnover is even delayed. This unexpected finding coincides with a biallelic mutation of the suppressor of cytokine signaling-1 (SOCS-1) gene in this cell, which abrogates SOCS box function of the protein. Ectopic expression of wild-type (wt) SOCS-1 in MedB-1 leads to growth arrest and dramatic reduction of phospho-JAK2 and its downstream partner phospho–signal transducer and activator of transcription-5 (phospho-STAT5). Ultimately, the target gene cyclin D1 is repressed in transfectants while RB1, which is silenced in MedB-1, is induced. We conclude that, in MedB-1, action of phospho-JAK2 is sustained due to defective SOCS-1. Hence, SOCS-1 qualifies as a novel tumor suppressor. Of note, SOCS-1 mutations are also present in the parental tumor of MedB-1 and were detected in 9 of 20 PMBLs.
Introduction
Primary mediastinal (thymic) B-cell lymphoma (PMBL) is a locally aggressive tumor with clinical, histopathological, immunologic, and molecular cytogenetic characteristics that classify PMBL as a subtype of diffuse large B-cell lymphoma.1
PMBL exhibits characteristic genetic abnormalities including frequent gains of 9p and distinct high-level amplifications with a defined consensus region on 9p24 involving the JAK2 locus.2-4 Expression profiling of PMBL revealed high JAK2 transcription5,6 and supported the concept of JAK2 as a candidate oncogene.
JAK2 codes for a protein that is part of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway and is induced by cytokines and several growth factors.7 Ligand binding to its cognate receptor leads to receptor oligomerization and transauto-phosphorylation of JAKs in their activating loop.8 Activated JAKs phosphorylate the receptor and the substrates (eg, STAT proteins), which dimerize and translocate in the nucleus, inducing the transcription of genes like cyclin D1,9 oncostatin M,10 and BCLxL,11 which mediate the cellular responses of cytokine stimulation. Kinase activity of JAK2 and subsequent activation of downstream targets are negatively regulated by suppressor of cytokine signaling (SOCS) proteins.12 SOCS proteins are a family of 8 members that share a central Scr-homology 2 (SH2) domain and a highly conserved C-terminal domain termed SOCS box.13,14 This domain associates with the elongins B and C to the elongin BC complex, which, in turn, couples SOCS proteins and their substrates to the proteasomal protein degradation pathway.15
Recently, JAK2 was found to be overexpressed and constitutively phosphorylated in MedB-1 mediastinal B lymphoma cells.16 This observation raises the question of the functional status of SOCS-1 in MedB-1 cells that allows JAK2 to be continuously active. Confirming overexpression at the transcriptional level and the high rate of phospho-JAK2 in MedB-1 cells, we now show that JAK2 is not overexpressed at the protein level and that its constitutive activation is due to delayed protein degradation. This unexpected setting is caused by a biallelic loss of function mutation in the SOCS box region of SOCS-1 in MedB-1. Restoring the wild-type (wt) SOCS-1 function in MedB-1 cells leads to growth arrest, rapid dephosphorylation of JAK2, silencing of cyclin D1, and induction of retinoblastoma 1 (RB1) expression. These data extend the concept of JAK-2 deregulation as the growth-promoting event and argue for a ancillary if not alternative role of mutated SOCS-1 in the activation of the JAK/STAT signaling pathway in PMBL.
Materials and methods
Cell culture
The MedB-1 cell line was established in our laboratory as previously described.17 Hodgkin lymphoma cell line L428 and Ramos Burkitt lymphoma cell line were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkultur (DSMZ; Braunschweig, Germany). Highly purified peripheral blood B lymphocytes were obtained from the blood bank of the German Red Cross (Ulm, Germany). Culture conditions have been previously described.17
RNA isolation, reverse transcriptase–polymerase chain reaction (RT-PCR), and real-time PCR
Total RNA was isolated using the RNeasy kit (Qiagen, Hilden, Germany) and treated with RNase-free DNAse (Roche, Mannheim, Germany). Complementary DNA synthesis and PCR were done as previously described.18 Real-time PCR was performed to determine relative DNA content and JAK2 expression using the Bio-Rad iCycler Detection System (BioRad Laboratories, Hercules, CA). PCR was carried out using 12.5 ng cDNA in a total of 20 μL containing 10 μL SYBR Green PCR Supermix (Bio-Rad Laboratories). Samples and water as negative control were run in triplicates on a 96-well plate. Thermal cycling conditions were as follows: 3 minutes at 95°C, 40 cycles at 95°C for 25 seconds, 60°C for 15 seconds, 72°C for 15 seconds. As reference, β2-microglobulin was used.
Relative expression of JAK2 was calculated using the 2-ΔΔCT method19 and compared with JAK2 gene dosage of autologous fibroblasts.
Analysis of cell proliferation
Cell proliferation assays were performed in complete culture medium, starting with a cell density of 1 × 105/mL in a total volume of 5 mL. Cells were cultured with or without 20 μM tyrphostin AG 490 (Sigma, St Louis, MO) for 8 days. Cell proliferation measurement of wt SOCS-1 and mock-transfected MedB-1 cells was done 2 days after transfection together with a control for wt SOCS-1 expression by reverse transcriptase–polymerase chain reaction (RT-PCR). Dead cells were removed using magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Every day, 3 aliquots of 50 μL cell suspension were counted in a Neubauer chamber.
Cell lysis and Western blotting
Cells (1 × 106 to 2 × 106) were washed 3 times with ice-cold phosphate buffered saline (PBS) containing 1 mM NaF and lysed for 15 minutes at 75°C in 2 × NuPAGE-LDS sample buffer (Invitrogen, Carlsbad, CA) containing 5% vol/vol 2-mercaptoethanol. Protein concentration was determined according to Chapdelaine et al20 and quantified using the ImageMaster VDS (Amersham Biosciences, Freiburg, Germany) and bovine serum albumin (BSA) as standard. Proteins (20 μg per lane) were separated on 2% to 8% NuPAGE Tris-Acetate or 4% to 12% NuPAGE Novex Bis-Tris gradient gels (Invitrogen) and transferred on polyvinylidene difluoride membranes (PVDF; Millipore, Germany) by semidry blotting; blocked in Tris (tris(hydroxymethyl)aminomethane)–buffered saline/0.1% Tween 20 with 5% BSA, 1 mM NaF, and 1 mM Na3VO4; and incubated with the primary antibodies in blocking solution overnight at 4°C. After incubation with appropriate secondary antibodies, proteins were visualized on Hyperfilm ECL using the enhanced chemiluminiscence (ECL) system (Amersham).
Antibodies
Primary antibodies used include anti-JAK2 antibody (06-255; Upstate Biotechnology, Charlottesville, VA), anti-STAT5B and anti-STAT5A+B antibodies (SC-1656, SC-836; Santa Cruz Biotechnology, Santa Cruz, CA), antiphospho-STAT5 antibody and antiphospho-JAK2 antibody (Cell Signaling Technology, Beverly, MA), and anti–β-actin antibody (Sigma).
Electrophoretic mobility shift assay (EMSA)
Nuclear protein extracts from MedB-1 cells, AG490-treated cells, and cells transfected with the SOCS-1 expression vector were prepared according to Dignam.21 A β-casein promoter sequence was used as specific probe for STAT5-DNA binding (Table 1). End labeling was done with α-32P–deoxycytidine triphosphate (α-32P-dCTP) in the Klenow enzyme reaction. Electrophoretic mobility shift assays (EMSAs) were carried out for 30 minutes at room temperature including 50 000 cpm of the probe, 10 mM Tris-HCl pH 7.5, 50 mM KCl, 0.1 mM EDTA (ethylenediaminetetraacetic acid), 1 mM dithiothreitol, 5% vol/vol glycerol, 0.1% Nonidet P-40 (NP-40), 1 μg/μL poly(dIdC), 1 mg/mL bovine serum albumin, and 1 μg nuclear protein extract in a volume of 10 μL. Competitors were added in 10- or 100-fold molar excess. In supershift or competition assays anti-STAT5 antibodies were incubated for 15 minutes before DNA probes were added. Samples were loaded on 4.5% polyacrylamide gels and run at 10 V/cm for 2 hours in 0.5 × Tris-borate-EDTA (TBE) buffer. Dried gels were exposed to an x-ray film.
Oligonucleotides used for PCR and EMSA
. | 5′-sequence-3′ . |
|---|---|
| PCR primers | |
| JAK2-f | AAGCCACTGCCAGAAACTTG |
| JAK2-r | ACTGAATTCCACCGTTTCCA |
| β2-M-f | TGTCTTTCAGCAAGGACTGG |
| β2-M-r | GATGCTGCTTACATGTCTCG |
| SOCS-1-f | AGAGCTTCGACTGCCTCTTC |
| SOCS-1-r | AGGGGAAGGAGCTCAGGTAG |
| Cyclin D1-f | CGTGGCCTCTAAGATGAAGG |
| Cyclin D1-r | CTGGCATTTTGGAGAGGAAG |
| RB1-f | GGAAGCAACCCTCCTAAACC |
| RB1-r | TTTCTGCTTTTGCATTCGTG |
| HPRT-1-f | GACCAGTCAACAGGGGACAT |
| HPRT-1-r | CTTGCGACCTTGACCATCTT |
| SOCS-1-mRNAf | CACCATGGTAGCACACAACCAGGTGG |
| SOCS-1-mRNAr | TCAAATCTGGAAGGGGAAGGAGCTC |
| EMSA probe | |
| β-casein-f | AGGAGATTTCTAGGAATTCAATCC |
| β-casein-r | AGGGGATTGAATTCCTAGAAATCT |
. | 5′-sequence-3′ . |
|---|---|
| PCR primers | |
| JAK2-f | AAGCCACTGCCAGAAACTTG |
| JAK2-r | ACTGAATTCCACCGTTTCCA |
| β2-M-f | TGTCTTTCAGCAAGGACTGG |
| β2-M-r | GATGCTGCTTACATGTCTCG |
| SOCS-1-f | AGAGCTTCGACTGCCTCTTC |
| SOCS-1-r | AGGGGAAGGAGCTCAGGTAG |
| Cyclin D1-f | CGTGGCCTCTAAGATGAAGG |
| Cyclin D1-r | CTGGCATTTTGGAGAGGAAG |
| RB1-f | GGAAGCAACCCTCCTAAACC |
| RB1-r | TTTCTGCTTTTGCATTCGTG |
| HPRT-1-f | GACCAGTCAACAGGGGACAT |
| HPRT-1-r | CTTGCGACCTTGACCATCTT |
| SOCS-1-mRNAf | CACCATGGTAGCACACAACCAGGTGG |
| SOCS-1-mRNAr | TCAAATCTGGAAGGGGAAGGAGCTC |
| EMSA probe | |
| β-casein-f | AGGAGATTTCTAGGAATTCAATCC |
| β-casein-r | AGGGGATTGAATTCCTAGAAATCT |
f indicates forward; r, reverse; β2-M, β2-microglobulin.
Pulse-chase experiments
MedB-1 and L428 cells (2 × 107 each) were harvested and incubated at 37°C for 15 minutes in methionine and cysteine-free RPMI 1640 medium (ICN Biomedical, Costa Mesa, CA) supplemented with 2.5% dialyzed fetal calf serum (FCS). Cells were pulsed with 0.5 mCi (18.5 MBq) TRANS 35S-LABEL (ICN Biomedical) in 2 mL methionine and cysteine-free RPMI 1640 medium for 30 minutes. Cells were washed in PBS and incubated in chase medium (RPMI 1640 supplemented with 10% FCS and 10-fold molar excess of Cys and Met) for various times.
Immunoprecipitation and determination of JAK2 degradation
Cells were rinsed twice with ice-cold PBS and lysed in ice-cold lysis buffer (50 mM Tris-HCl pH 7.4; 150 mM NaCl; 1 mM EDTA; 0.5% NP-40; 0.25% sodium desoxycholate; 1 mM NaF; 1 mM Na3VO4; 1 mM phenylmethanesulfonyl fluoride; 1 μg/mL leupeptin, pepstatin, aprotinin) for 1 hour and centrifuged for 15 minutes at 20 000g and 4°C. Lysates were precleared with 100 μL protein A–agarose bead slurry (Amersham Biosciences) for 1 hour, and 35S incorporation into total proteins was determined by liquid scintillation counting (LS 6000; Beckman Coulter, Fullerton, CA). Aliquots of 500 μg total protein were incubated with 5 μL anti-JAK2 antibody and 100 μL protein A–agarose bead slurry overnight at 4°C. Agarose beads were collected by pulsing 5 seconds at 14 000 rpm and washed 3 times in ice-cold lysis buffer. Samples were heated for 10 minutes at 75°C in 2 × NuPAGE-LDS sample buffer containing 5% vol/vol 2-mercaptoethanol and proteins subjected to electrophoresis. The gels were fixed in 40% methanol, 10% acetic acid, 50% water for 15 minutes, saturated with Amplify solution (Amersham), dried, and autoradiographed. For quantification of JAK2 synthesis and degradation, labeled bands were analyzed using the ImageMaster VDS.
Analysis of SOCS-1 cDNA sequence
PCR was performed on cDNAs of MedB-1 and L428 cells with SOCS-1–specific primers (Table 1). For high resolution analysis 1 μL of the reaction was loaded onto a DNA 1000 LabChip (Agilent Technologies, Palo Alto, CA) and run on the Bioanalyser 2001 (Agilent Technologies). Remaining product was subjected to agarose gel electrophoresis and extracted bands inserted for sequencing into a pGEM-T Easy vector (Promega, Madison, WI). Plasmids were isolated using the Promega plasmid kit and custom sequenced with standard sequencing primers (MWG Biotech, Ebersbach, Germany).
Analysis of SOCS-1 mutations in PMBL
DNA was isolated from frozen tumor tissue obtained from 20 PMBLs using the DNeasy Tissue Kit (Qiagen). The coding sequence of SOCS-1 was amplified by PCR, inserted into a pGEM-T Easy vector, and propagated in Escherichia coli JM109. Five clones each were picked and plasmids subjected to sequencing. As controls, DNA from 2 nonneoplastic spleens, 3 tonsils, and peripheral blood lymphocytes of 10 healthy blood donors was used. The 10 healthy blood donors were members of the institution, including coauthors, who gave their informed consent according to the Declaration of Helsinki. The DNAs from hyperplastic tonsils, traumatic spleens, and PMBL tumors were drawn from the tissue bank of snap-frozen tissues. This material was pseudonymized to comply with the German law for correct usage of archival tissue for clinical research (Deutsches Ärzteblatt 2003; 100 A1632). Approval for this procedure was obtained from the local ethics committee.
Wild-type SOCS-1 expression plasmid preparation and transfection
The SOCS-1 coding sequence was amplified by PCR on DNA from human spleen tissue and inserted in the pcDNA 3.1D/V5-His-TOPO vector (Invitrogen). The plasmid was purified using the Endofree Plasmid Purification MaxiKit (Qiagen). SOCS-1 wt sequence was verified by sequencing. The pcDNA 3.1 vector was used as control plasmid.
Transient transfections were done using the Nucleofector device (Amaxa, Cologne, Germany). In short, 2 × 106 MedB-1 cells were suspended in 100 μL Cell line Nucleofector solution V, 5 μg plasmid was added, and the cells were electroporated (program U 01), resuspended in 2.5 mL prewarmed cell culture medium, and transferred to an 37°C incubator. The survival rate was measured by trypan blue exclusion and found to be higher than 80%. Transfection efficiency was monitored by fluorescence using pmaxGFP plasmid of the Nucleofector kit.
Results
JAK2 and STAT5 in MedB-1 cells are highly phosphorylated
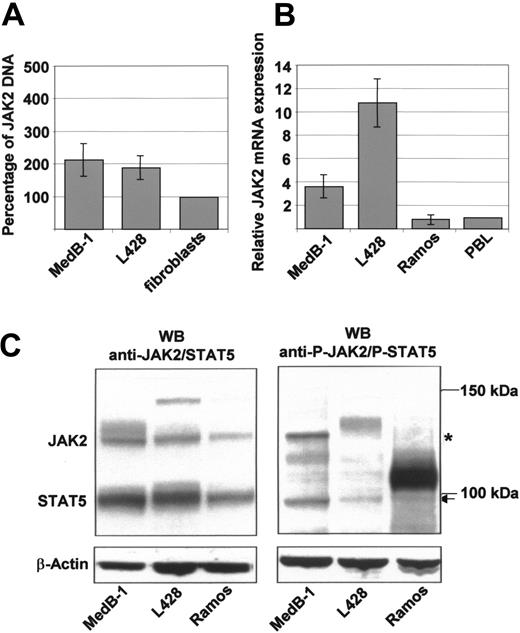
Comparative genomic hybridization (CGH) analysis data on PMBL indicate frequent gains of chromosomal material on 9p24 including the JAK2 gene.2,3 To delineate a possible relationship between the gene dosage and the constitutive activation of JAK2 observed in MedB-1 cells,2,16 we analyzed the JAK2 DNA content, the JAK2 mRNA expression, JAK2 protein content, and the extent of its phosphorylation in this cell line, which harbors a trisomy 9.17 Further, we examined the L428 Hodgkin lymphoma cell line known to contain a gain on the telomeric part of 9p.3 Compared with autologous fibroblasts obtained from the parental tumor tissue of MedB-1, we found an about 2-fold JAK2 amplification in MedB-1 DNA and an about 2-fold amplification in L428 (Figure 1A). This led to an about 4-fold increase of JAK2 mRNA compared with normal peripheral B lymphocytes and the Burkitt cell line Ramos (Figure 1B). Still, JAK2 transcript levels in MedB-1 cells were 3 times lower than those in L428 (Figure 1B). To determine whether there is an effect of the high amount of JAK2 transcripts on the JAK2 protein expression in these cells, we performed Western blot analyses using total cell lysates. To avoid protein loss and degradation during stripping processes of blots, a simultaneous incubation with anti-JAK2 and anti-STAT5 antibodies was used. Despite marked differences in JAK2 mRNA levels in MedB-1, L428, and Ramos, JAK2 protein contents differed much less (Figure 1C). By contrast, the antiphospo-JAK2 antibody, specific for JAK2 phosphorylated at tyrosine Y1007 and Y1008, detected a strong band located at 130 kDa in MedB-1 whereas in L428 and Ramos cells JAK2 phosphorylation was virtually absent (Figure 1C). Because the STAT5 proteins are downstream targets of JAK2,22 we analyzed their expression and phosphorylation status in the 3 cell lines. Using an anti-STAT5A/B antibody in the immunoblot, STAT5 proteins were found to be highly expressed in MedB-1 and L428 and less so in Ramos cells. The antiphospho-STAT5 antibody detected high phosphorylation of the STAT5 proteins, migrating at approximately 95 kDa in MedB-1, whereas the STAT5 phosphorylation was definitively lower in L428 and Ramos cells (Figure 1C).
JAK2 gene amplification, JAK2 mRNA expression, and Western blot analysis. (A) JAK2 gene amplification in MedB-1 cells. DNA prepared from MedB-1 cells, L428 cells, and autologous fibroblasts from the parental tumor of MedB-1 was analyzed by real-time PCR with PCR primers specific for an DNA fragment within exon 5 of the JAK2 gene and normalized on β2-microglobulin gene exon 2. JAK2 DNA contents in MedB-1 and L428 are related to those in fibroblasts calculated as 100%. (B) JAK2 mRNA expression in MedB-1 cells, L428 Hodgkin cells, and Ramos cells. Expression is related to that in peripheral B-lymphocytes. Error bars indicate the standard deviation of 3 independent experiments. (C) Western blot analysis of JAK2/STAT5 and its phosphorylated forms. JAK2 and STAT5 protein expression and its phophorylation were measured in total protein extracts of MedB-1, L428, and Ramos cells by immunoblotting; 20 μg total protein was subjected per lane. JAK2/STAT5 and phospho-JAK2 (*)/phospho-STAT5 (←), respectively, were detected simultaneously with the appropriate antibodies; β-actin was used as loading control.
JAK2 gene amplification, JAK2 mRNA expression, and Western blot analysis. (A) JAK2 gene amplification in MedB-1 cells. DNA prepared from MedB-1 cells, L428 cells, and autologous fibroblasts from the parental tumor of MedB-1 was analyzed by real-time PCR with PCR primers specific for an DNA fragment within exon 5 of the JAK2 gene and normalized on β2-microglobulin gene exon 2. JAK2 DNA contents in MedB-1 and L428 are related to those in fibroblasts calculated as 100%. (B) JAK2 mRNA expression in MedB-1 cells, L428 Hodgkin cells, and Ramos cells. Expression is related to that in peripheral B-lymphocytes. Error bars indicate the standard deviation of 3 independent experiments. (C) Western blot analysis of JAK2/STAT5 and its phosphorylated forms. JAK2 and STAT5 protein expression and its phophorylation were measured in total protein extracts of MedB-1, L428, and Ramos cells by immunoblotting; 20 μg total protein was subjected per lane. JAK2/STAT5 and phospho-JAK2 (*)/phospho-STAT5 (←), respectively, were detected simultaneously with the appropriate antibodies; β-actin was used as loading control.
MedB-1 cells and their parental tumor contain biallelic mutations in the coding region of SOCS-1
To detect the cause of high constitutive JAK2 phosphorylation in MedB-1 cells, we examined the mRNA expression of known regulators of JAK2 activity: phosphatases SHP1 and SHP2, the leukocyte common antigen CD45, and the suppressors of cytokine signaling SOCS-1 and SOCS-3 by RT-PCR. SHP1, SHP2, and SOCS-3 expression proved normal, corresponding in length to gene bank data and in abundance compared with the housekeeper HPRT 1, whereas CD45 expression was undetectable (not shown). The PCR product amplified with SOCS-1 primers showed, after electrophoresis, 2 truncated DNA bands instead of the expected 201 base pair (bp) band resulting with wt SOCS-1. These abnormal bands were extracted and sequenced.
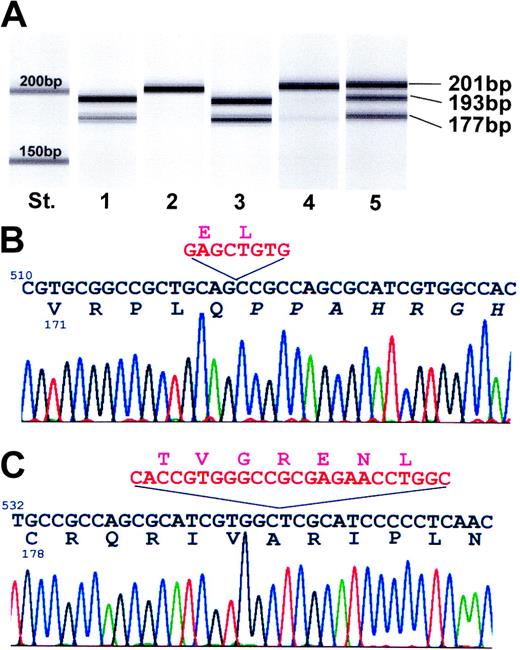
In Figure 2A, the gel view of the separated PCR products run on an DNA 1000 LabChip assay is shown. Because the coding sequence of SOCS-1 is located on exon 2, PCR products of 193 and 177 bp length were obtained using cDNA (Figure 2A, lane 1) or DNA templates of MedB-1 (Figure 2A, lane 3). The identical intensities of the 2 bands formed on DNA reveal a deletion mutation of 8 bp on allele A and a 24 bp deletion on allele B. SOCS-1 mutations are restricted to the lymphoma because SOCS-1 amplification using cDNA and DNA from autologous fibroblasts yielded wt sequences (Figure 2A, lanes 2, 4). In DNA extracted from parental tumor tissue (Figure 2A, lane 5), both the deleted and wt SOCS-1 DNA fragments were detectable. Sequence analysis of the 8 bp deleted allele (Figure 2B), which is dominantly expressed in MedB-1 cells, proved a frame shift mutation at nucleotide 525 of the coding sequence leading to a C-terminal nonsense protein sequence up to amino acid 176. The 24 bp inframe deletion resulted in a loss of 7 amino acids from position 184 to 190 in the SOCS-1 protein sequence (Figure 2C). The biallelic mutation in the SOCS-1 gene of MedB-1 cells and its lymphoma of origin involves the C-terminal region of the protein including the SOCS box domain. SOCS box is important for the degradation of SOCS-1–associated proteins.
Mutational analysis of the SOCS-1 gene. (A) Gel view of a DNA 1000 LabChip biosizing assay. SOCS-1 mRNA–specific PCR products from RT-PCR assays on RNA derived from MedB-1 cells (lane 1) and autologous fibroblasts (lane 2). Lanes 3, 4, and 5 represent specific SOCS-1 gene fragments amplified by PCR on DNA separated from MedB-1 cells (lane 3), autologous fibroblasts (lane 4), and tumor tissue (lane 5). Wild-type SOCS-1 sequence appeared as the 201-bp band. St, standard 200 bp and 150 bp, respectively. (B) Detailed sequencing chromatogram of the 193-bp PCR product indicates an 8-bp deletion (red sequence) causing a reading frame shift (altered protein sequence is given in italics). (C) Detailed sequencing chromatogram of the 177 bp PCR product indicating an inframe deletion of 24 nucleotides (red sequence) and loss of 7 amino acids. DNA and protein sequences are numbered relative to the start as 1.
Mutational analysis of the SOCS-1 gene. (A) Gel view of a DNA 1000 LabChip biosizing assay. SOCS-1 mRNA–specific PCR products from RT-PCR assays on RNA derived from MedB-1 cells (lane 1) and autologous fibroblasts (lane 2). Lanes 3, 4, and 5 represent specific SOCS-1 gene fragments amplified by PCR on DNA separated from MedB-1 cells (lane 3), autologous fibroblasts (lane 4), and tumor tissue (lane 5). Wild-type SOCS-1 sequence appeared as the 201-bp band. St, standard 200 bp and 150 bp, respectively. (B) Detailed sequencing chromatogram of the 193-bp PCR product indicates an 8-bp deletion (red sequence) causing a reading frame shift (altered protein sequence is given in italics). (C) Detailed sequencing chromatogram of the 177 bp PCR product indicating an inframe deletion of 24 nucleotides (red sequence) and loss of 7 amino acids. DNA and protein sequences are numbered relative to the start as 1.
Constitutive JAK2 activation in MedB-1 is due to its impaired degradation
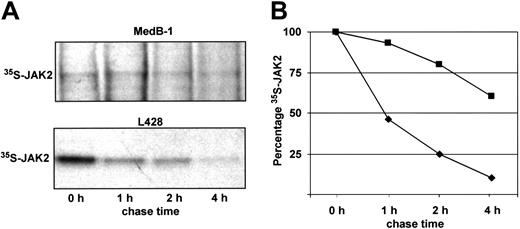
To examine whether the elevated JAK2 activity in MedB-1 cells is due to delayed degradation, we pulsed MedB-1 and L428 cells as control for 30 minutes with 35S-methionine/cysteine and chased the cells in complete medium. At time points indicated in Figure 3, protein extracts were made and JAK2 was immunoprecipitated and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. Immediately after the pulse and after 4 hours of chase, no significant difference of 35S incorporation into the total proteins of MedB-1 and L428 cells was measured (data not shown). However, comparing the signals of 35S-JAK2 in the autoradiograms at time zero, a striking difference in JAK2 synthesis by L428 and MedB-1 cells emerged, because de novo JAK2 synthesis was abnormally low in the MedB-1 but not in L428 cells. Further, the turnover of 35S-JAK2 in MedB-1 cells was strikingly protracted compared with that in L428 cells (Figure 3A). Analysis of the intensities of the 35S-JAK2 bands revealed that the half-life of JAK2 was more than 4 hours in MedB-1 while being around 1 hour in L428 cells (Figure 3B).
Turnover of JAK2 protein. MedB-1 and L428 cells were pulsed with 35S-methionine/cysteine for 30 minutes and chased for the times indicated. Cell protein extracts were used for immunoprecipitation with anti-JAK2 antibodies and subjected to SDS-PAGE and autoradiography. (A) Details of the x-ray films. (Top) 35S-JAK2 signals obtained from MedB-1 protein extracts indicating degree of synthesis (0 hours) and dynamics of degradation. (Bottom) Signals obtained from an experiment with L428 cells after. (B) Time-dependent degradation of JAK2 in MedB-1 (▪) and L428 cells ( ). Intensities of the bands were analyzed using the ImageMaster VDS hardware and software, and the integrated optical densities of the bands at time zero were set as 100% synthesis.
). Intensities of the bands were analyzed using the ImageMaster VDS hardware and software, and the integrated optical densities of the bands at time zero were set as 100% synthesis.
Turnover of JAK2 protein. MedB-1 and L428 cells were pulsed with 35S-methionine/cysteine for 30 minutes and chased for the times indicated. Cell protein extracts were used for immunoprecipitation with anti-JAK2 antibodies and subjected to SDS-PAGE and autoradiography. (A) Details of the x-ray films. (Top) 35S-JAK2 signals obtained from MedB-1 protein extracts indicating degree of synthesis (0 hours) and dynamics of degradation. (Bottom) Signals obtained from an experiment with L428 cells after. (B) Time-dependent degradation of JAK2 in MedB-1 (▪) and L428 cells ( ). Intensities of the bands were analyzed using the ImageMaster VDS hardware and software, and the integrated optical densities of the bands at time zero were set as 100% synthesis.
). Intensities of the bands were analyzed using the ImageMaster VDS hardware and software, and the integrated optical densities of the bands at time zero were set as 100% synthesis.
Ectopic SOCS-1 expression down-regulates cell proliferation of MedB-1 cells
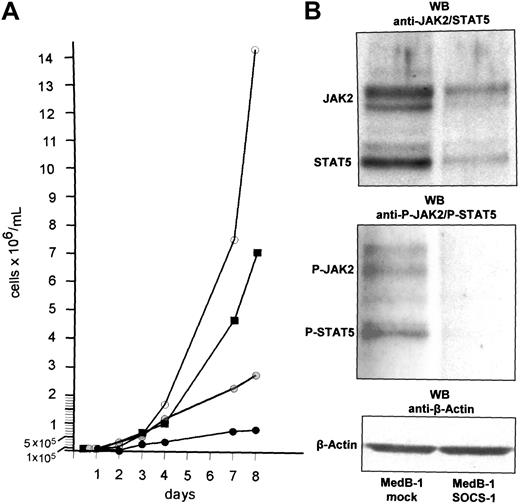
To evaluate the influence of ectopic wt SOCS-1 expression on MedB-1 cell proliferation and JAK2/STAT5 activation, we performed proliferation assays and Western blot analyses using protein extracts derived from MedB-1 cells 48 hours after SOCS-1 transfection. In Figure 4A we show the proliferation characteristics of MedB-1 cells within a culture time of 8 days. In complete medium, MedB-1 cells grew exponentially with a doubling time of about 24 hours. In medium supplemented with 20 μM Tyrphostin AG490, a JAK2 inhibitor,23 proliferation rate was decreased with a doubling time extended to 48 hours. An even more striking effect on cell proliferation was achieved in MedB-1 cells transiently transfected with SOCS-1 expression plasmid. These cells grew very slowly and linearly with a doubling time of about 4 days, whereas transfection with the empty vector did not significantly influence proliferation of MedB-1 cells. As shown in Western blots (Figure 4B), this effect went along with diminished JAK2 and STAT5 protein contents and very low phosphorylation of JAK2 and STAT5 in MedB-1 cells 48 hours after transfection with wt SOCS-1. Thus, SOCS-1 fulfills the criteria of a tumor suppressor.
Effect of wt SOCS-1 transfection on MedB-1 cells. (A) MedB-1 cell proliferation assay. The cell densities at the beginning of the experiments were adjusted to 1 × 105 cells/mL. Cell numbers were determined in triplicates every day in 50 μL cell suspension from MedB-1 cells cultured in complete medium (○), in medium supplemented with 20 μM Tyrphostin AG490 ( ), and in MedB-1 cells transfected with wt SOCS-1 plasmid (•) and mock plasmid (▪). (B) Western blot analysis of wt SOCS-1–transfected MedB-1 cells. Twenty-four hours after transfection with a mock plasmid (lane 1) and the wt SOCS-1 plasmid (lane 2), proteins were extracted and subjected to Western blot analyses as described; β-actin is shown as loading control. While the content of JAK2 and STAT5 proteins is at comparable levels, phosphorylation is barely detectable in transfectants.
), and in MedB-1 cells transfected with wt SOCS-1 plasmid (•) and mock plasmid (▪). (B) Western blot analysis of wt SOCS-1–transfected MedB-1 cells. Twenty-four hours after transfection with a mock plasmid (lane 1) and the wt SOCS-1 plasmid (lane 2), proteins were extracted and subjected to Western blot analyses as described; β-actin is shown as loading control. While the content of JAK2 and STAT5 proteins is at comparable levels, phosphorylation is barely detectable in transfectants.
Effect of wt SOCS-1 transfection on MedB-1 cells. (A) MedB-1 cell proliferation assay. The cell densities at the beginning of the experiments were adjusted to 1 × 105 cells/mL. Cell numbers were determined in triplicates every day in 50 μL cell suspension from MedB-1 cells cultured in complete medium (○), in medium supplemented with 20 μM Tyrphostin AG490 ( ), and in MedB-1 cells transfected with wt SOCS-1 plasmid (•) and mock plasmid (▪). (B) Western blot analysis of wt SOCS-1–transfected MedB-1 cells. Twenty-four hours after transfection with a mock plasmid (lane 1) and the wt SOCS-1 plasmid (lane 2), proteins were extracted and subjected to Western blot analyses as described; β-actin is shown as loading control. While the content of JAK2 and STAT5 proteins is at comparable levels, phosphorylation is barely detectable in transfectants.
), and in MedB-1 cells transfected with wt SOCS-1 plasmid (•) and mock plasmid (▪). (B) Western blot analysis of wt SOCS-1–transfected MedB-1 cells. Twenty-four hours after transfection with a mock plasmid (lane 1) and the wt SOCS-1 plasmid (lane 2), proteins were extracted and subjected to Western blot analyses as described; β-actin is shown as loading control. While the content of JAK2 and STAT5 proteins is at comparable levels, phosphorylation is barely detectable in transfectants.
Wild-type SOCS-1 modifies expression of cell cycle regulatory proteins
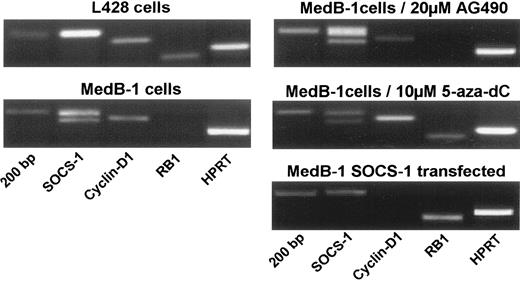
To find genes involved in maintaining the high proliferation rate in MedB-1 cells, we performed RT-PCR experiments on cDNA derived from MedB-1 cells cultured in complete medium, cells treated for 5 days in medium containing 20 μM AG490 or 10 μM 5-aza-deoxycytosine, and MedB-1 cells transiently transfected with a wt SOCS-1 expression vector. Complementary DNA prepared from L428 cells was included as control in these experiments. In Figure 5, the expression pattern obtained by RT-PCR of selected genes SOCS-1, cyclin D1, retinoblastoma 1 (RB1), and, as housekeeping gene, hypoxanthine guanine phosphoribosyltransferase 1 (HPRT-1) is shown. In L428 cells, SOCS-1 expression was similarly high as that of HPRT-1 while cyclin D1 and RB1 transcripts were present at low levels. In PCRs using MedB-1 cDNA, the 2 truncated SOCS-1 fragments of 193 bp and 177 bp length were amplified. Cyclin D1 message was clearly present whereas RB1 transcripts were absent. AG490 treatment of MedB-1 cells led to a slight reduction of cyclin D1 expression without marked effects on the other genes. In MedB-1 cells cultured with the DNA demethylating agent 5-aza-deoxycytosine, RB1 transcription was switched on and cyclin D1 expression increased slightly. Striking responses on gene expression were achieved in transiently SOCS-1–transfected MedB-1 cells. In this experiment the main PCR product with SOCS-1 primers was the expected 201 bp fragment of wt SOCS-1 transcript. Cyclin D1 expression was undetectable in the transfectants. Conversely, RB1 gene expression was induced to levels exceeding those obtained by DNA demethylation.
Comparative RT-PCR analysis of SOCS-1, cyclin D1, RB1, and HPRT expression. RT-PCR using specific primer pairs to detect the expression of the indicated genes was performed on RNA extracts derived from MedB-1 cells cultured in complete medium or cultured for 5 days in medium supplemented with 20 μM Tyrphostin AG490 or 10 μM 5-aza-deoxycytosine (5-aza-dC) and on RNA from MedB-1 cells transiently transfected with a SOCS-1 expression plasmid. AG490 slightly decreases cyclin D1 expression, and 5-aza-dC induces RB1 expression. Wild-type SOCS-1 induces RB1 expression and abolishes cyclin D1 transcription.
Comparative RT-PCR analysis of SOCS-1, cyclin D1, RB1, and HPRT expression. RT-PCR using specific primer pairs to detect the expression of the indicated genes was performed on RNA extracts derived from MedB-1 cells cultured in complete medium or cultured for 5 days in medium supplemented with 20 μM Tyrphostin AG490 or 10 μM 5-aza-deoxycytosine (5-aza-dC) and on RNA from MedB-1 cells transiently transfected with a SOCS-1 expression plasmid. AG490 slightly decreases cyclin D1 expression, and 5-aza-dC induces RB1 expression. Wild-type SOCS-1 induces RB1 expression and abolishes cyclin D1 transcription.
Wild-type SOCS-1 inhibits STAT5 DNA binding
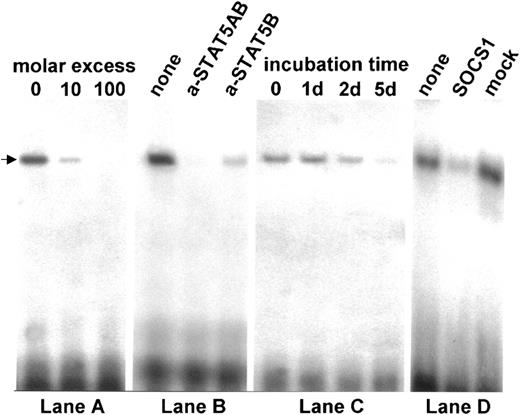
The activation of STAT5 can be shown both by its high phosphorylation ratio and by its interaction with the cognate DNA motifs. To investigate the influence of wt SOCS-1 expression in MedB-1 cells on the JAK2/STAT5 signaling cascade, we applied EMSAs to detect STAT5 binding ability toward a 32P-labeled oligonucleotide containing the STAT5 binding site of the β-casein promoter (Table 1). Nuclear protein extract from MedB-1 cells formed a strong protein DNA complex (Figure 6) in which the labeled probe was totally replaced by a 100-fold molar excess of competitor (Figure 6, lane A). Preincubation of protein extracts with anti-STAT5 antibodies did not result in a complex shift but inhibited complex formation (Figure 6, lane B). DNA binding was totally inhibited by antibody against STAT5A/B. The fact that anti-STAT5B antibody extensively diminished the probe-DNA complex formation indicates that, in MedB-1 cells, STAT5B is the dominant DNA binding partner. The prolonged lifetime of activated JAK2 on STAT5 phophorylation was also, albeit indirectly, shown by the action of AG490. No immediate inhibitory response on STAT5-DNA complex formation was observed using protein extracts from MedB-1 cells incubated with 20 μM AG490. After 2 days of treatment with this JAK2 inhibitor, the reduction in complex formation was minimal while a clearly diminished STAT5 probe binding occurred after 5 days of treatment (Figure 6, lane C). An earlier effect on STAT5 DNA probe binding was seen with protein extracts from MedB-1 cells 2 days after transfection with wt SOCS-1. The intensities of the complex dropped to about one third of those formed by protein extracts from nontransfected or mock vector–transfected MedB-1 cells (Figure 6, lane D). Taking into account that the transfection efficiency was around 80% (as monitored by the green fluorescent protein [GFP] reporter), the remaining protein-DNA complex was most likely due to activated STAT5 in the nontransfected minority of MedB-1 cells.
DNA binding of STAT5 in MedB-1 cells. EMSAs were performed using a 52P-labeled β-casein gene promoter region containing the STAT5 DNA binding motif and nuclear protein extracts from MedB-1 cells. Competition with unlabeled probe (lane A) and inhibition of complex formation by anti-STAT5 antibodies (lane B) demonstrate specificity of protein-DNA binding. (Lane C) EMSA on nuclear extracts from MedB-1 cells treated with 20 μM Tyrphostin AG490 indicates a very late effect on STAT5-DNA binding. (Lane D) EMSA on nuclear extracts from MedB-1 cells (none) and cells transfected with wt SOCS-1 (SOCS1) expression plasmid or mock plasmid (mock) for 48 hours. Wild-type SOCS-1 specifically reduces STAT5-DNA binding.
DNA binding of STAT5 in MedB-1 cells. EMSAs were performed using a 52P-labeled β-casein gene promoter region containing the STAT5 DNA binding motif and nuclear protein extracts from MedB-1 cells. Competition with unlabeled probe (lane A) and inhibition of complex formation by anti-STAT5 antibodies (lane B) demonstrate specificity of protein-DNA binding. (Lane C) EMSA on nuclear extracts from MedB-1 cells treated with 20 μM Tyrphostin AG490 indicates a very late effect on STAT5-DNA binding. (Lane D) EMSA on nuclear extracts from MedB-1 cells (none) and cells transfected with wt SOCS-1 (SOCS1) expression plasmid or mock plasmid (mock) for 48 hours. Wild-type SOCS-1 specifically reduces STAT5-DNA binding.
In conclusion, MedB-1 cells and their lymphoma of origin harbor a biallelic loss of function mutation of the SOCS-1 gene. The mutated SOCS-1 proteins critically interfere with the JAK2/STAT5 signaling pathway in impairing JAK2 degradation and thus sustaining JAK2 phosphorylation. This, in turn, leads via constitutive STAT5 phosphorylation to the induction of cyclin D1 and by a yet elusive downstream mechanism to RB1 repression. Collectively, these data definitively define SOCS-1 as a novel tumor suppressor.
SOCS-1 mutations in PMBL
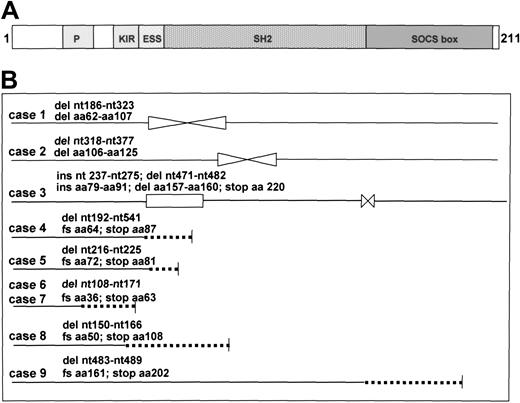
To find out whether SOCS-1 mutations occur in PMBL, we analyzed the coding sequence of SOCS-1 in 20 cryostored tumors. As normal controls we isolated DNA from 2 nonneoplastic spleens, 3 tonsils, and peripheral blood lymphocytes of 10 healthy blood donors. In this control panel only wt SOCS-1 was found. In 9 of 20 PMBLs, deletions in the sequence were detectable (Figure 7). Two lymphomas (cases 6 and 7) had an identical mutation, whereas in all other cases the positions and sizes of mutations were different. One case (no. 1) had an inframe deletion, predicting a loss of 46 amino acids (aa) in the protein sequence affecting the extended SH2 (ESS) domain and in the SH2 region. Case no. 2 had a deletion predicting a loss of 20 aa in the SH2 domain. In case no. 3, an inframe insertion of 39 nucleotides and a 12-bp deletion were found affecting the ESS/SH2 domain and the SOCS box, respectively. In 5 cases (nos. 4 to 8) deletion mutations downstream the start codon were found, predicting an abort of SOCS-1 protein synthesis in the N-terminal region. Very reminiscent to the situation found in MedB-1, case no. 9 had a deletion leading to a nonsense sequence in the SOCS box region. Thus, we provide evidence that in about half of our series of PMBL, SOCS-1 is mutated. To find out whether SOCS1 mutation in PMBL is correlated with an overrepresentation of JAK2, we compared the JAK2 status with CGH and fluorescence in situ hybridization (FISH) data2 of the lymphomas included in this study (Table 2). Seven of 9 lymphomas with a SOCS-1 mutation had a gain of chromosome 9 including the JAK2 locus. In comparison, 6 of 8 lymphomas without SOCS-1 mutation had gains of the JAK2 locus. Thus, these genomic events were frequent in our series of PMBL but are neither correlated nor mutually exclusive.
SOCS-1 protein structure and SOCS-1 mutations. (A) Structure of SOCS-1 protein. The N-terminal domain of SOCS-1 contains a proline-rich region (P), a kinase inhibitory region (KIR) including aa 55 to 66, and an extended SH2 subdomain (ESS) (aa 67 to 78). KIR, ESS, and SH2 domains are required for JAK inactivation. The SOCS box (aa 161 to 210) contains a 10 aa consensus sequence for interaction with elongin BC complex. (B) SOCS-1 mutations in PMBL. The coding sequence of the SOCS-1 gene was amplified by PCR using DNA from PMBLs as template and subjected to sequencing. The protein sequences related from the DNA sequences are schematically shown. The positions of mutation in SOCS-1 DNA and its consequences on protein sequence are indicated. del and  indicate deletion; ins and □, insertion; dotted line, nonsense sequence; nt, position on cDNA sequence; aa, position on corresponding protein sequence; and fs, frame shift.
indicate deletion; ins and □, insertion; dotted line, nonsense sequence; nt, position on cDNA sequence; aa, position on corresponding protein sequence; and fs, frame shift.
SOCS-1 protein structure and SOCS-1 mutations. (A) Structure of SOCS-1 protein. The N-terminal domain of SOCS-1 contains a proline-rich region (P), a kinase inhibitory region (KIR) including aa 55 to 66, and an extended SH2 subdomain (ESS) (aa 67 to 78). KIR, ESS, and SH2 domains are required for JAK inactivation. The SOCS box (aa 161 to 210) contains a 10 aa consensus sequence for interaction with elongin BC complex. (B) SOCS-1 mutations in PMBL. The coding sequence of the SOCS-1 gene was amplified by PCR using DNA from PMBLs as template and subjected to sequencing. The protein sequences related from the DNA sequences are schematically shown. The positions of mutation in SOCS-1 DNA and its consequences on protein sequence are indicated. del and  indicate deletion; ins and □, insertion; dotted line, nonsense sequence; nt, position on cDNA sequence; aa, position on corresponding protein sequence; and fs, frame shift.
indicate deletion; ins and □, insertion; dotted line, nonsense sequence; nt, position on cDNA sequence; aa, position on corresponding protein sequence; and fs, frame shift.
Molecular cytogenetic data regarding chromosome 9 and/or chromosomal bands 9p23-p24 evaluated by CGH and FISH of PMBL with SOCS-1 mutations
Case no. . | CGH . | FISH: YAC clone 776a11, 9p23-p24, % gain . |
|---|---|---|
| 1 | No gain | 46.5 |
| 2 | + 9p | 33 |
| 3 | + 9p | 42 |
| 4 | + 9 | ND |
| 5 | + 9 | 21 |
| 6 | No gain | 0 |
| 7 | No gain | 27.5 |
| 8 | No gain | ND |
| 9 | + 9 | 43.5 |
Case no. . | CGH . | FISH: YAC clone 776a11, 9p23-p24, % gain . |
|---|---|---|
| 1 | No gain | 46.5 |
| 2 | + 9p | 33 |
| 3 | + 9p | 42 |
| 4 | + 9 | ND |
| 5 | + 9 | 21 |
| 6 | No gain | 0 |
| 7 | No gain | 27.5 |
| 8 | No gain | ND |
| 9 | + 9 | 43.5 |
CGH indicates comparative genomic hybridization; FISH, fluorescence in situ hybridisation; ND, not done. Source: Bentz et al.2
Discussion
We confirm and extend data published by Guiter et al16 that in MedB-1 mediastinal lymphoma cells JAK2 protein is highly phosphorylated. This leads to a high level of activated STAT5 evidenced by Western blotting with phosphospecific antibodies against JAK2 and STAT5 and by formation of STAT5-DNA complexes in EMSAs using a STAT5-specific DNA probe and nuclear MedB-1 protein extracts.
The concept that JAK2 might be a primary oncogenic key player in PMBL arose from molecular cytogenetic studies2 and arbitrarily primed PCR fingerprinting24 revealing that chromosome 9 has extra chromosomal material in up to 75% of cases and that the consensus region at 9p24 contains the JAK2 locus.4 We confirm these data by showing that MedB-1, which has a trisomy 9,2 has about the double dose of JAK2 DNA compared with that of autologous fibroblasts, which were subcultured and stored at the time we established the MedB-1 line from the patient's tumor. Further support for this view came from expression profiling data on PMBL published by 2 independent groups.5,6 We show that the JAK2 expression in MedB-1 cells is clearly higher than in resting peripheral B cells and Ramos cells. In contrast, MedB-1 cells have lower transcript levels compared with L428 Hodgkin cells, even though both cell lines have an additional JAK2 copy number, as shown by FISH analysis.3
First indications that the constitutively high JAK2 phosphorylation in MedB-1 cells might not be due to a simple gene dosage effect came from our protein data indicating that the high JAK2 transcript levels in MedB-1 and L428 translate in only a slightly elevated JAK2 protein content when compared with control cells. Further, JAK2 protein of MedB-1 and L428 was at comparable levels. However, L428 had a definitely lower phospho-JAK2 ratio compared with MedB-1. Therefore, we examined the neosynthesis and turnover rate of JAK2 in these 2 cell lines. In pulse and chase experiments we observed a strikingly low JAK2 synthesis accompanied by a rather retarded degradation in MedB-1 not seen in L428 cells. L428 cells featured a robust de novo JAK2 protein synthesis and a half-life consistent with the literature.25 The half-life of JAK2 was calculated by Siewert et al25 in a study determining different protein turnovers of interleukin-6–type cytokine signaling components to be 1.9 hours, which is in line with the half-life measured in L428 cells, while it was over 4 hours in MedB-1. Thus, a possible gene dosage effect of overrepresented JAK2 seemed, at least in MedB-1 cells, to be completely overrun at the functional level.
These unexpected results raised the question: What causes the constitutive activation of the JAK2/STAT5 signaling pathway in MedB-1 cells?
Three different classes of negative regulators are known to contribute to cytokine signaling. These are protein inhibitors of activated STATs (PIAS),26 Src-homology 2 (SH2)–containing protein tyrosine phosphatases (SHPs),27 and the protein family of suppressors of cytokine signaling (SOCS).28 Transcripts of SHP 1, a cytoplasmatic protein tyrosine phosphatase associated with JAK2 dephosphorylation,29 was clearly detectable in MedB-1 cells, whereas the cell membrane–spanning phosphatase CD45, involved in negative regulation of cytokine receptor signaling,30 was undetectable by RT-PCR (data not shown). SOCS-1 is located on chromosome 16p13.13. The product, also termed JAK-binding protein (JAB), is a member of 8 SOCS proteins containing a variable N-terminus, a central SH2 domain, and a conserved C-terminus termed SOCS box14 (Figure 7A). SOCS-1 is bound via its SH2 region to the catalytic domain of phospho-JAK2 and inhibits the intrinsic kinase activity of JAK2. As a consequence, tyrosine-phosphorylated JAK2 and STATs are greatly reduced.12,13
First hints that aberrant SOCS-1 might promote tumor genesis came from microsatellite analyses of hepatocellular carcinomas. Using markers covering the chromosomal arm 16p.13.13 including the SOCS-1 locus, the authors observed an allelic loss in 48% of the tumors.31 In addition, silencing of SOCS-1 gene expression by CpG methylation was found in hepatocellular carcinomas32 and in myeloid leukemia, mantle cell lymphoma, follicular lymphoma, and myeloma,33-35 and tumor progression was shown to be related to constitutive activation of JAK/STAT signaling. To prove whether aberrant SOCS-1 is functional in MedB-1 cells, we performed PCR and RT-PCR experiments on DNA and cDNA derived from MedB-1 cells using SOCS-1–specific primers. Our central new finding is a biallelic deletion mutation of SOCS-1 in MedB-1. The deletion in both alleles affects the C-terminal region of SOCS-1 protein leading to a predicted nonsense sequence up to amino acid 176 in one allele and a loss of amino acids 184 to 190 in the other. Both deletions involve the conserved SOCS box, spanning from amino acid 161 to 210 of the SOCS-1 protein consisting of 211 amino acids. As evidenced by comparing the SOCS-1 DNAs, neither of the 2 different mutations was present in autologous fibroblasts, indicating that the patient did not carry a silent, monoallelic SOCS-1 mutation in his germ line. Importantly, the mutations were already present in the patient's lymphoma tissue and hence were both acquired in vivo. The C-terminal domain of SOCS-1 associates with elongins B and C. The elongin BC complex subsequently couples SOCS proteins and their substrates to the proteosomal protein degradation pathway.15 Thus, the assumption is that mutated, loss-of-function SOCS-1 might be the reason for sustained and low turnover of phospho-JAK2.
Our transfectant studies with wt SOCS-1 yielded evidence that the active state of transducers and late protein degradation of JAK2 in MedB-1 is maintained by the absence of SOCS box function caused by the SOCS-1 mutation.
As mentioned, the high ratio of phospho-JAK2 leads to secondary activation of downstream signaling targets. Wt SOCS-1 abolished the constitutive activation of JAK2/STAT5 pathway while the rate of phospho-JAK2 and phospho-STAT5 was dramatically reduced.
As a functional readout we probed the expression of cyclin D1, which is under the transcriptional control of STAT5.36 Ectopic expression of wt SOCS-1 leads to abrogation of cyclin D1 expression. Furthermore, we found an induction of RB1 gene expression in wt SOCS-1 transfectants. RB1 protein is an important negative cell cycle regulator not yet known to be functionally linked to SOCS-1. MedB-1 cells constitutively lack RB1 expression. Because silencing of RB1 by CpG methylation in a wide range of cancers is well established,37,38 we assumed that this mechanism is also operative in MedB-1. In fact, after incubation of MedB-1 cells with the demethylating agent 5-aza-deoxycytosine, RB1 was expressed. Because RB1 expression was clearly detectable in MedB-1 cells transiently transfected with wt SOCS-1, intact SOCS-1 protein may influence, besides its known function, epigentic events of gene regulation as shown here by the induction of RB1. Anyway, this event, combined with the loss of cyclinD 1 expression, may contribute to the growth arrest observed in wt SOCS-1–transfected MedB-1 cells.
High JAK2 expression leading to enhanced tumor growth has been proposed as a consequence of genomic overpresentation of JAK2. Our data presented here do not exclude an additive effect of JAK2 overexpression but collectively challenge the concept of JAK2 deregulation as the primary growth-promoting event in PMBL and establish SOCS-1 as a novel tumor suppressor, which is mutated in MedB-1, its tumor of origin, and in about half of PMBL tested so far.
Prepublished online as Blood First Edition Paper, November 30, 2004; DOI 10.1182/blood-2004-09-3701.
Supported by a grant of the Deutsche Krebshilfe, Mildred Scheel Stiftung (70-22859-Ba2) (T.F.E.B., P.M.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal