Abstract
Human telomerase uses a specific cellular RNA, called hTERC, as the template to synthesize telomere repeats at chromosome ends. Approximately 10% to 15% of patients with aplastic anemia or other bone marrow failure syndromes are carriers of hTERC sequence variants whose functional significance, in most cases, is unknown. We screened 10 reported and 2 newly discovered hTERC variants from such patients and found that 10 of these negatively affected telomerase enzymatic function when they were used to reconstitute telomerase enzymatic function in human cells. Most functional deficits were due to perturbations of hTERC secondary structure and correlated well with the degrees of telomere shortening and reduced telomerase activity observed in peripheral blood lymphocytes of the representative patients. We also found no evidence of dominant-negative activity in any of the mutants. Therefore, loss of telomerase activity and of telomere maintenance resulting from inherited hTERC mutations may limit marrow stem cell renewal and predispose some patients to bone marrow failure.
Introduction
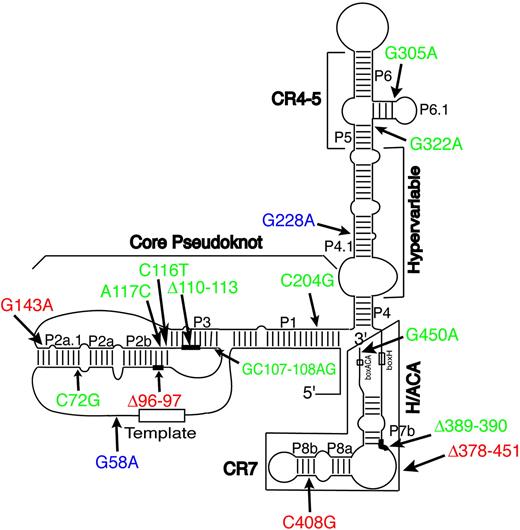
Telomerase is a specialized DNA polymerase that adds long, repetitive stretches of simple telomeric DNA sequence (ie, [TTAGGG]n in the vertebrates) onto the chromosomal termini.1 The minimal telomerase holoenzyme is a ribonucleoprotein (RNP) complex with 2 components: a protein (called TERT) that has RNA-dependent DNA polymerase (ie, reverse transcriptase) activity, and an associated RNA (called TERC) that serves as its template.1 Vertebrate TERCs are believed to adopt a complex, folded structure due to extensive intramolecular base-pairing that forms an ordered series of paired (P) or helical regions. This phylogenetically conserved structure, depicted for human TERC (hTERC) in Figure 1, is viewed as comprising 4 separate conformational domains, termed the core, CR4-CR5, box H/ACA, and CR7 domains, respectively, together with an interposed region of variable length and sequence called the hypervariable paired region.2 Site-directed mutagenesis studies of hTERC have validated much of this predicted structure and confirmed that each of the 4 conserved domains contributes features necessary for telomerase function. At certain locations, including the single-stranded templating region that is copied into telomeric DNA, specific RNA base sequences are required for biologic activity. As is true of many biologically active RNA molecules, however, most of the internally base-paired regions of hTERC can be extensively mutated without loss of function provided that the normal base-pairing pattern is preserved.3,4 Those findings, together with the strict conservation of this base-pairing pattern in evolution, imply that telomerase activity depends on specific features of both base sequence and secondary structure in hTERC, some of which have yet to be fully elucidated. The core domain includes the templating region along with a conserved pseudoknot structure that serves both to delineate the templating region and to mediate dimerization of hTERC molecules during telomerase assembly.5,6 The core and CR4-CR5 domains each contribute to binding of the TERT protein,4 though the specific requirements for binding are unknown. The box H/ACA domain is required for proper processing, stability, and trafficking of hTERC within cells, whereas the CR7 domain provides signals that target hTERC into a specialized nuclear compartment where telomerase RNP assembly occurs.7-10
Schematic depiction of the predicted secondary structure of hTERC. This schematic depiction is as proposed by Chen et al.2 The 8-base template sequence (rectangle) and other structural features are indicated, including the core, CR4-CR5, box H/ACA, and CR7 domains, and the hypervariable paired region. Mutations associated with DC are indicated in red; those associated with AA, MDS, or PNH are in green. The G228A and G58A variants (in blue) are fully active in vitro (Fu and Collins,17 Marrone et al,27 and the present study) and also found in healthy individuals, and may be functionally inconsequential. Δ denotes nucleotide deletion (see Figure 2A for details). Nucleotide deletions are indicated by thick lines, whereas boxed regions show a large deletion that completely removes the sequences of the box H/ACA and CR7 domains.
Schematic depiction of the predicted secondary structure of hTERC. This schematic depiction is as proposed by Chen et al.2 The 8-base template sequence (rectangle) and other structural features are indicated, including the core, CR4-CR5, box H/ACA, and CR7 domains, and the hypervariable paired region. Mutations associated with DC are indicated in red; those associated with AA, MDS, or PNH are in green. The G228A and G58A variants (in blue) are fully active in vitro (Fu and Collins,17 Marrone et al,27 and the present study) and also found in healthy individuals, and may be functionally inconsequential. Δ denotes nucleotide deletion (see Figure 2A for details). Nucleotide deletions are indicated by thick lines, whereas boxed regions show a large deletion that completely removes the sequences of the box H/ACA and CR7 domains.
Inherited mutations in hTERC underlie one form of a rare human disorder, known as dyskeratosis congenita (DC) that involves hematopoietic failure. This disorder is characterized by abnormal skin pigmentation, nail dystrophy, and oral leukoplakia, and is often complicated by life-threatening bone marrow failure and immunodeficiency.11 The autosomal dominant form of DC results from germ line inheritance of mutations of the hTERC gene, and all affected individuals studied to date are heterozygous (ie, carry one mutant and one wild-type allele) for the gene. Lymphocytes from affected individuals show decreased hTERC expression, decreased telomerase activity, and significantly reduced telomere lengths as compared with those from age-matched controls.12,13
Building on those findings, we and others have investigated whether telomerase mutations also contribute to other, more common disorders that involve bone marrow failure, such as aplastic anemia (AA), paroxysmal nocturnal hemoglobinuria (PNH), or myelodysplastic syndrome (MDS).14-16 Surveys of patients with apparently acquired forms of those disorders have revealed an assortment of hTERC sequence variants, each of them individually rare and heterozygous, but collectively occurring in up to 10% to 15% of affected patients-a markedly higher incidence than in controls. Moreover, leukocytes from some heterozygous patients have significantly shorter telomeres than those of either controls or patients carrying only wild-type hTERC alleles. Recently, we and other researchers have tested 5 of the reported AA-associated hTERC alleles for their ability to support telomerase biologic activity, and found that 4 were at least partially defective.3,17 These results together suggest that hTERC sequence variants may be linked to the pathogenesis of AA and other marrow failure disorders in certain cases. Because only a handful of hTERC variants have been tested, however, the generality of these findings is unknown.
In this study, we systematically examine the functional properties of 10 previously reported DC-, AA-, and MDS-associated variants of hTERC in a cell-based telomerase reconstitution assay. We also report here for the first time 2 new hTERC variants identified in patients with AA or essential thrombocythemia. Our findings indicate that most of these disease-associated hTERC variants have significantly impaired telomerase biologic activity. We show that most of these defects are attributable to alterations in hTERC secondary structure, as compensatory mutations designed to restore the normal base-pairing pattern also restore enzymatic activity. Our data provide support for the hypothesis that mutations in the RNA component of telomerase may cause or predispose to these disorders in a significant subset of patients.
Materials and methods
Cloning of novel hTERC variants
Peripheral blood leukocytes were collected from patients with acquired AA or essential thrombocythemia, each of whom had given informed consent according to protocols approved by the institutional review board of the National Heart, Lung, and Blood Institute. The hTERC gene was amplified from genomic DNA isolated from these samples as described previously.15
Telomere length measurements
Engineering VA13+hTERT stable cell line
DNA encoding the wild-type hTERT gene was cloned into the plasmid pBabe-Puro at the EcoRI and SalI restriction sites. The resulting vectors were transfected transiently into the PA317 packaging cell line, which produces amphotropic murine leukemia virus (MuLV) pseudotyped particles, via the calcium phosphate transfection technique (Gibco-BRL, Carlsbad, CA). Pseudotyped particles containing the hTERT expression cassette were harvested 48 hours after transfection and were used to infect VA13 (ATCC, Manassas, VA), a human lung fibroblast cell line transformed by SV40 large T-Ag, that expresses neither the hTERT nor the hTERC component of the human telomerase complex. Puromycin-resistant colonies were selected by growth in Dulbecco modified Eagle medium supplemented with glucose (4.5 g/L), 10% bovine calf serum, and 2.0 μg/mL puromycin. About 100 drug-resistant colonies of the VA13+hTERT cells were pooled and expanded for use in the subsequent experiments.
In vivo reconstitution of telomerase activity
Wild-type or mutant pcDNA3-hTERC DNAs (1 μg) were transfected into VA13+hTERT cells (at approximately 70% confluency) in 6-well polystyrene dishes using SuperFect transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions. In the cases where 2 different versions of the hTERC gene were expressed simultaneously in the VA13+hTERT cells, the genes were cloned into the pBud-CE 4.1 vectors (Stratagene, La Jolla, CA) to be expressed from either the cellular EF1α promoter or the viral cytomegalovirus (CMV) promoter. Transfection efficiency was monitored by scoring green fluorescent protein expression under confocal microscopy in parallel transfection reactions supplemented with the reporter vector pEGFP (Stratagene). Approximately 48 hours after transfection, cells were scraped from the dish in the presence of 1 mL cold phosphate-buffered saline. Cellular extracts were then prepared in 1 × CHAPS (3-3[(C-cholamidopropyl)di-methylammonio]-1-propane sulfonate) lysis buffer as suggested by the manufacturer (Chemicon International, Temecula, CA). Telomerase activity of the cellular extract from 2 × 104 cells was assayed using the TRAPeze Telomerase Detection Kit following the manufacturer's directions (Chemicon International), except that polymerase chain reaction (PCR) was performed as follows: 95°C for 2 minutes; 25 cycles of 94°C for 10 seconds, 50°C for 30 seconds, 72°C for 30 seconds; and 72°C for 5 minutes. Products were analyzed on a 12% native polyacrylamide gel and examined by phosphor imaging (Molecular Dynamics, Sunnyvale, CA).
Telomerase activity in primary cells
Peripheral blood cells collected from patients or healthy (control) individuals were cultured in RPMI-1640 with l-glutamine and 10% fetal calf serum in the presence of 5 μg/mL phytohemagglutinin and 40 IU/mL interleukin-2 for 4 days at 37°C under 5% CO2. An aliquot of cells was then dually stained with anti-CD19-fluorescein isothiocyanate (FITC) and anti-CD3- phycoerythrin (PE) (BD Biosciences, San Diego, CA) for analysis in an LSRII flow cytometer (BD Biosciences). Protein extract was prepared from 2 × 106 cells, and serial dilutions containing 1.0 μg, 0.5 μg, and 0.1 μg total protein (BioRad, Hercules, CA), respectively, were assayed for telomerase activity in the TRAPeze Kit (Chemicon International).
Northern blotting analysis
Wild-type or mutant pcDNA3-hTERC vectors (6 μg) were transfected into VA13+hTERT cells (at approximately 70% confluency) in 100-mm polystyrene dishes using SuperFect transfection reagent. Approximately 48 hours after transfection, Trizol reagent was used to extract total cellular RNA as suggested by the manufacturer (Invitrogen, Carlsbad, CA). Northern blot analysis was performed essentially as described.20
Immunoprecipitation Northern blotting analysis
FLAG-tagged hTERT protein was expressed in vitro from the pCR3-FLAG-hTERT vector using the TnT quick-coupled transcription-translation system (Promega, Madison, WI) in the presence of 200 ng in vitro-transcribed, gel-purified CR4-CR5 fragment from hTERC (spanning hTERC nucleotides 239 to 332) at 37°C for 2 hours. The resulting telomerase complexes were affinity-enriched on anti-FLAG agarose beads (Sigma, St Louis, MO). To detect hTERT-bound telomerase RNAs, Northern blotting was performed on the enriched telomerase preparations as described in our previous report.3
Results
Our study focused on 12 distinct hTERC variant sequences identified in recent surveys of patients with DC, AA, PNH, or MDS.15,16,21 The variants and their associated clinical and laboratory findings are summarized briefly in Table 1. Only one hTERC variant (G228A) analyzed here was observed in healthy controls as well as in patients with AA (Figure 1 and Table 1). The 11 remaining variants were each detected only in affected individuals or their relatives. The latter group includes 2 novel hTERC variants (A117C and Δ389-390) that we discovered in 2 unrelated patients; the former exhibited typical AA along with idiopathic hepatic cirrhosis, whereas the latter was diagnosed with essential thrombocythemia. As essential thrombocythemia is a clonal disorder, the germ line origin of the hTERC variant was established by concurrent results obtained from buccal mucosa specimens from this patient as well as from his son. All of the patients whose variant hTERC alleles were the subject of this study were heterozygous (ie, they carried one wild-type hTERC gene in addition to the variant).
Clinical and laboratory data from patients with hTERC variants
hTERC sequence variant . | Clinical diagnosis . | Telomere length . | Telomerase activity* . |
|---|---|---|---|
| Core domain | |||
| C116T | AA (severe pancytopenia) | + | - |
| C204G | AA (moderate pancytopenia) | + | - |
| A117C | AA (severe pancytopenia) | + | - |
| G143A | DC | + | - |
| Δ96-97 | DC | + | - |
| CR4-CR5 domain | |||
| G305A | AA (moderate) | ++ | + |
| G322A | MDS | ND | + |
| H/ACA domain | |||
| Δ389-390 | Essential thrombocythemia | ND | - |
| C408G | DC | + | - |
| G450A | AA (severe) | +++ | +++ |
| CR7 domain | |||
| Δ378-451 | DC | ++ | - |
| Hypervariable paired region | |||
| G228A | AA (moderate) or healthy | +++ | +++ |
hTERC sequence variant . | Clinical diagnosis . | Telomere length . | Telomerase activity* . |
|---|---|---|---|
| Core domain | |||
| C116T | AA (severe pancytopenia) | + | - |
| C204G | AA (moderate pancytopenia) | + | - |
| A117C | AA (severe pancytopenia) | + | - |
| G143A | DC | + | - |
| Δ96-97 | DC | + | - |
| CR4-CR5 domain | |||
| G305A | AA (moderate) | ++ | + |
| G322A | MDS | ND | + |
| H/ACA domain | |||
| Δ389-390 | Essential thrombocythemia | ND | - |
| C408G | DC | + | - |
| G450A | AA (severe) | +++ | +++ |
| CR7 domain | |||
| Δ378-451 | DC | ++ | - |
| Hypervariable paired region | |||
| G228A | AA (moderate) or healthy | +++ | +++ |
Sources for the original descriptions, associated clinical findings, and telomere length data are as follows: C116T and C204G22 ; G143A and Δ96-9724 ; A117C and Δ389-390, present study; G305A and G322A23 ; and C408G and Δ378-451.25 Original descriptions, associated clinical findings, and telomere length data for A117C, G305A, and G322A are from the present study. Telomere lengths of peripheral blood lymphocytes are expressed in comparison to those of age-matched healthy individuals assayed simultaneously, as reported in the indicated references: +++, within reference range; ++, 2 kb to 3 kb shorter than the reference range; +, 3 kb to 6 kb shorter than the reference range; ND, not determined.
The telomerase activity of each variant was determined in this study using reconstituted VA13+hTERT cells, and is expressed in comparison to that of wild-type hTERC (+++, 20%-100%; ++, 2%-20%; +, 1%-2%; -, undetectable), based on 2 or 3 independent measurements
Disease-associated mutations in the core domain abolish telomerase activity
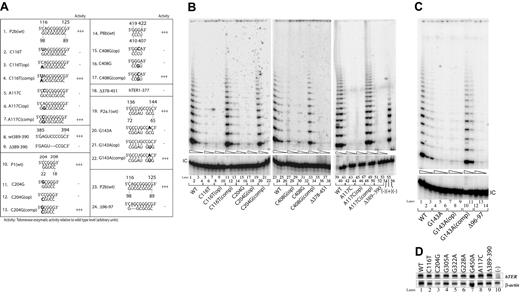
Five of the disease-associated sequence variants we examined affected bases within the core domain of hTERC. Each represented either a point substitution of an individual base or a 2-nucleotide deletion in the presumptive helical regions of the RNA (Figure 1): 3 natural variants (C116T, A117C, and Δ96-97) involved residues in the distal paired portion of the pseudoknot (a region called the P2b helix), whereas the other 2 (C204G and G143A) fell within the nearby P1 and P2a.1 helices, respectively. We reconstituted telomerase function with each of these variant sequences individually in the normally telomerase-negative VA13+hTERT cells, and found that each was severely deficient in reconstituting telomerase function, yielding less than 1% of the catalytic activity achieved by wild-type hTERC (Figure 2B, lanes 5-7, 14-16, 42-44; Figure 2C, lanes 4-6, 12-14).
Disease-associated mutations located in the core domain or the box H/ACA and CR7 domains abolish telomerase activity. (A) Telomerase enzymatic activities as determined in VA13+hTERT cells for naturally occurring hTERC mutations and their derivatives involving the core, box H/ACA, and CR7 domains. When the natural sequence variants are located within a paired region (eg, C116T), the nucleotides that are predicted to base-pair with them are also mutated to the complementary bases (eg, C116T(op)). The compensatory mutations (eg, C116T(comp)) are created in order to restore the helical structures. The sequence changes are indicated in bold. Telomerase activity of each mutant is expressed in comparison to that of the wild-type (wt) (+++, 20%-100%; ++, 2%-20%; +, 1%-2%; −, undetectable), based on 2 or 3 independent determinations. (B, C) Representative TRAP gels showing the relative telomerase enzymatic activities obtained from the representative substitution or deletion mutations and compensatory mutations. Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. Lane 54 shows a negative control composed of wild-type (WT) cell lysate denatured at 95°C for 5 minutes prior to assay. Lane 55 shows PCR products amplified from the non-hTERC control TSR8 DNA template supplied in the TRAP kit. Lane 56 shows cells transfected with the pcDNA3.1 vector control lacking the hTERC coding sequence. “IC” indicates PCR products amplified from an unrelated DNA template, which is included as an internal control for PCR amplification efficiency in each reaction. (D) Northern blot analysis of selected naturally occurring hTERC sequence variants expressed in transfected VA13+hTERT cells (top). Lane 10 shows RNA prepared from cells that were transfected with the pcDNA3.1 vector lacking the hTERC coding sequence. Cellular β-actin mRNA (bottom) was assayed in parallel.
Disease-associated mutations located in the core domain or the box H/ACA and CR7 domains abolish telomerase activity. (A) Telomerase enzymatic activities as determined in VA13+hTERT cells for naturally occurring hTERC mutations and their derivatives involving the core, box H/ACA, and CR7 domains. When the natural sequence variants are located within a paired region (eg, C116T), the nucleotides that are predicted to base-pair with them are also mutated to the complementary bases (eg, C116T(op)). The compensatory mutations (eg, C116T(comp)) are created in order to restore the helical structures. The sequence changes are indicated in bold. Telomerase activity of each mutant is expressed in comparison to that of the wild-type (wt) (+++, 20%-100%; ++, 2%-20%; +, 1%-2%; −, undetectable), based on 2 or 3 independent determinations. (B, C) Representative TRAP gels showing the relative telomerase enzymatic activities obtained from the representative substitution or deletion mutations and compensatory mutations. Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. Lane 54 shows a negative control composed of wild-type (WT) cell lysate denatured at 95°C for 5 minutes prior to assay. Lane 55 shows PCR products amplified from the non-hTERC control TSR8 DNA template supplied in the TRAP kit. Lane 56 shows cells transfected with the pcDNA3.1 vector control lacking the hTERC coding sequence. “IC” indicates PCR products amplified from an unrelated DNA template, which is included as an internal control for PCR amplification efficiency in each reaction. (D) Northern blot analysis of selected naturally occurring hTERC sequence variants expressed in transfected VA13+hTERT cells (top). Lane 10 shows RNA prepared from cells that were transfected with the pcDNA3.1 vector lacking the hTERC coding sequence. Cellular β-actin mRNA (bottom) was assayed in parallel.
As each of the foregoing mutations would be predicted to disrupt one or more base pairs in hTERC (Figure 1), we next created additional mutations designed to test the importance of those base pairs per se (Figure 2A). For each of the 5 natural mutants, we first designed a corresponding mutant (denoted “op”) in which the same putative base pairs were instead abolished by point mutations in the opposite RNA strand. We then created a second group of mutants (denoted “comp”) in which the natural base mutations were accompanied by complementary mutations on the opposite strand that were designed to restore the predicted base-pairing pattern. In each case, we found that mutation of the opposite strand alone abolished telomerase activity to the same degree as the natural mutation (Figure 2B, lanes 8-10, 17-19, 45-47; Figure 2C, lanes 7-9), but that activity was fully restored by compensatory mutations that re-established normal base-pairing (Figure 2B, lanes 11-13, 20-22, 48-50; Figure 2C, lanes 10-11). These results were not attributable to differences in hTERC synthesis, processing, or stability, as Northern blotting verified that each construct produced comparable steady-state levels of hTERC expression in the transfected cells (Figure 2D, lanes 1, 2, and 8). These findings, summarized in Figure 2A, together indicate that the disease-associated hTERC core variants are functionally defective and that their defects result from altering conserved secondary structures in the RNA rather than from changing the identities of specific bases.
Mutations of essential RNA sequences and structures in the CR4-CR5 domain
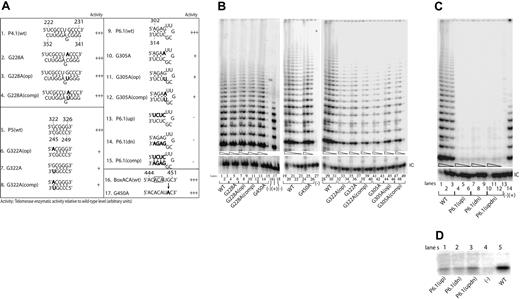
We next examined the properties of 2 separate sequence variants of the conserved CR4-CR5 domain. These variants, G305A and G322A, had been identified in patients with AA and MDS, respectively, and each involved separate regions of this domain whose structure is ambiguous. Although residue G305 was originally predicted to reside in a single-stranded region of hTERC,2 subsequent mutational studies of its murine orthologue (mTERC) have suggested that the corresponding residue is paired,4 forming part of a short helical region denoted P6.1 (Figure 1). The possible existence and function of a P6.1 helix in hTERC has not been explored in detail, however. Similarly, recent studies using chemical and enzymatic probes of RNA structure have suggested that residue G322 may pair with residue C247 as part of an elongated P5 helix in hTERC (Figure 1 and Figure 3A, line 1), but no functional evidence for such interaction has been reported.
Mutations of the hypervariable region or the CR4-CR5 or box H/ACA domains show variable degrees of telomerase activity. (A) Telomerase enzymatic activity in VA13+hTERT cells expressing various hTERC variants involving the hypervariable paired region or the CR4-CR5 or box H/ACA domains. Natural sequence variants and engineered mutations are indicated in bold. The telomerase activity for each variant is expressed in comparison to that of the wild-type (+++, 20%-100%; ++, 2%-20%; +, 1%-2%; −, undetectable) based on 2 or 3 independent determinations. (B, C) Representative gels showing the relative telomerase enzymatic activities obtained from informative variants. Serial fivefold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. Lanes 16 and 27 of panel B and lane 13 of panel C indicate negative controls in which the wild-type cell lysates were denatured at 95°C for 5 minutes before analysis. Lanes 17 of panel B and 14 of panel C show PCR products amplified from the control TSR8 DNA template supplied in the kit. Lanes 18 of panel B and 15 of panel C show cells transfected with the pcDNA3.1 vector lacking the hTERC coding sequence. (D) Northern blotting analysis of affinity-enriched telomerase complexes assembled in vitro using wild-type P6.1 stem (spanning TERC nucleotides 239 to 332) or its mutants. Telomerase RNA-protein complexes were first assembled in the rabbit reticulocyte lysates. The negative control (lane 4) was a lysate that received no hTERT expression vector.
Mutations of the hypervariable region or the CR4-CR5 or box H/ACA domains show variable degrees of telomerase activity. (A) Telomerase enzymatic activity in VA13+hTERT cells expressing various hTERC variants involving the hypervariable paired region or the CR4-CR5 or box H/ACA domains. Natural sequence variants and engineered mutations are indicated in bold. The telomerase activity for each variant is expressed in comparison to that of the wild-type (+++, 20%-100%; ++, 2%-20%; +, 1%-2%; −, undetectable) based on 2 or 3 independent determinations. (B, C) Representative gels showing the relative telomerase enzymatic activities obtained from informative variants. Serial fivefold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. Lanes 16 and 27 of panel B and lane 13 of panel C indicate negative controls in which the wild-type cell lysates were denatured at 95°C for 5 minutes before analysis. Lanes 17 of panel B and 14 of panel C show PCR products amplified from the control TSR8 DNA template supplied in the kit. Lanes 18 of panel B and 15 of panel C show cells transfected with the pcDNA3.1 vector lacking the hTERC coding sequence. (D) Northern blotting analysis of affinity-enriched telomerase complexes assembled in vitro using wild-type P6.1 stem (spanning TERC nucleotides 239 to 332) or its mutants. Telomerase RNA-protein complexes were first assembled in the rabbit reticulocyte lysates. The negative control (lane 4) was a lysate that received no hTERT expression vector.
Testing of the disease-associated G305A and G322A variants indicated that each was expressed at normal concentrations within transfected cells (Figure 2D, lanes 4 and 5) but was nevertheless functionally defective, supporting no more than 1% of wild-type hTERC activity (Figure 3B, lanes 32-37, 41-46). Compensatory mutations failed to restore activity to an appreciable degree in either instance (Figure 3B, lanes 38-40, 47-49). Because these findings appeared, in the case of G305A, to be at odds with the reported properties of the P6.1 helix in mTER,4 we extended our analysis by testing additional hTERC mutants designed either to disrupt all 4 base pairs of the putative P6.1 orthologue simultaneously (mutants P6.1[up] and P6.1[dn]) or to replace them with alternative paired bases (mutant P6.1[comp]). Telomerase enzymatic function was completely abrogated by any of these mutations (Figure 3C, lanes 4-12), a finding that provides no information regarding the existence of a P6.1 structural counterpart in humans, but suggests that the specific base sequence in this region of the CR4-CR5 domain is critical for hTERC function.
In mTERC, integrity of the P6.1 stem structure is required for interaction with the catalytic mTERT protein.4 Because the requirements for human hTERT protein binding to hTERC have not been precisely mapped, we therefore asked whether mutations of the residues corresponding to P6.1 in hTERC would affect this interaction in cells. To that end, we compared telomerase RNP complexes assembled in cells expressing hTERC fragments (spanning nucleotides 239 to 332) that contained either the wild-type CR4-CR5 domain or its mutants. Telomerase RNP complexes were reconstituted in vitro using the rabbit reticulocyte lysates to express a FLAG-tagged hTERT protein in the presence of the synthetic RNA molecule containing various sequences of the CR4-CR5 domain (“Materials and methods”). Telomerase RNP complexes were then immunoprecipitated from the lysates using an anti-FLAG antibody against the FLAG-tagged hTERT protein, and were then probed for hTERC by Northern blotting. As illustrated in Figure 3D, all 3 of the engineered CR4-CR5 mutants we tested showed substantially impaired binding to hTERT protein, although the single mutant that retained a P6.1-like pattern of base-pairing bound to a modestly greater extent than those that did not. The data on the P6.1(up) and P6.1(dn) mutants are consistent with those published recently by Moriarty and colleagues.26 Taken together, these findings suggest that specific sequences in the CR4-CR5 region of hTERC are needed for optimal hTERT protein binding, though it remains unclear whether a P6.1-like helical structure contributes to this binding as it does in the murine telomerase.
Divergent phenotypes of sequence variants in the box H/ACA domain
We tested 2 different hTERC sequence variants that map to the box H/ACA domain (Figure 1). This domain is suspected to interact with the cellular protein dyskerin and to help direct the nucleolytic processing of hTERC from a larger RNA precursor, and it is also required for subsequent stability and localization of hTERC within cells.12 When tested in our assay, one of these sequence variants (Δ389-390), which we recently identified in a patient with essential thrombocythemia, completely abolished telomerase activity, though it had no apparent effect on stability of the RNA (Figure 2A, lines 8-9; Figure 2B, lanes 51-53). Surprisingly, by contrast, the second variant (G450A), which lies near the nucleolytic processing site and alters the penultimate base in mature hTERC, was indistinguishable from wild-type hTERC in size, steady-state concentration, or catalytic function (Figure 1; Figure 2D, lane 7; Figure 3A, lines 16 and 17; Figure 3B, lanes 13-15, and 24-26). Despite its association with severe AA in a single patient, the G450A variant thus appears to represent a fully functional variant of hTERC. Indeed, this patient possessed normal telomere length and showed a sustained remission of hematologic disease when treated with immunosuppressive therapy. As controls for our assays, we also tested 2 previously characterized natural hTERC mutations (C408G and Δ378-451) that affect the conserved box H/ACA and CR7 domains, respectively (Figure 1).13,27 Consistent with previous reports,17,27 we found that these 2 mutations each substantially reduced the levels of telomerase enzymatic activity (Figure 2A, lines 14-18; Figure 2B, lanes 30-32, and 36-38). In addition to the naturally occurring C408G variant, we introduced mutations targeting the corresponding base in the upper strand of this predicted P8b stem (G408A(op)) and the compensatory mutant C408G(comp) (Figure 2A, lines 15 and 17). As shown in Figure 3B, while the C408G(op) mutation abolished telomerase enzymatic function (lanes 27-29), the compensatory mutation successfully rescued the activity (lanes 33-35), suggesting that this short stem structure of the CR7 domain forms and is required for optimal telomerase function. This is consistent with recent nuclear magnetic resonance data supporting the formation of this small stem structure using synthetic hTERC RNA fragment.28
A functionally intact sequence variant of the hypervariable paired region
We examined one additional natural variant hTERC with a point substitution (G228A) in the 5′ strand of the hypervariable paired region (Figure 1), a region whose base sequence has diverged relatively widely among vertebrate species though its base-pairing has tended to be preserved. As illustrated in Figure 3B (lanes 4-6), we found that the G228A variant retained fully wild-type catalytic activity, as did the related G228A(op) single substitution or G228A(comp) compensatory mutation (Figure 3A, lines 1-4; Figure 3B, lanes 7-12). Like G450A, the naturally occurring G228A variant is a polymorphism of hTERC with no functional consequences in telomerase activity. In accord with that interpretation, this latter polymorphism has not only been identified in a patient with AA, but also in at least one healthy individual in the control cohort of an earlier study15,29 ; its prevalence in the population at large remains to be determined.
A lack of dominant-negative effect in cells simultaneously expressing both the wild-type hTERC gene and the individual sequence variants
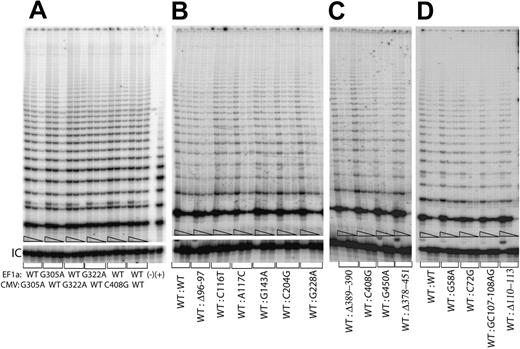
Since previous studies have shown that human telomerase enzyme functions as a dimeric or multimeric complex, consisting of at least 2 hTERC RNAs and 2 hTERT proteins,6,26,30-33 it is possible that some of the disease-associated hTERC sequence variants may act as dominant-negatives to prevent the proper formation of a functional telomerase RNP. To test this possibility, we forced the expression of 2 different versions of the hTERC gene simultaneously in the same cell by expressing both genes from the same vector (Figure 4). Vectors that expressed either 2 wild-type copies of hTERC or a wild-type and the mutated copy of the gene were transfected into the VA13+hTERC cells, and cell lysates were prepared for the TRAP assay as outlined above. No major differences were observed in samples that expressed the individual mutated hTERC variants together with the wild-type copy as compared with the samples that expressed the wild-type hTERC copies (Figure 4), regardless of the promoters from which the genes were being expressed (Figure 4A). Similar observations were made for the 4 different disease-related hTERC variants described in our previous study (Figures 4D and 1). Taken together, these data suggest that the disease-associated hTERC sequence variants tested thus far do not function as dominant-negatives in human cells.
Telomerase enzymatic activity in VA13+hTERT cells simultaneously expressing various hTERC variants and the wild-type copy from a single pBud-CE 4.1 vector series. (A) While the wild-type hTERC copies are expressed from both the cellular EF1α and the viral CVM promoters, the mutated hTERC genes (ie, either the G305A or the G322A) are alternatively expressed from either the EF1α or the CMV promoter with respect to the wild-type hTERC copy. Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. (−) Telomerase function of an aliquot of the sample that expressed both wild-type copies was inactivated by denaturing the sample at 95°C for 3 minutes prior to assaying for telomerase enzymatic activity. (+) indicates PCR products amplified from the control TSR8 template supplied in the kit. Panels B, C, and D show telomerase activities of the wild-type hTERC gene expressed from the EF1α promoter in the same lysates that contained the individual hTERC variants expressed from the CMV promoter. “IC” indicates PCR products amplified from an internal control DNA template, which is used as internal control for PCR amplification efficiency in each reaction.
Telomerase enzymatic activity in VA13+hTERT cells simultaneously expressing various hTERC variants and the wild-type copy from a single pBud-CE 4.1 vector series. (A) While the wild-type hTERC copies are expressed from both the cellular EF1α and the viral CVM promoters, the mutated hTERC genes (ie, either the G305A or the G322A) are alternatively expressed from either the EF1α or the CMV promoter with respect to the wild-type hTERC copy. Serial 5-fold dilutions of the transfected cell lysates (indicated by triangles) were assayed for each sample to ensure linearity of the assay. (−) Telomerase function of an aliquot of the sample that expressed both wild-type copies was inactivated by denaturing the sample at 95°C for 3 minutes prior to assaying for telomerase enzymatic activity. (+) indicates PCR products amplified from the control TSR8 template supplied in the kit. Panels B, C, and D show telomerase activities of the wild-type hTERC gene expressed from the EF1α promoter in the same lysates that contained the individual hTERC variants expressed from the CMV promoter. “IC” indicates PCR products amplified from an internal control DNA template, which is used as internal control for PCR amplification efficiency in each reaction.
Telomerase enzymatic function of primary cells collected from patients
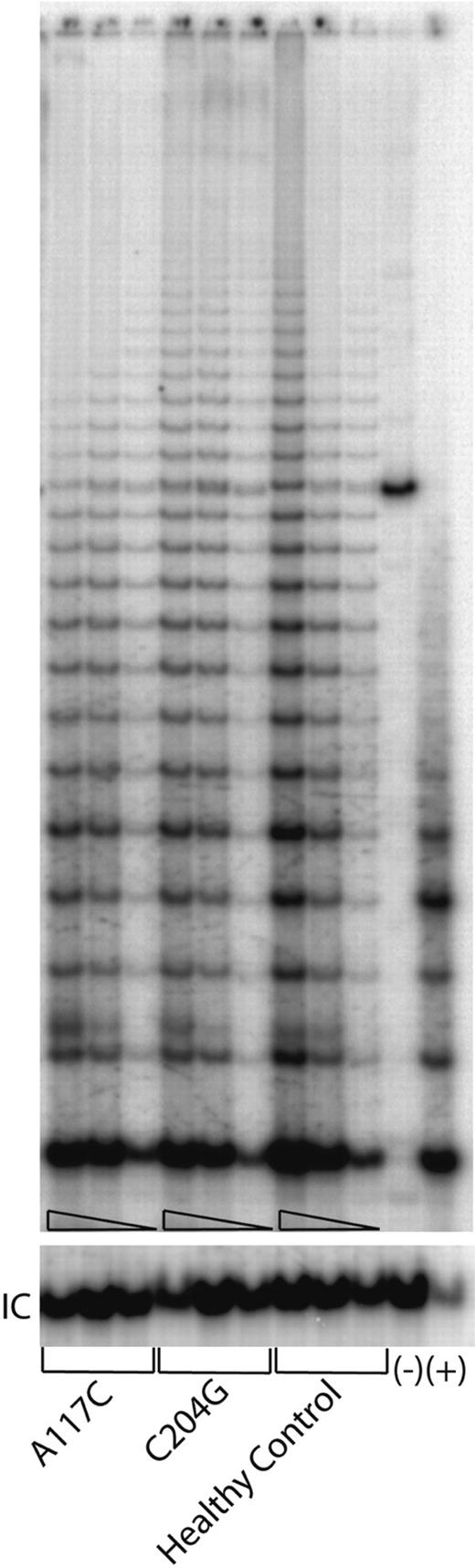
To determine whether haploinsufficiency can also be observed directly in primary cells, we analyzed the effect of the hTERC mutations in the T lymphocytes collected from either a healthy individual or the patients with AA who carry either the A117C or the C204G that were available to us. By using an equal amount of the protein extracts prepared from the primary cells, which have been expanded in tissue cultures, we compared the ability of the extracts to add telomeric repeats into a DNA substrate using the TRAPeze kit as described above. Compared with the sample of a healthy individual, both samples collected from the patients with AA showed moderately reduced telomerase enzymatic function (Figure 5), which is compatible with what might be expected in cells with one wild-type and one inactive hTERC allele. These data confirm the observations that were made with the transient reconstitution of the telomerase RNPs in the VA13+hTERT cells for the corresponding hTERC sequence alterations (Figure 4), further emphasizing the haploinsufficiency effect of the mutations and the validity of our in vivo reconstitution study.
Telomerase enzymatic activities expressed in primary blood cells from the representative patients and healthy individuals. Serial dilutions containing 1.0 μg, 0.5 μg, and 0.1 μg total protein, respectively, from cell lysates for each sample were tested (triangles). Lanes 1 to 9 are from a patient who carries the A117C, or C204, or G305A mutations, respectively. Lanes 10 to 12 are from an unrelated healthy control individual. Lane 13 shows a negative control in which 1 μg lysate from the healthy control sample was denatured at 95°C for 5 minutes prior to assay. (+) indicates PCR products amplified from a control TRS8 template provided in the kit (lane 14). “IC” indicates PCR products amplified from an internal control DNA template, which is used as internal control for PCR amplification efficiency in each reaction.
Telomerase enzymatic activities expressed in primary blood cells from the representative patients and healthy individuals. Serial dilutions containing 1.0 μg, 0.5 μg, and 0.1 μg total protein, respectively, from cell lysates for each sample were tested (triangles). Lanes 1 to 9 are from a patient who carries the A117C, or C204, or G305A mutations, respectively. Lanes 10 to 12 are from an unrelated healthy control individual. Lane 13 shows a negative control in which 1 μg lysate from the healthy control sample was denatured at 95°C for 5 minutes prior to assay. (+) indicates PCR products amplified from a control TRS8 template provided in the kit (lane 14). “IC” indicates PCR products amplified from an internal control DNA template, which is used as internal control for PCR amplification efficiency in each reaction.
Discussion
This study provides functional evidence that variant telomerase RNA alleles found in some patients with bone marrow failure are unable to support normal levels of telomerase enzymatic activity. Together with earlier reports,3,13,17 our data are consistent with the hypothesis that inherited defects in telomerase function and telomere maintenance contribute to the pathogenesis of such disorders in a subset of patients, even when no familial tendency is apparent. The levels of telomerase enzymatic activity supported by individual disease-associated hTERC variants in the cell-based assay we used correlated very well with independent measurements of telomere lengths in lymphocytes from patients who carried those variant alleles (Table 1). In particular, telomeres in patients harboring the C116T, A117C, C204G, or G305A variants have previously been reported to be markedly shorter (averaging 3.7-4.6 kb) than those of healthy age-matched individuals (8-12 kb),15 and none of these 4 variants proved able to support detectable telomerase activity in our cell-based vivo telomerase reconstitution assay. Primary cells harvested from representative patients who were heterozygous for either the A117C or the C204G allele, moreover, showed correspondingly lower levels of telomerase activity (Figures 2B, 3B, and 4). Conversely, the normal telomerase activities of the G228A and G450A variants are consistent with the essentially wild-type average telomere lengths observed in individuals heterozygous for these alleles, some of whom are hematologically normal.15 These correlations help support the validity of the assay we employed in this study, and strengthen the view that the telomere shortening seen in some patients with bone marrow failure may result directly from telomerase dysfunction.
The naturally occurring mutations observed in this study are distributed throughout the hTERC molecule. Their effects confirm and extend earlier findings from site-directed mutagenesis, indicating that all 4 domains of hTERC contribute to function and that the intricately base-paired structure of this RNA is critical for its biologic activity.3,5,34,35 Indeed, it is striking that a high proportion of the seemingly minor point mutations can severely compromise telomerase function by perturbing RNA structure. These include 3 adjacent mutations in the P2b stem, a single point substitution in the P2a.1 stem, and another point substitution located within the P1 stem, all of which are involved in the proper folding of the pseudoknot in the hTERC core domain. Our finding that compensatory mutations can restore activity to most of these hTERC pseudoknot variants, coupled with evidence from structural analyses using NMR spectroscopy,28,34 highlights the importance of the normal base-pairing pattern in this region of the RNA. It remains to be determined whether this reflects a role of the pseudoknot in global folding of the hTERC monomer, in dimerization of hTERC, or in some other aspect of telomerase holoenzyme assembly and function. While this manuscript was in preparation, a similar study appeared,27 which provided a comprehensive functional analysis of 4 disease-associated mutations (ie, C408G, G228A, Δ96-97, Δ378-451) that overlapped with some of those analyzed in our current work. This study also suggests that optimal telomerase function depends on the proper folding of the hTERC RNA molecule.27
Our finding that one of 2 variants in the box H/ACA domain was functionally defective is in full accord with results from earlier studies that used site-directed mutagenesis,12,17,27 though we detected no accompanying abnormalities in the RNA processing, localization, or stability characteristics that have been associated with this domain. The defect of the G305A mutant, which maps within a putative P6.1 stem-loop in the CR4-CR5 domain, is more difficult to interpret since the conformation of this region is less well understood than that of its murine counterpart. Although our efforts to restore function of this mutant through compensatory mutations (Figure 3A, lines 9-15) were unsuccessful, this may simply indicate that the local conformation in hTERC differs from that of mTERC, as suggested by a recent report of a possible long-range interaction that may involve some base-pairing interaction in this region.36 Nevertheless, NMR structural analysis of a short RNA fragment of the P6.1 stem suggests that it folds correctly into a helical structure.37 Our results suggest, moreover, that mutations in this region may inactivate hTERC by compromising its interaction with the hTERT protein (Figure 3D), a phenotype that is consistent with a recent observation made by Moriarty and colleagues.26
All of the patients whose variant hTERC alleles were the subject of this study were heterozygous; indeed, to our knowledge no person with inactivating mutations in both hTERC loci has yet been described. This may be relevant to the mechanism and temporal course of disease in these patients. The consequences of telomere shortening have been demonstrated in mice, where biallelic inactivation of the mTERC gene leads to a gradual decline in average telomere length over sequential generations, culminating eventually in a loss of hematopoietic activity and in other somatic disorders.38,39 In humans, whose telomeres are normally much shorter than those of mice, a similar process might occur when only a single hTERC allele is lost, leading to progressive losses of telomere length and stem-cell replicative pools over the lifespan of an affected individual. This would place some proportion of hTERC heterozygotes at risk of exhausting the regenerative capacity of their marrow stem cells during adulthood, especially when challenged by environmental insults.16,21 If the clonal lifespans of those stem cells were limited by telomere shortening, even minor stochastic variations in the initial numbers and replicative histories of marrow stem cells could dictate which heterozygous individuals develop symptomatic marrow failure and the age at which it occurs. Additional variability in disease phenotype could be imparted by the specific hTERC variant that an individual inherits, as some variants we tested appear able to support intermediate levels of telomerase activity whereas others are completely inactive (Table 1). Finally, because telomerase deficiency would also affect chromosomes transmitted through the germ line, the frequency and severity of its manifestations might also be expected to increase over successive generations.21 Although no such trend has yet been recognized in AA, MDS, or PNH, which are generally regarded as acquired disorders, the familial forms of DC do typically occur with increasing severity and earlier age-of-onset as they are transmitted through affected kindreds.11,21 By tipping the normal balance between telomere shortening and elongation, a partial loss of telomerase activity in hTERC heterozygotes could thus account for the variable penetrance and sporadic nature of AA and other marrow-failure disorders, while masking their underlying hereditary predisposition.
Prepublished online as Blood First Edition Paper, November 18, 2004; DOI 10.1182/blood-2004-09-3659.
Supported in part by the Leukemia and Lymphoma Society of America, the American Cancer Society-Emory's Winship Cancer Institutional Grant (H.L.) and by National Institutes of Health grants AI36636 and AI40317 (T.G.P.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Xiaoying Yang is acknowledged for excellent technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal