Abstract
Diffuse large B-cell lymphoma (DLBCL) is a heterogeneous disease with recognized variability in clinical outcome, genetic features, and cells of origin. To date, transcriptional profiling has been used to highlight similarities between DLBCL tumor cells and normal B-cell subtypes and associate genes and pathways with unfavorable outcome. To identify robust and highly reproducible DL-BCL subtypes with comprehensive transcriptional signatures, we used a large series of newly diagnosed DLBCLs, whole genome arrays, and multiple clustering methods. Tumors were also analyzed for known common genetic abnormalities in DLBCL. There were 3 discrete subsets of DLBCL—“oxidative phosphorylation,” “B-cell receptor/proliferation,” and “host response” (HR)—identified characterized using gene set enrichment analysis and confirmed in an independent series. HR tumors had increased expression of T/natural killer cell receptor and activation pathway components, complement cascade members, macrophage/dendritic cell markers, and inflammatory mediators. HR DLB-CLs also contained significantly higher numbers of morphologically distinct CD2+/CD3+ tumor-infiltrating lymphocytes and interdigitating S100+/gamma interferon-induced lysosomal transferase–positive (GILT+) CD1a–/CD123– dendritic cells. The HR cluster shared features of histologically defined T-cell/histiocyte-rich B-cell lymphoma, including fewer genetic abnormalities, younger age at presentation, and frequent splenic and bone marrow involvement. These studies identify tumor microenvironment and host inflammatory response as defining features in DLBCL and suggest rational treatment targets in specific DLBCL subsets.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid malignancy in adults, comprising almost 40% of all lymphoid tumors. Although a subset of DLBCL patients can be cured with standard adriamycin-containing combination chemotherapy, the majority die of their disease. Robust clinical prognostic models such as the International Prognostic Index can be used to identify patients who are less likely to be cured with standard therapy.1 However, such models do not provide specific insights regarding tumor cell biology, novel therapeutic targets, or more effective treatment strategies. Furthermore, recent studies suggest that subsets of DLBCL may differ with respect to normal cell of origin and genetic bases for transformation as well as clinical outcome.
DLBCLs are thought to arise from normal antigen-exposed B cells that have migrated to or through germinal centers (GCs) in secondary lymphoid organs.2 Like normal GC B cells and their descendents, DLBCLs have somatic mutations of immunoglobulin receptor variable (v) region genes.2 These tumors also exhibit genetic changes that may be related to normal GC functions. For example, normal GC B cells undergo vigorous clonal expansion and editing of the immunoglobulin receptor via processes that require DNA strand breaks. In small subsets of DLBCL, several translocations into the immunoglobulin locus have been described, including t(8;14), t(3;14), and t(14;18).3 A subset of DLBCLs also exhibits aberrant somatic hypermutation of genes that are not targeted by this editing process in normal GC B cells.4 However, a significant percentage of DLBCLs lack known genetic abnormalities.
Given the documented clinical and genetic heterogeneity of DLBCLs, it would be useful to have comprehensive molecular signatures of tumors that share similar features. In addition to highlighting potential pathogenetic mechanisms, such signatures might identify promising subtype-specific targets and pathways for therapeutic intervention. With the advent of gene expression profiling, it is now possible to obtain signatures of DLBCL subtypes.
To date, transcriptional profiling of DLBCLs has been used to highlight similarities between subsets of tumors and normal B cells and to identify features associated with unfavorable responses to empiric combination chemotherapy. For example, a series of molecular models have been described that relate DLBCL subsets to normal GC B cells, in vitro–activated peripheral blood B cells, or an unspecified, third group.5,6 In these studies, DLBCLs with features common to normal GC B cells responded more favorably to standard empiric combination chemotherapy. In additional profiling studies, the molecular signatures of DLBCLs with different responses to standard chemotherapy were examined.7 Of the pathways associated with poor responses to current regimens, 2 have already been credentialed and targeted for possible therapeutic intervention (Su et al8 ; Smith et al63 ).
However, DLBCLs are not a homogeneous group of tumors that differ only with respect to outcome or possible cell of origin. Given the genetic heterogeneity in DLBCL, there are likely to be subsets of tumors with different pathogenetic mechanisms and possible treatment targets. With a more extensive series of primary tumors and arrays with increasing genome coverage, it is now possible to identify robust subsets of large-cell lymphoma with unique, comprehensive transcriptional profiles. For example, we and others recently found that the molecular signature of primary mediastinal large B-cell lymphoma (MLBCL) differs from that of DLBCL and shares important features with that of a clinically similar disorder, classical Hodgkin lymphoma (nodular sclerosis subtype).9,10 In the current study, we address the more difficult question of unrecognized biologic heterogeneity within DLBCLs, using multiple clustering methods and comprehensive genetic analyses to identify discrete subsets of tumors.
Patients, materials, and methods
Case selection and histologic classification
Tumor specimens and retrospective clinical data from 176 DLBCL patients were analyzed according to the Dana Farber/Partners Cancer Care institution review board–approved protocol. All tumor specimens were nodal biopsies from newly diagnosed, previously untreated patients. The histopathology and immunophenotype of each DLBCL was reviewed by expert hematopathologists to confirm the diagnosis. Clinical variables included in the full International Prognostic Index (IPI) (age, stage, number of extranodal sites, lactate dehydrogenase, and performance status) were obtained; an IPI score was available for 144 patients (supplementary information on the Blood website; see the Supplemental Document link at the top of the online article). Overall survival (OS) and freedom from progression (FFP) were determined by the Kaplan-Meier method in 130 study patients who received full-dose CHOP-based (cyclophosphamide, adriamycin, vincristine, prednisone) therapy (eg, 3-4 cycles + radiation therapy for localized disease or minimum of 6 cycles for advanced disease) and had long-term clinical follow-up or disease progression during or following induction therapy.
Target cRNAs of oligonucleotide microarrays
Target cRNAs were prepared as previously described.7 For 17 randomly selected tumors, 2 separate aliquots of RNA were used for target preparation and analysis. Samples were hybridized to Affymetrix U133A and U133B oligonucleotide microarrays (Affymetrix, Santa Clara, CA) that include probe sets from approximately 33 000 genes. Arrays were subsequently developed and scanned as previously described (Shipp et al7 and Supplemental Document).
Gene expression analysis
A statistical analysis of the duplicate samples was used to identify genes with high reproducibility within duplicates and high variation across patient tumors (Supplemental Document). Genes were ranked using a robust F statistic, and the top 5% (2118 genes) were included in the final gene set. Similar analyses were performed using the top 10% of ranked genes (Supplemental Document).
Unsupervised analysis by consensus clustering
In the analysis, 3 unsupervised clustering algorithms were used: hierarchical clustering (HC),11 self-organizing maps (SOMs),12 and model-based probabilistic clustering (PC)13 (Supplemental Document). The stability of the identified clusters (ie, sensitivity of the cluster boundaries to sampling variability) was assessed using consensus clustering.14 With this method, perturbations of the original dataset are simulated by resampling techniques. The clustering algorithm of choice is applied to each of the perturbed datasets, and the agreement, or consensus, among multiple runs is assessed and summarized in a consensus matrix (Supplemental Document).
Data-set perturbations were obtained by randomly selecting 80% of the samples (141/176 tumors) at each iteration. There were 200 subsampling iterations performed for each clustering algorithm (HC, SOM, and PC). Consensus matrices were built and evaluated for structures including 2 to 9 clusters (Supplemental Document). Confusion matrices were used to measure the agreement between clusters produced by different algorithms and determine the number of samples assigned to similar clusters by any 2 algorithms. A meta-consensus was used to identify the tumors that were similarly assigned by all 3 clustering algorithms (Supplemental Document).
Gene expression differential analysis
From the top 5% (2118 genes) pool, genes associated with each of the DLBCL clusters were identified using the binary distinction “cluster X vs NOT cluster X.” Genes were ranked according to the signal-to-noise ratio (SNR) (Supplemental Document).
Gene set enrichment analysis (GSEA)
GSEA was performed as previously described15 using a total of 281 gene sets from 4 independent sources: (1) Biocarta, an internet resource (Biocarta, San Diego, CA) that includes 169 biologic pathways involved in adhesion, apoptosis, cell activation, cell cycle regulation, cell signaling, cytokines/chemokines, developmental biology, hematopoiesis, immunology, metabolism, and neuroscience; (2) GenMAPP (Gene MicroArray Pathway Profiler), a set of web-accessible pathways (www.GenMAPP.org) and gene families including 45 gene sets involved in metabolic and cell signaling processes; (3) 64 manually curated pathways involved in mitochondrial function and metabolism and additional gene sets that are coregulated in normal murine tissues15 (Supplemental Document); and (4) 3 recently described coregulated gene sets in DLBCL.5
Enrichment was assessed by: (1) ranking the 2118 genes in the top 5% pool with respect to the phenotype “cluster X versus not cluster X”; (2) locating the represented members of a given gene set within the ranked 2118 genes; (3) measuring the proximity of the gene set to the overexpressed end of the ranked list with a Kolmogorov-Smirnoff (KS) score (with a higher score corresponding to a higher proximity); and (4) comparing the observed KS score to the distribution of 1000 permuted KS scores for all gene sets (Supplemental Document). A P < .005, corrected for multiple hypothesis testing (MHT-p), was used to identify highly significant associations between specific gene sets and DLBCL clusters.
Fluorescence in-situ hybridization (FISH)
Air-dried touch preparations were prepared from fresh frozen tumor specimens. Interphase nuclei were hybridized to commercially available probes flanking or spanning the IGH, BCL2, and BCL6 loci: LSI IGH/BCL2 Dual Color, Dual Fusion Translocation Probe for detection of t(14;18) and LSI BCL6 Dual Color, Break Apart Rearrangement Probe for detection of any rearrangement involving 3q24 (t(3;); Vysis, Downers Grove, IL). Translocations were detected by fluorescence microscopy after nuclear counterstaining with DAPI (4,6 diamidino-2-phenylindole).
Morphologic analysis of tumor-infiltrating lymphocytes (TILs)
All study DLBCLs with available hematoxylin and eosin (H&E)–stained diagnostic specimens (119 tumors) were independently assessed for the presence of TILs by an expert morphologist (M.M.) who had no previous information regarding the DLBCL transcriptional profiles. For the majority of tumors, anti-CD2 stained specimens were also available for review. Tumors were initially scanned at high power (× 640) to identify morphologically normal, CD2+ lymphocytes with round or oval nuclei and delicately dispersed chromatin; such lymphocytes were scored only when they directly infiltrated the tumor (TILs).16 There were 20 to 30 representative fields of the tumor independently scored for TILs at × 400 and an average TILs/× 400 score was obtained. DLBCLs were classified as having either less than or more than 20 TILs/× 400 field.
Immunohistochemistry (IHC)
There were 2 representative 0.6-mm cores obtained from diagnostic areas of available paraffin-embedded, formalin- or B5-fixed DLBCLs (80 tumors) and inserted into a tissue array. Tissue array sections were analyzed using: 1) mouse monoclonals anti-CD2 (LFA-2; Novocastra Laboratories, Newcastle upon Tyne, United Kingdom), anti-CD123 (Bioscience, San Diego, CA), and anti-CD1a (Dako, Carpintera, CA); 2) rabbit polyclonals anti-CD3 and anti-S100 (Dako) and anti–gamma interferon-induced lysosomal transferase (GILT, Gift from Peter Cresswell, Yale University School of Medicine, New Haven, CT17 ) (Supplemental Document); and 3) horseradish peroxidase–conjugated secondary antibodies (antimouse or antirabbit, Envision detection kit; Dako). Slides were developed with diaminobenzidine (DAB; Dako), counterstained with harris hematoxylin, and analyzed in blinded fashion by 2 expert hematopathologists, without information regarding cluster designations.
The numbers of CD2+ and CD3+ cells/core were separately recorded for duplicate samples and represented in 5 categories: (1) fewer than 50 cells/core; (2) 50 to 150 cells/core; (3) 150 to 250 cells/core; (4) 250 to 500 cells/core; (5) more than 500 cells/core. Separate analyses of GILT-stained dendritic cells and tumor cells were performed. The number of GILT+ dendritic cells/core was assessed in duplicate samples and represented in 3 categories: (1) 0 to 25 cells/core; (2) 25 to 100 cells/core; and (3) more than 100 cells/core. The number of S100+ dendritic cells/core was assessed in duplicate samples and represented in 4 categories: (1) 0 to 25 cells; (2) 25 to 50 cells; (3) 50 to 100 cells; and (4) more than 100 cells.
Slides were viewed on an Olympus BX41 microscope with an objective lens of 40 × 10.75 Olympus UPlan FL (Olympus, Melville, NY). The pictures were taken using Olympus QColor3 and analyzed with acquisition software QCapture v2.60 (QImaging, Burnaby, BC, Canada) and Adobe Photoshop 6.0 (Adobe, San Jose, CA).
Cluster validation
An independent group of 221 newly diagnosed DLBCLs with available cDNA microarray (“lymphochip”) profiles5 was used for cluster validation. This dataset represented the originally described 240 tumors5 following removal of 19 subsequently identified primary MLBCLs (A. Rosenwald and L. Staudt, National Cancer Institute, written communication, June 2004). Of the top 5% (2118) of genes, 703 were also represented on the lymphochip platform. These overlapping lymphochip probes were used in HC, SOM, PC, and metaconsensus to identify the dominant structure in the independent DLBCL dataset (Supplemental Document).
The level of agreement between the consensus clusters in our dataset and the independent series was determined by comparing the gene markers for each of the respective clusters. Cluster markers were defined as the set of genes with the highest SNR for the corresponding one-versus-all distinction (Supplemental Document). The overlap between respective pairs was represented in a 2-dimensional contingency table and assessed with a Fisher exact test. Similar analyses were also performed using the entire set of genes represented on the lymphochip (7K+) or the top 50% of genes selected with a median absolute deviation (MAD) filter (Supplemental Document).
Cell-of-origin signature
DLBCLs from our dataset were sorted according to the most recent cell-of-origin (COO) signature6 (germinal center B cell [GCB], activated B cell [ABC], and other [not otherwise specified]), using linear predictive scores (LPS) and the 23 (of 27) COO probes represented on the oligonucleotide arrays (Supplemental Document). Confusion matrices were used to measure the agreement between the LPS-defined COO signatures and our meta-consensus defined comprehensive clusters (Supplemental Document).
Results
Identification of DLBCL consensus clusters
To identify biologically meaningful subsets of DLBCL with similar transcription profiles, we used a large series of tumors from highly representative, newly diagnosed patients (Supplemental Document). We were interested in DLBCL subsets that were sufficiently robust to be captured by multiple methods. For this reason, we used 3 different clustering algorithms (hierarchical clustering [HC], self-organizing maps [SOMs], and probabilistic clustering [PC]) and the top 5% of genes with the highest reproducibility across duplicate samples and largest variation across patient tumors. In addition, we used a resampling-based method (consensus clustering) that automatically selects the most stable numbers of clusters with each algorithm.
With all 3 clustering algorithms, the most robust substructure included 3 discrete clusters (Figure 1A, left panel). There was a high level of agreement between clusters produced by the individual algorithms, with more than 84% of DLBCLs assigned to the same clusters by any 2 algorithms (Figure 1A, right panel). A meta-consensus confirmed that 141 of the 176 tumors were assigned to the same clusters by all 3 methods (Figure 1B). We predicted the cluster membership of the remaining 35 tumors using a naive-Bayes model trained on the 141 DLBCLs with concordant cluster labels (Supplemental Document). Similar results were obtained when the clustering analysis was performed with the top 10%, rather than the top 5%, of genes, indicating that the results were not dependent upon the initial gene selection. The top 50 genes associated with each DLBCL group are visually represented in Figure 1C.
Identification of consensus clusters. (A) Left panels: Consensus matrices produced by hierarchical clustering (HC, K = 3), self-organizing maps (SOM, K = 3), and probabilistic clustering (PC). Right panels: Comparisons of the cluster assignments of the different algorithms (PC vs HC, HC vs SOM, and PC vs SOM). More than 84% of DLBCLs were assigned to the same clusters by any 2 algorithms. (B) Left panel: Consensus matrix comparing the assignments made by all 3 clustering algorithms (Meta Consensus [PC vs HC] vs [PC vs SOM]). Right panel: Comparisons of the meta-consensus cluster assignments. Of the 176 tumors, 141 were assigned to the same clusters by all 3 algorithms. (C) Expression profiles of the 3 DLBCL clusters. The top 50 genes associated with each DLBCL cluster are shown. Each column is a sample, and each row is a gene. Color scale at bottom indicates relative expression and standard deviations from the mean. Red indicates high-level expression; blue, low-level expression.
Identification of consensus clusters. (A) Left panels: Consensus matrices produced by hierarchical clustering (HC, K = 3), self-organizing maps (SOM, K = 3), and probabilistic clustering (PC). Right panels: Comparisons of the cluster assignments of the different algorithms (PC vs HC, HC vs SOM, and PC vs SOM). More than 84% of DLBCLs were assigned to the same clusters by any 2 algorithms. (B) Left panel: Consensus matrix comparing the assignments made by all 3 clustering algorithms (Meta Consensus [PC vs HC] vs [PC vs SOM]). Right panel: Comparisons of the meta-consensus cluster assignments. Of the 176 tumors, 141 were assigned to the same clusters by all 3 algorithms. (C) Expression profiles of the 3 DLBCL clusters. The top 50 genes associated with each DLBCL cluster are shown. Each column is a sample, and each row is a gene. Color scale at bottom indicates relative expression and standard deviations from the mean. Red indicates high-level expression; blue, low-level expression.
Characterization of the DLBCL consensus clusters
Having defined the expression profiles of 3 discrete DLBCL clusters, the next challenge was to interpret them objectively. We first asked whether previously characterized, coregulated sets of genes were more abundant in specific clusters using GSEA (Table 1; Patients, materials, and methods; and Mootha et al15 ).
Gene set enrichment analysis of the DLBCL consensus clusters
. | Ox Phos . | . | BCR/proliferation . | . | Host response . | . | |||
|---|---|---|---|---|---|---|---|---|---|
. | KS . | MHT P . | KS . | MHT P . | KS . | MHT P . | |||
| Mitochondrial pathways | |||||||||
| PGC | 130.9 | .004 | 13.2 | .763 | 3.8 | .931 | |||
| VOXPHOS | 156.1 | .001 | 13.3 | .760 | 2.9 | .948 | |||
| Human mito DB | 152.6 | .002 | 11.8 | .790 | 0.6 | .987 | |||
| Mitochondrial | 157.5 | .001 | 16.2 | .703 | 0.4 | .991 | |||
| OXPHOS | 141.0 | .003 | 13.7 | .753 | 2.9 | .946 | |||
| Gen MAPP | |||||||||
| Electron transport | 148.1 | .000 | 14.4 | .641 | 3.1 | .839 | |||
| Cell cycle | 33.4 | .298 | 104.8 | .004 | 0.8 | .873 | |||
| Complement activation-classical | 11.9 | .685 | 7.4 | .766 | 105.9 | .004 | |||
| Bio Carta | |||||||||
| Complement | 12.0 | .745 | 6.4 | .846 | 91.7 | .004 | |||
| T cytotoxic | 8.5 | .809 | 7.0 | .835 | 113.5 | .000 | |||
| T helper | 8.5 | .809 | 7.0 | .835 | 113.5 | .000 | |||
| T ob 1 | 27.0 | .440 | 2.8 | .902 | 103.2 | .002 | |||
| Coregulated gene sets | |||||||||
| C7 | 26.2 | .494 | 7.4 | .870 | 164.6 | .001 | |||
| C10 | 130.5 | .004 | 1.4 | .98 | 19.7 | .62 | |||
| Additional DLBCL gene sets | |||||||||
| Proliferation | 120.4 | .103 | 142.0 | .064 | 3.1 | .854 | |||
| Lymph node | 24.3 | .577 | 1.7 | .902 | 279.7 | .000 | |||
. | Ox Phos . | . | BCR/proliferation . | . | Host response . | . | |||
|---|---|---|---|---|---|---|---|---|---|
. | KS . | MHT P . | KS . | MHT P . | KS . | MHT P . | |||
| Mitochondrial pathways | |||||||||
| PGC | 130.9 | .004 | 13.2 | .763 | 3.8 | .931 | |||
| VOXPHOS | 156.1 | .001 | 13.3 | .760 | 2.9 | .948 | |||
| Human mito DB | 152.6 | .002 | 11.8 | .790 | 0.6 | .987 | |||
| Mitochondrial | 157.5 | .001 | 16.2 | .703 | 0.4 | .991 | |||
| OXPHOS | 141.0 | .003 | 13.7 | .753 | 2.9 | .946 | |||
| Gen MAPP | |||||||||
| Electron transport | 148.1 | .000 | 14.4 | .641 | 3.1 | .839 | |||
| Cell cycle | 33.4 | .298 | 104.8 | .004 | 0.8 | .873 | |||
| Complement activation-classical | 11.9 | .685 | 7.4 | .766 | 105.9 | .004 | |||
| Bio Carta | |||||||||
| Complement | 12.0 | .745 | 6.4 | .846 | 91.7 | .004 | |||
| T cytotoxic | 8.5 | .809 | 7.0 | .835 | 113.5 | .000 | |||
| T helper | 8.5 | .809 | 7.0 | .835 | 113.5 | .000 | |||
| T ob 1 | 27.0 | .440 | 2.8 | .902 | 103.2 | .002 | |||
| Coregulated gene sets | |||||||||
| C7 | 26.2 | .494 | 7.4 | .870 | 164.6 | .001 | |||
| C10 | 130.5 | .004 | 1.4 | .98 | 19.7 | .62 | |||
| Additional DLBCL gene sets | |||||||||
| Proliferation | 120.4 | .103 | 142.0 | .064 | 3.1 | .854 | |||
| Lymph node | 24.3 | .577 | 1.7 | .902 | 279.7 | .000 | |||
GSEA was performed using gene sets from (1) Biocarta; (2) GenMapp; and (3) a series of manually curated pathways involved in mitrochondrial function and metabolism (Mitrochondrial pathways).15 Additional coregulated gene sets from normal murine tissues (Coregulated gene sets)15 and DLBCLs (DLBCL gene sets)5 were also used. KS indicates Kolmogorov-Smirnoff score, and MHT P connotes a P value corrected for multiple hypothesis testing.
The first DLBCL cluster was significantly enriched in genes involved in oxidative phosphorylation, mitochondrial function, and the electron transport chain (Table 1). More detailed analysis of this DLBCL cluster, termed “OxPhos,” revealed increased expression of members of the nicotinamide adenine dinucleotide dehydrogenase (NADH) complex and cytochrome c/cytochrome c oxidase (COX) complex as well as adenosine triphosphate (ATP) synthase components and additional mitochondrial membrane enzymes (Table 2, “OxPhos cluster”).18 OxPhos tumors also had higher levels of the antiapoptotic BCL2 related family member, BFL-1/A1.19 Given the known consequences of mitochondrial membrane perturbation—cytochrome release and caspase-mediated apoptosis—and the regulation of mitochondrial membrane potential and cytochrome c release by BCL2 family members, these results are of particular interest. OxPhos tumors also had increased expression of multiple components of the 26S proteasome and general and mitochondrial ribosomal subunits (Table 2, “OxPhos cluster”).20
DLBCL consensus cluster signatures
Function . | Gene . |
|---|---|
| OxPhos cluster | |
| NADH dehydrogenase complex | NADH dehydrogenase (ubiquinone) 1:α/β subcomplex 1, 8 kDa; β subcomplex 1, 7 kDa; β subcomplex 2, 8 kDa Ubiquinol-cytochrome c reductase hinge protein |
| Cytochrome c/cytochrome oxidase (COX) complex | Cytochrome c COX 5b, 6a1, 7a2L, 7b, 7c, 11 |
| ATP synthase, mitochrondrial | F0 complex, subunit c |
| F1 complex β polypeptide, γ polypeptide 1 and O subunit | |
| ATP other | ATP binding protein |
| ATP binding cassette subfamily D (ALD, member 3) | |
| ATPase H+ transporting, lysosomal: 21 kDa, VO subunit c and 9 kDa | |
| Other, mitochondrial | Translocases of inner mitochondrial membrane (TIMM): nos. 8B and 23 |
| Translocases of outer mitochondrial membrane (TOMM): nos. 7 and 20 | |
| Diazepam binding inhibitor | |
| Apoptosis | BFL-1/A1 |
| MIHC (BIRC3) | |
| TNFA1P8 (SCC-S2) | |
| TNFRSF6 (FAS) | |
| Apoptosis-related protein (APR-3) | |
| Proteosome | Proteosome subunits: α types 2, 5, 6, and 7; and β type 4 |
| Proteosome 26s subunits: ATPase 2 and non-ATPase 4 | |
| Ribosome, mitochondrial | L3, L15, L39, S17, S31, and S36 |
| Ribosome, other | L3, L4, L5, L10, L13a, L15, L17, L27, L30, L31, L36A, L36, large PO, S12, S17, S20, and S21 |
| Other | X-ray repair complementing defective repair 5 (XRCC5) |
| Superoxide dismutase 1 (SOD1) | |
| Jumping translocation breakpoint | |
| CDW52 | |
| H2AZ | |
| PTEN | |
| BCR/proliferation cluster | |
| BCR signaling cascade | CD22 |
| CD19 | |
| Igμ | |
| CD79a | |
| BLK | |
| SYK | |
| PLC γ 2 | |
| Inositol 1, 4, 5 triphosphate receptor type 3 | |
| Inositol 1, 4, 5 triphosphate 3 kinase B | |
| Inositol polyphosphate-5-phosphatase 145 kDa | |
| MAP4K1 | |
| Class II molecules | CD74 (invariant polypeptide MHC class II) |
| Transcription factors | PAX5 |
| FOXO1A | |
| BCL6 | |
| POU2AF1 (BOB-1/OBF-1/OCAB) | |
| STAT6 | |
| TCF3 (E2A) | |
| NFAT | |
| SPI-B | |
| ETS-1 (E26 homolog) | |
| Ikaros | |
| MYC | |
| Other B-cell markers | CD37 |
| BC11A | |
| Proliferation | KI67 |
| CDK2 | |
| Signal-induced proliferation-associated 1 like 3 | |
| Replication/repair | DEAD/H box polypeptides 11 and 39 |
| Postmeiotic segregation increased (PMS) 2-like 2, 6, and 9 | |
| Minichromosome maintenance deficient (MCM) 2, 4, 5, and 7 | |
| p53 | |
| H2AX | |
| PAX transactivation domain-interacting protein (PTIP) | |
| MUTL homolog 6 | |
| Motility/cytoskeletal | Villin 2 (Ezrin) |
| Transcription modulators | SMARC A4, B1, and F1 |
| Posttranscriptional modification | HDAC1 |
| MYST histone acetyltransferase 4 | |
| Ubiquitin-activating enzyme E1 | |
| Other | Heat shock protein 90 beta |
| Host response cluster | |
| T/NK cell | T-cell receptor α and β |
| CD2 | |
| CD3 δ, ϵ and γ | |
| CD6 | |
| CD28 | |
| GATA3 | |
| cMAF | |
| CXCR6 | |
| LST (NKp30) | |
| Zap70 | |
| Linker for activation of T-cells (LAT) | |
| FYN | |
| FYN binding protein (SLAP) | |
| Lymphocyte activation gene 3 (LAG3)* | |
| CD100 (Sema 4D) | |
| Perforin | |
| NK transcript 4* | |
| T-cell immune regulator (TIRC7) | |
| Leukocyte-associated Ig-like receptor 1 (LAIR-1) | |
| Complement | Complement 1qB, 1S, and 4A |
| Complement 3α receptor | |
| Clade G (C1 inhibitor) | |
| Monocyte/macrophage | CD14* |
| CD163 | |
| B lymphocyte activator, macrophage expressed (BLAME) | |
| FGR | |
| SHPS-1 (BIT/SIRPα) | |
| Granulin | |
| Allograft inflammatory factor | |
| Antigen processing | Lysosomal-associated membrane protein 1 (LAMP1)* |
| Cathepsins B and D | |
| IFN—γ inducible protein 30 (GILT)* | |
| Interferon (IFN) | IFN-induced transmembrane proteins 1 and 2 |
| Guanylate binding proteins 1** and 2, IFN-inducible | |
| STAT1* | |
| Interferon regulatory factors (IRFs) 1 and 7 | |
| MHC Class I | HLA A, C, E and F |
| TNF family members | TNFRSF 1A and B |
| TNFSF10 (TRAIL) | |
| TNFSF13 (APRIL)* | |
| Cytokine receptors | IL 2 receptor γ |
| IL 6 receptor | |
| IL 15 receptor α* | |
| TGF β receptor II | |
| CSF-1 receptor* | |
| ECM/adhesion | LFA-1 (integrins α L and β 2)* |
| PECAM1 (CD31) | |
| P-selectin glycoprotein ligand (PSGL-1) | |
| Collagens: type XVIII α 1 and type IV α 2 | |
| Stromal-derived factor 1 | |
| Apoptosis | Caspase 4, apoptosis-related cysteine protease* |
| Other | Notch 2 |
| Disabled homolog 2 (DAB2) | |
| FOXO3A | |
| MAF B | |
| Prostaglandin E receptor 4 | |
| S100A4 |
Function . | Gene . |
|---|---|
| OxPhos cluster | |
| NADH dehydrogenase complex | NADH dehydrogenase (ubiquinone) 1:α/β subcomplex 1, 8 kDa; β subcomplex 1, 7 kDa; β subcomplex 2, 8 kDa Ubiquinol-cytochrome c reductase hinge protein |
| Cytochrome c/cytochrome oxidase (COX) complex | Cytochrome c COX 5b, 6a1, 7a2L, 7b, 7c, 11 |
| ATP synthase, mitochrondrial | F0 complex, subunit c |
| F1 complex β polypeptide, γ polypeptide 1 and O subunit | |
| ATP other | ATP binding protein |
| ATP binding cassette subfamily D (ALD, member 3) | |
| ATPase H+ transporting, lysosomal: 21 kDa, VO subunit c and 9 kDa | |
| Other, mitochondrial | Translocases of inner mitochondrial membrane (TIMM): nos. 8B and 23 |
| Translocases of outer mitochondrial membrane (TOMM): nos. 7 and 20 | |
| Diazepam binding inhibitor | |
| Apoptosis | BFL-1/A1 |
| MIHC (BIRC3) | |
| TNFA1P8 (SCC-S2) | |
| TNFRSF6 (FAS) | |
| Apoptosis-related protein (APR-3) | |
| Proteosome | Proteosome subunits: α types 2, 5, 6, and 7; and β type 4 |
| Proteosome 26s subunits: ATPase 2 and non-ATPase 4 | |
| Ribosome, mitochondrial | L3, L15, L39, S17, S31, and S36 |
| Ribosome, other | L3, L4, L5, L10, L13a, L15, L17, L27, L30, L31, L36A, L36, large PO, S12, S17, S20, and S21 |
| Other | X-ray repair complementing defective repair 5 (XRCC5) |
| Superoxide dismutase 1 (SOD1) | |
| Jumping translocation breakpoint | |
| CDW52 | |
| H2AZ | |
| PTEN | |
| BCR/proliferation cluster | |
| BCR signaling cascade | CD22 |
| CD19 | |
| Igμ | |
| CD79a | |
| BLK | |
| SYK | |
| PLC γ 2 | |
| Inositol 1, 4, 5 triphosphate receptor type 3 | |
| Inositol 1, 4, 5 triphosphate 3 kinase B | |
| Inositol polyphosphate-5-phosphatase 145 kDa | |
| MAP4K1 | |
| Class II molecules | CD74 (invariant polypeptide MHC class II) |
| Transcription factors | PAX5 |
| FOXO1A | |
| BCL6 | |
| POU2AF1 (BOB-1/OBF-1/OCAB) | |
| STAT6 | |
| TCF3 (E2A) | |
| NFAT | |
| SPI-B | |
| ETS-1 (E26 homolog) | |
| Ikaros | |
| MYC | |
| Other B-cell markers | CD37 |
| BC11A | |
| Proliferation | KI67 |
| CDK2 | |
| Signal-induced proliferation-associated 1 like 3 | |
| Replication/repair | DEAD/H box polypeptides 11 and 39 |
| Postmeiotic segregation increased (PMS) 2-like 2, 6, and 9 | |
| Minichromosome maintenance deficient (MCM) 2, 4, 5, and 7 | |
| p53 | |
| H2AX | |
| PAX transactivation domain-interacting protein (PTIP) | |
| MUTL homolog 6 | |
| Motility/cytoskeletal | Villin 2 (Ezrin) |
| Transcription modulators | SMARC A4, B1, and F1 |
| Posttranscriptional modification | HDAC1 |
| MYST histone acetyltransferase 4 | |
| Ubiquitin-activating enzyme E1 | |
| Other | Heat shock protein 90 beta |
| Host response cluster | |
| T/NK cell | T-cell receptor α and β |
| CD2 | |
| CD3 δ, ϵ and γ | |
| CD6 | |
| CD28 | |
| GATA3 | |
| cMAF | |
| CXCR6 | |
| LST (NKp30) | |
| Zap70 | |
| Linker for activation of T-cells (LAT) | |
| FYN | |
| FYN binding protein (SLAP) | |
| Lymphocyte activation gene 3 (LAG3)* | |
| CD100 (Sema 4D) | |
| Perforin | |
| NK transcript 4* | |
| T-cell immune regulator (TIRC7) | |
| Leukocyte-associated Ig-like receptor 1 (LAIR-1) | |
| Complement | Complement 1qB, 1S, and 4A |
| Complement 3α receptor | |
| Clade G (C1 inhibitor) | |
| Monocyte/macrophage | CD14* |
| CD163 | |
| B lymphocyte activator, macrophage expressed (BLAME) | |
| FGR | |
| SHPS-1 (BIT/SIRPα) | |
| Granulin | |
| Allograft inflammatory factor | |
| Antigen processing | Lysosomal-associated membrane protein 1 (LAMP1)* |
| Cathepsins B and D | |
| IFN—γ inducible protein 30 (GILT)* | |
| Interferon (IFN) | IFN-induced transmembrane proteins 1 and 2 |
| Guanylate binding proteins 1** and 2, IFN-inducible | |
| STAT1* | |
| Interferon regulatory factors (IRFs) 1 and 7 | |
| MHC Class I | HLA A, C, E and F |
| TNF family members | TNFRSF 1A and B |
| TNFSF10 (TRAIL) | |
| TNFSF13 (APRIL)* | |
| Cytokine receptors | IL 2 receptor γ |
| IL 6 receptor | |
| IL 15 receptor α* | |
| TGF β receptor II | |
| CSF-1 receptor* | |
| ECM/adhesion | LFA-1 (integrins α L and β 2)* |
| PECAM1 (CD31) | |
| P-selectin glycoprotein ligand (PSGL-1) | |
| Collagens: type XVIII α 1 and type IV α 2 | |
| Stromal-derived factor 1 | |
| Apoptosis | Caspase 4, apoptosis-related cysteine protease* |
| Other | Notch 2 |
| Disabled homolog 2 (DAB2) | |
| FOXO3A | |
| MAF B | |
| Prostaglandin E receptor 4 | |
| S100A4 |
Gene lists derived from the top 250 genes with higher levels of expression in the indicated consensus cluster.
Genes included in previously described LN signature.5
The second DLBCL cluster, termed “BCR/proliferation,” had more abundant expression of cell-cycle regulatory genes (Table 1), including CDK2 and MCM (minichromosome maintenance deficient) family members (Table 2).21 These tumors also had increased expression of DNA repair genes including postmeiotic segregation increased 2 (PMS2) family members,22 H2AX,23 PTIP,24 and p53 (Table 2, “BCR/proliferation cluster”). This DLBCL cluster also had higher levels of many components of the B-cell receptor (BCR) signaling cascade (CD19, Ig, CD79a, BLK, SYK, PLCγ2, and MAP4K) and additional B-cell–specific or essential transcription factors (including PAX5, OBF-1, E2A, BCL6, STAT6, and MYC; Table 2, “BCR/proliferation cluster”).25,26
Unlike the other 2 DLBCL subsets, the third DLBCL cluster had a signature that was largely defined by the associated host response rather than the tumor itself (Table 1). By GSEA, this cluster was enriched for markers of T-cell–mediated immune responses and the classical complement pathway (Table 1). These tumors also had increased expression of an overlapping set of coregulated inflammatory mediators and connective tissue components (C7; Table 1 and Supplemental Document).
Detailed analysis of the third cluster, termed “host response” (HR), revealed increased expression of multiple components of the T-cell receptor (TCR) (TCRα and β and CD3 subunits), CD2, and additional molecules associated with T/NK-cell activation27 and the complement cascade (Table 2, “Host response cluster”). HR tumors also had more abundant monocyte/macrophage and dendritic cell transcripts, molecules required for efficient antigen processing, and certain HLA class I antigens28-35 (Table 2). Consistent with the signature of an ongoing inflammatory/immune response, HR tumors had increased expression of interferon-induced genes, certain tumor necrosis family (TNF) ligands and receptors, cytokine receptors, adhesion molecules, and extracellular matrix components36-38 (Table 2).
Of note, patients in the 3 consensus clusters had similar 5-year survivals (OxPhos, 53%; BCR/proliferation, 60%; and HR, 54%; P = .53), suggesting that the clusters may be more useful for identifying potential pathogenetic mechanisms and cluster-specific rational therapeutic targets than predicting responses to empiric combination chemotherapy.
Genetic abnormalities and clinical features in the newly identified DLBCL clusters
Having identified 3 subclasses of DLBCL, we asked whether these subgroups differed with respect to known chromosomal translocations in the disease (t(14;18) and t(3;), involving the BCL6 locus; Table 3). The distribution of t(14;18) and t(3;) was examined in the 116 tumors with available data and no more than one translocation (one OxPhos tumor with both translocations was omitted from the analysis). There was an association between cluster membership and the examined genetic abnormalities (P = .059, Fisher exact test; Table 3). BCL2 translocations were more common in the Oxphos cluster, whereas BCL6 translocations were more frequent in the BCR/proliferation cluster. Translocations of either type were uncommon in the HR cluster (Table 3).
Genetic abnormalities in the DLBCL consensus clusters
. | OxPhos . | . | BCR/proliferation . | . | HR . | . | Total . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic abnormality* . | n = 37 . | % . | n = 50 . | % . | n = 29 . | % . | n = 116 . | % . | ||||
| t(14;18) | 8 | (22) | 5 | (10) | 1 | (3) | 14 | (12) | ||||
| t(3;...) | 2 | (5) | 8 | (16) | 1 | (3) | 11 | (9) | ||||
| None | 27 | (73) | 37 | (74) | 27 | (93) | 91 | (78) | ||||
. | OxPhos . | . | BCR/proliferation . | . | HR . | . | Total . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic abnormality* . | n = 37 . | % . | n = 50 . | % . | n = 29 . | % . | n = 116 . | % . | ||||
| t(14;18) | 8 | (22) | 5 | (10) | 1 | (3) | 14 | (12) | ||||
| t(3;...) | 2 | (5) | 8 | (16) | 1 | (3) | 11 | (9) | ||||
| None | 27 | (73) | 37 | (74) | 27 | (93) | 91 | (78) | ||||
116 tumors had available data and no more than one translocation. One OxPhos tumor with both translocations was omitted from the analysis. The distribution of genetic abnormalities across clusters was represented in a 3 × 3 contingency table and analyzed with a Fisher exact test (P = .059).
The increased incidence of t(14;18) in OxPhos tumors was of particular interest given this cluster's oxidative phosphorylation/mitochondrial gene expression signature and overexpression of additional antiapoptotic BCL2 family members (Tables 1 and 2, “OxPhos cluster”).
The near absence of known cytogenetic abnormalities and the prominent inflammatory/immune infiltrate in HR DLBCLs prompt speculation regarding other, as-yet-uncharacterized mechanisms of transformation in these tumors. In this regard, it is noteworthy that patients with HR DLBCLs were significantly younger than those with OxPhos or BCR/proliferation tumors (P = .04, Kruskal-Wallis test; Supplemental Document). Patients with HR tumors also had a significantly higher incidence of splenic and bone marrow (BM) involvement (P = .02 and P = .03, respectively).
Immunohistochemical and morphologic analysis of HR tumors
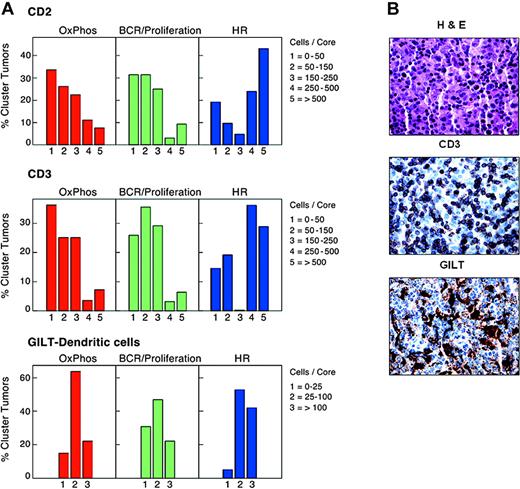
The unique characteristics of the HR cluster—fewer known genetic abnormalities and prominent host immune and inflammatory cell transcripts—prompted us to assess host immune cells in study tumors using morphologic and immunohistochemical approaches. Hematoxylin and eosin–and CD2-stained slides of study DLBCLs were evaluated for the presence of tumor-infiltrating lymphocytes (TILs) by an expert morphologist who had no information regarding the DLBCL transcriptional profiles. HR tumors contained significantly higher numbers of TILs than DLBCLs in the other clusters (P < .0001, Fisher exact test; Supplemental Document).
Since HR tumors had more abundant CD2 and CD3ϵ transcripts (Table 2, “Host response cluster”), we also used CD2 and CD3 immunostaining to quantify infiltrating T cells in study DLBCLs. HR tumors contained significantly higher numbers of CD2+ and CD3+ T cells than DLBCLs in the other clusters (P = .005 and P = .003, respectively, Kruskal-Wallis exact test; Figure 2A). Consistent with these observations, 8 of the 10 tumors initially diagnosed as T-cell/histiocyte-rich DLBCLs39 were included in the HR cluster (49 tumors total). Additional components of the HR signature—ZAP70 and its substrate, LAT (linker for the activation of T cells)40 ; the T helper 2 (TH2) transcription factors, GATA3 and c-MAF41 ; the T helper 1 (TH1) and T cytotoxic 1 (TC1) cytokine receptor, CXCR642 ; the natural killer (NK) cell triggering receptor, LST (NKp30)43 ; perforin 1; and the CD28 costimulatory molecule44 —suggest that these tumors include a mixed population of activated T/NK cells (Table 2, “Host response cluster”).
T- and dendritic cell infiltrates in study DLBCLs. (A) Numbers of normal infiltrating CD2+ and CD3+ cells and GILT-positive dendritic cells in primary DLBCLs in each cluster. HR tumors included significantly higher numbers of CD2+ and CD3+ T cells than DLBCLs in the other clusters (P = .005 and P = .003, respectively; Kruskal-Wallis exact test). HR tumors also contained higher numbers of GILT+ dendritic cells (P = .06, Kruskal-Wallis exact test). (B) Hematoxylin and eosin staining and CD3 and GILT immunostaining of a representative HR tumor.
T- and dendritic cell infiltrates in study DLBCLs. (A) Numbers of normal infiltrating CD2+ and CD3+ cells and GILT-positive dendritic cells in primary DLBCLs in each cluster. HR tumors included significantly higher numbers of CD2+ and CD3+ T cells than DLBCLs in the other clusters (P = .005 and P = .003, respectively; Kruskal-Wallis exact test). HR tumors also contained higher numbers of GILT+ dendritic cells (P = .06, Kruskal-Wallis exact test). (B) Hematoxylin and eosin staining and CD3 and GILT immunostaining of a representative HR tumor.
In addition to having higher numbers of infiltrating T and NK cells, HR tumors had increased levels of likely macrophage and dendritic cell transcripts, including the gamma interferon–induced lysosomal thiol reductase, GILT17,34 (Table 2, “Host response cluster”). Since GILT is required for effective peptide processing and optimal antigen presentation,34,45,46 we used GILT immunostaining to both identify and characterize the dendritic cells in study tumors. When compared with the other clusters, HR tumors contained increased numbers of GILT+ dendritic cells (P = .06, Kruskal-Wallis test; Figure 2A-B).
For this reason, we further characterized tumor dendritic cells (DCs) with S100, CD1a, and CD123. These markers distinguish interdigitating DCs (S100+CD1a–CDC123–) that interact with antigen-specific T cells in secondary lymphoid organs from other DC subtypes.47,48 There was no detectable CD1a expression in study DLBCLs, and only 2 tumors (non-HR) contained CD123+ cells. In marked contrast, S100+ DCs were readily detectable and significantly more abundant in HR tumors than DLBCLs from other clusters (P = .009, Kruskal Wallis test). In addition, the numbers of CD2+/CD3+ infiltrating T cells and GILT+/S100+ DCs were highly correlated in individual tumors (P < .0001, Jonckheere-Terpstra test; Figure 2B and Supplemental Document). Therefore, HR tumors contain interdigitating DCs and associated infiltrating T cells, likely capable of participating in a coordinated immune response. Consistent with this interpretation, the HR signature also includes adhesion molecules such as LFA-1 that strengthen T-cell/DC contact and T-cell surface molecules, such as Sema4D/CD100 and LAG3/CD223, that promote DC maturation and activation (Table 2, “Host response cluster”).44,49-51
Validation of DLBCL consensus clusters in an independent dataset
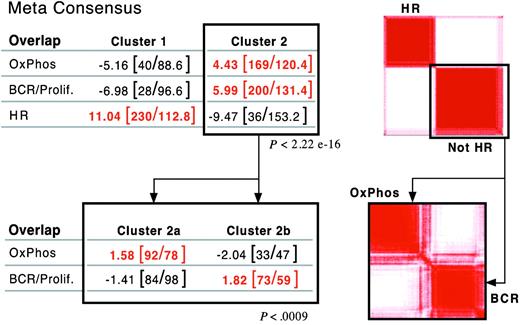
After defining 3 consensus clusters in our own DLBCL series, we asked whether there were similar clusters in an independent group of newly diagnosed DLBCLs with available gene expression profiles.5 Using the overlapping set of highly reproducible/highly variable genes (703 common genes), our clustering procedure subdivided the independent DLBCL series into 2, rather than 3, major groups (Figure 3, right panel; and Supplemental Information). The signature for one of the independent clusters was highly enriched for HR transcripts (overlap, P < 2.2 × 10–16; Figure 3A, top left panel). We further analyzed the “non-HR” tumors by clustering this group in the space of non-HR markers. Non-HR tumors separated into 2 discrete clusters with highly significant enrichment for either BCR/proliferation or OxPhos transcripts (overlap, P < .0009; Figure 3B, bottom panels).
Validation of DLBCL consensus clusters in an independent dataset. Application of consensus clustering and meta-consensus (as in Figure 1B) to the independent DLBCL series (top right panel). One of the identified consensus clusters was highly enriched for HR transcripts (P < 2.2 × 10–16, top left panel). Application of consensus clustering and meta-consensus to the non-HR cluster (bottom right panel). The non-HR tumors sorted into 2 discrete clusters with highly significant enrichment for either BCR/proliferation or OxPhos transcripts (P = < .0009, bottom left panel).
Validation of DLBCL consensus clusters in an independent dataset. Application of consensus clustering and meta-consensus (as in Figure 1B) to the independent DLBCL series (top right panel). One of the identified consensus clusters was highly enriched for HR transcripts (P < 2.2 × 10–16, top left panel). Application of consensus clustering and meta-consensus to the non-HR cluster (bottom right panel). The non-HR tumors sorted into 2 discrete clusters with highly significant enrichment for either BCR/proliferation or OxPhos transcripts (P = < .0009, bottom left panel).
Similar structure was also identified when tumors were clustered using less restricted sets of genes (either the top 50% of genes ranked by a MAD-based variation filter or all genes), indicating that the structure was not dependent upon a highly selected gene set (Supplemental Document). Taken together, these results confirm the presence of similar consensus clusters in an independent DLBCL dataset.
Relationship of consensus clusters to the cell-of-origin signature
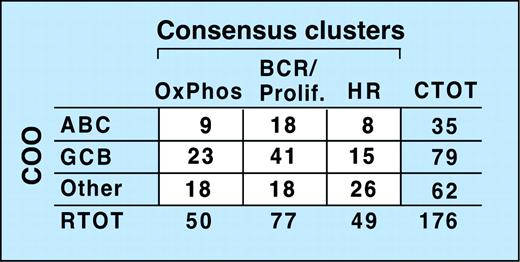
Recent studies suggest that subsets of DLBCL share elements of the transcriptional profile of normal purified germinal center B cells (GCBs) or in vitro–activated peripheral blood B cells (ABCs), while other DLBCLs lack these features (Other).5,6 To compare the newly defined consensus clusters (CCs) with these cell-of-origin (COO) subsets, we first classified our tumors with respect to COO (Wright et al,6 “Patients, materials, and methods,” and Supplemental Document). Of note, tumors identified as GCBs were associated with significantly longer overall survivals (P = .003).
Comparison of the CC and COO assignments indicates that the 2 classification schema are capturing largely different aspects of DLBCL biology (Figure 4 and Supplemental Document). Although 53% of tumors in the BCR/proliferation cluster and 46% of tumors in the OxPhos cluster were classified as GC-like, the remainder were designated ABC or Other (Figure 4). In the HR cluster, there were relatively more unspecified (Other) DLBCLs (Figure 4), likely because unspecified (Other) DLBCLs have less striking B-cell signatures and HR tumors have prominent inflammatory infiltrates.
Relationship of consensus clusters to cell-of-origin (COO) signature. Comparison of study DLBCLs sorted into consensus clusters with the same tumors classified by COO. The lack of a clear correlation between the 2 clustering systems is reflected by the absence of a matrix diagonal structure (ie, large numbers along the diagonal and numbers close to 0 in the off-diagonal entries). CTOT indicates total number of tumors in a row.
Relationship of consensus clusters to cell-of-origin (COO) signature. Comparison of study DLBCLs sorted into consensus clusters with the same tumors classified by COO. The lack of a clear correlation between the 2 clustering systems is reflected by the absence of a matrix diagonal structure (ie, large numbers along the diagonal and numbers close to 0 in the off-diagonal entries). CTOT indicates total number of tumors in a row.
In DLBCLs, additional sets of coregulated genes (proliferation, major histocompatibility complex [MHC] class II, and lymph node [LN]) were previously reported to be expressed independently of the COO signature.5 For these reasons, we asked whether these additional coregulated gene sets contributed to consensus cluster signatures using GSEA. Not surprisingly, the BCR/proliferation signature had some evidence of enrichment with the previously described proliferation genes5 (MHT, P = .06; Table 1). There was also highly significant enrichment of the LN gene set in our HR signature (P =< .001; Table 1). Given the composition of the LN gene set—T/NK activation antigens, complement components, monocyte markers, interferon-inducible genes, HLA class I molecules, additional cytokines, and connective tissue components5 —these results are in keeping with the broader definition of a DLBCL cluster characterized by a concomitant host immune/inflammatory response.
Discussion
Using 3 different clustering methods and whole genome arrays, we identified 3 robust subsets of DLBCL and confirmed their presence in an independent series. The characteristics of these clusters—OxPhos, BCR/proliferation, and HR—suggest that these tumors may have novel pathogenetic mechanisms and possible treatment targets. In addition, the signatures identify the tumor microenvironment as a defining feature.
The current study indicates that additional, nonoverlapping information can be obtained by sorting DLBCLs with respect to consensus clusters and putative COO. In fact, other features of DLBCLs that track independently of COO (“proliferation” and “lymph node signature”5 ) were captured by the comprehensive clusters. The updated COO signature identifies a subset of “GC-like” DLBCLs that responded more favorably to empiric combination chemotherapy. Although the comprehensive consensus clusters were less predictive of response to empiric combination chemotherapy, the clusters reproducibly defined major groups of tumors that may be amenable to targeted intervention. For example, OxPhos tumors have increased expression of proteosomal subunits and molecules regulating mitochondrial membrane potential and apoptosis. These DLBCLs may be particularly sensitive to proteosome blockade20 or BCL2 family inhibition. In contrast, HR tumors may be more sensitive to immunomodulatory approaches.
Thus far, the HR cluster has been most extensively characterized. HR tumors were largely defined by their inflammatory/immune cell infiltrate, including CD2+/CD3+ TILs and interdigitating S100+/GILT+ CD1a–CD123– dendritic cells and suggesting a coordinated immune response. HR tumors had less frequent genetic abnormalities and occurred in younger patients, prompting speculation regarding an alternative pathogenetic mechanism. Patients with HR tumors also had unique clinical features, presenting more commonly with splenomegaly and bone marrow involvement.
The T-cell/dendritic cell infiltrates in HR tumors resemble those of a smaller provisional (WHO) subtype of DLBCL, T-cell/histiocyte-rich B-cell lymphoma (T/HRBCL), which includes abundant nonneoplastic T cells and associated macrophages (“histiocytes”).39,52-55 Like HR DLBCLs, T/HRBCLs are reported to have fewer known genetic lesions and occur in slightly younger patients who often have splenomegaly and BM involvement.53,54 However, histologically defined T/HRBCLs represent a smaller subset of DLBCL than our HR cluster. It is likely that the comprehensive transcriptional profiles identify additional DLBCL patients with more subtle, related signatures.
In addition to providing insights regarding the nature of the associated immune response in HR tumors, the newly identified molecular and immunohistochemical features of these DLBCLs may increase diagnostic accuracy. For example, histologically defined T/HRLCL is a “gray zone” lymphoma that may resemble lymphocyte-predominant Hodgkin lymphoma, a more indolent disease with different recommended therapy.56,57
The current HR signature contains more information regarding the infiltrating immune cells and associated inflammatory response than the associated malignant B cells. Microdissected tumor cells from T/HRBCL were previously shown to have clonal Ig gene rearrangements, somatic hypermutation, and a mutation pattern suggestive of antigen selection.52 In the current study, HR tumors expressed higher levels of Notch 2, a molecule implicated in specific stages of mature B-cell development.58 HR tumors also expressed higher levels of TNF receptors and additional TNF costimulatory molecules (such as APRIL) known to protect malignant B cells from apoptosis.59,60 At present, the antigen specificity of HR malignant B cells remains undefined. It is possible that HR malignant B cells and the associated infiltrating T cells are directed against the same antigen; if so, the TILs and interdigitating dendritic cells may actually support tumor growth.61 Alternatively, TILs might be directed against the malignant B cells in HR tumors. However, patients in the HR cluster did not have better outcomes following empiric chemotherapy, suggesting that their immune responses were ineffective and/or inhibited by counter-regulatory mechanisms,62 or their tumors were inherently less responsive to CHOP-based treatment.
For these reasons, it will be important to identify HR tumors with pre-existing abundant T- and dendritic cell infiltrates and further characterize their associated underlying immune response. Such directed approaches to HR tumors and the other newly identified DLBCL consensus clusters will likely define more rational treatment targets in this heterogeneous disease.
Prepublished online as Blood First Edition Paper, November 18, 2004; DOI 10.1182/blood-2004-07-2947.
S.M. and K.J.S. contributed equally to the work. T.R.G. and M.A.S. contributed equally to the work.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Supplemental data
This document provides supplementary and detailed analysis information not included in the paper. Our sources of information and the original data sets can be found among the Broad Institute's Cancer Program Publications/Projects page at http://www.broad.mit.edu/cgi-bin/cancer/publications/pub_paper.cgi?mode=view&paper_id=102.

![Figure 1. Identification of consensus clusters. (A) Left panels: Consensus matrices produced by hierarchical clustering (HC, K = 3), self-organizing maps (SOM, K = 3), and probabilistic clustering (PC). Right panels: Comparisons of the cluster assignments of the different algorithms (PC vs HC, HC vs SOM, and PC vs SOM). More than 84% of DLBCLs were assigned to the same clusters by any 2 algorithms. (B) Left panel: Consensus matrix comparing the assignments made by all 3 clustering algorithms (Meta Consensus [PC vs HC] vs [PC vs SOM]). Right panel: Comparisons of the meta-consensus cluster assignments. Of the 176 tumors, 141 were assigned to the same clusters by all 3 algorithms. (C) Expression profiles of the 3 DLBCL clusters. The top 50 genes associated with each DLBCL cluster are shown. Each column is a sample, and each row is a gene. Color scale at bottom indicates relative expression and standard deviations from the mean. Red indicates high-level expression; blue, low-level expression.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/5/10.1182_blood-2004-07-2947/6/m_zh80050575200001.jpeg?Expires=1770862730&Signature=Z2GIqO2rTIh1CeEftxev59RNXWE6LESYUySmZ2BvEwA6BasfsaY6bGGzK3v7leXhPjJXoL-nxO7y~g2ieNO-Ho-ySHXaS2TLaRb79kR1PHe5Ey9wxQMOfOJUOUJ1VasK4a4TZrMVjE6AzeCSsFDu7e-QhiakGZRh~b9HNpuvxRNu6HgRPJLI8LuPC2WlRNEhAjz~MGOGUxfdL3t4RBfO2lY7nrgIXjqIjfQkQMia0uctvbvmboT6ReMCqzXgmNbRN1eTg8l5ar-k3wlMyghqBNmQKXzS3FEIYHYo~JPybcRM~h40eXubSUO90eoK~Nu6xliZN69dhVu6WQjw8oZAQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal