Abstract
To characterize the transcriptional program that governs terminal granulocytic differentiation in vivo, we performed comprehensive microarray analyses of human bone marrow populations highly enriched in promyelocytes (PMs), myelocytes/metamyelocytes (MYs), and neutrophils (bm-PMNs). These analyses identified 11 310 genes involved in differentiation, of which 6700 were differentially regulated, including previously unidentified effector proteins and surface receptors of neutrophils. Differentiation of PMs toward MYs was accompanied by a marked decline of proliferative and general cellular activity as defined by down-regulation of E2 promoter binding factor (E2F) target genes; cyclin dependent kinases 2, 4, and 6; and various metabolic, proteasomal, and mitochondrial genes. Expression patterns of apoptosis genes indicated death control by the p53 pathway in PMs and by death receptor pathways in bm-PMNs. Effector proteins critical for host defense were expressed successively throughout granulocytic differentiation, whereas receptors and receptor ligands essential for the activation of the host defense program were terminally up-regulated in bm-PMNs. The up-regulation of ligand-receptor pairs, which are defined inducers as well as target genes of nuclear factor-κB (NF-κB), suggests a constitutive activation of NF-κB in bm-PMNs by autocrine loops. Overall, these results define a granulocytic differentiation model governed by a highly coordinated fail-safe program, which promotes completion of differentiation before cells gain responsiveness toward activating stimuli that accompany infections. (Blood. 2005; 105:1785-1796)
Introduction
Polymorphonuclear neutrophilic granulocytes (neutrophils/PMNs) constitute the most abundant population of white blood cells and are essential players in innate immune defense of mammalian hosts against microorganisms. Once neutrophils have migrated to sites of infection, they recognize microorganisms and their products to initiate a first line of defense using a number of distinct mechanisms. These defense mechanisms include phagocytosis, generation of reactive oxygen intermediates, and the release of antimicrobial granule proteins for killing and degradation of microorganisms.1,2
Neutrophils are short-lived cells, which are continuously generated from hematopoietic stem cells (HSCs) in the bone marrow (BM) by a process called granulopoiesis. The hallmark of early granulopoiesis is the successive commitment of pluripotent HSCs via multipotent common myeloid progenitors (CMPs) and bipotent granulocyte-macrophage progenitors (GMPs) toward unipotent progenitors restricted to the granulocytic lineage.3,4 Once the progenitors are committed to the granulocytic lineage, they initiate terminal granulopoiesis and differentiate into mature neutrophils. Terminal granulopoiesis gives rise to a series of morphologically distinct stages, which are readily identified by their characteristic nuclear shape and their content of granules. At the myeloblast/promyelocyte (MB/PM) stages the cells still proliferate and generate primary granules with their constituting proteins. At the myelocyte/metamyelocyte (MC/MM) stages, cell proliferation and expression of primary granule proteins stop concomitantly with the successive generation of secondary and tertiary granules and their constituting granule proteins. Finally, the synthesis of granule proteins ceases, and the cells acquire their full antimicrobial potential when maturation proceeds toward the stages of neutrophils with band shaped (BCs) and segmented polymorphic nuclei (PMNs).1,2,5
The molecular mechanisms that govern differentiation of HSCs along the myeloid lineage toward granulocytes rather than monocytes depend on 2 key regulators.5 One key regulator is the CCAAT enhancer binding protein-α (C/EBPα), which is expressed in myeloid progenitors—that is, CMPs and GMPs—and is maintained during granulocytic differentiation but down-regulated during monocytic and erythroid differentiation.6,7 C/EBPα knock-out mice exhibit a block of granulocytic differentiation at the GMP to promyelocyte transition, resulting in a complete lack of promyelocytes and their progeny.8 Further studies have demonstrated that C/EBPα transactivates several myeloid- and granulocyte-specific genes, including the early appearing primary granule protein myeloperoxidase (MPO) and the granulocyte colony-stimulating factor receptor (G-CSFR).5,9 Another key regulator is the C/EBPϵ transcription factor, which is up-regulated by C/EBPα and by the granulocyte colony-stimulating factor (G-CSF) through the G-CSFR signaling pathway.10 Mice with a targeted disruption of C/EBPϵ fail to produce mature neutrophils and to express the late-appearing secondary/tertiary granule proteins such as lactoferrin (LF) and gelatinase.11 Hence, C/EBPα is critical for early granulocytic differentiation (that is, the commitment of HSCs toward promyelocytes), whereas C/EBPϵ, which acts downstream of C/EBPα, regulates terminal granulocytic differentiation (that is, the transition from promyelocytes to mature neutrophils).
To gain more insight into the molecular mechanisms that govern the transition from promyelocytes to mature neutrophils, we applied microarray technology to systematically profile the expression patterns of 17 020 genes (44 760 probe sets) in human BM populations highly enriched in promyelocytes (PMs), myelocytes/metamyelocytes (MYs), and neutrophils (bm-PMNs).
These analyses define a granulocytic differentiation model, which is characterized by a highly coordinated regulation of distinct genetic programs.
Material and methods
Isolation of bone marrow populations and peripheral blood neutrophils
BM and peripheral blood (PB) samples were collected in parallel from healthy individuals. All samples were obtained after informed consent had been given according to the guidelines established by the Ethics Committee of the Cities of Copenhagen and Frederiksberg.
Cell populations representing successive stages of terminal granulocytic differentiation were isolated from human BM samples by 2-layer density gradient centrifugation as described previously.12 PB samples were obtained by venipuncture immediately after BM aspiration, and neutrophils (pb-PMNs) isolated by 1-layer density gradient centrifugation.12 Subsequently, each population was depleted of nongranulocytic cells by immunomagnetic sorting using the magnetic cell sorting (MACS) system according to instructions of the manufacturer (MACS; Miltenyi, Bergisch Gladbach, Germany). The monoclonal antibodies (MoAbs) used for depletion of nongranulocytic cells are listed in Table 1.
Panel of MoAbs used for MACS depletion of nongranulocytic cells in BM and PB populations and for subsequent flow cytometry analysis of residual contaminating cells
. | . | . | . | . | MACS depletion cocktail . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Surface molecule . | Cell type . | FACS MoAb clone . | Fluorochrome-conjugated to MoAb . | MACS MoAb clone . | PM . | MY . | bm-PMN . | pb-PMN . | |||
| CD2 | T cells, NK cells | Not used | RPA-2.10 | X | X | X | X | ||||
| CD3 | T cells | SK7 | PE | UCHT1 | X | X | — | — | |||
| CD5 | T cells | DK23 | FITC | Not used | X | X | — | — | |||
| CD10 | B-, T-cell precursors, mature neutrophils | Not used | SS2/36 | X | X | — | — | ||||
| CD11b | NK cells, monocytes, neutrophils, myeloid precursors | ICRF44 | PE, APC | ICRF44 | X | — | — | — | |||
| CD14 | Myelomonocytic cells | TÛK4 | PE, FITC | M5E2 | X | X | X | X | |||
| CD15 | Neutrophils, monocytes, eosinophils | MMA | FITC, APC | HI98 | X | X | — | — | |||
| CD16 | NK cells, monocytes, neutrophils, myeloid precursors | NKP15 | FITC | DJ130c | X | X | — | — | |||
| CD19 | B cells | 4G7 | FITC | HIB19 | X | X | X | X | |||
| CD49d | Eosinophils | 9F10 | PE | 9F10 | — | — | X | X | |||
| CD56 | NK cells | My31 | PE | B159 | X | X | X | X | |||
| CD61 | Platelets, megakaryocytes, monocytes | Y2/51 | FITC, PE | VI-PL2 | X | X | X | X | |||
| GLY-A | Erythroid cells, erythroid precursors | JC159 | R-PE | GA-R2 | X | X | X | X | |||
. | . | . | . | . | MACS depletion cocktail . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Surface molecule . | Cell type . | FACS MoAb clone . | Fluorochrome-conjugated to MoAb . | MACS MoAb clone . | PM . | MY . | bm-PMN . | pb-PMN . | |||
| CD2 | T cells, NK cells | Not used | RPA-2.10 | X | X | X | X | ||||
| CD3 | T cells | SK7 | PE | UCHT1 | X | X | — | — | |||
| CD5 | T cells | DK23 | FITC | Not used | X | X | — | — | |||
| CD10 | B-, T-cell precursors, mature neutrophils | Not used | SS2/36 | X | X | — | — | ||||
| CD11b | NK cells, monocytes, neutrophils, myeloid precursors | ICRF44 | PE, APC | ICRF44 | X | — | — | — | |||
| CD14 | Myelomonocytic cells | TÛK4 | PE, FITC | M5E2 | X | X | X | X | |||
| CD15 | Neutrophils, monocytes, eosinophils | MMA | FITC, APC | HI98 | X | X | — | — | |||
| CD16 | NK cells, monocytes, neutrophils, myeloid precursors | NKP15 | FITC | DJ130c | X | X | — | — | |||
| CD19 | B cells | 4G7 | FITC | HIB19 | X | X | X | X | |||
| CD49d | Eosinophils | 9F10 | PE | 9F10 | — | — | X | X | |||
| CD56 | NK cells | My31 | PE | B159 | X | X | X | X | |||
| CD61 | Platelets, megakaryocytes, monocytes | Y2/51 | FITC, PE | VI-PL2 | X | X | X | X | |||
| GLY-A | Erythroid cells, erythroid precursors | JC159 | R-PE | GA-R2 | X | X | X | X | |||
MoAb clones used for MACS depletion and fluorescence activated cell sorting (FACS) analysis were directed against different epitopes of the same surface markers with exception of the anti-CD49d and anti-CD11b MoAbs.
— indicates not used.
To minimize changes in gene expression because of cellular activation all steps of the density gradient separation procedure and the immunomagnetic sorting were performed immediately after cell collection at 4°C or below—that is, on ice, in a cold room, or a cooled centrifuge, using nonpyrogenic reagents and plasticware.
The purity of each population was assessed by microscopy of Wright Giemsa-stained cytospins and by flow cytometry. Two- and 3-color flow cytometric analyses of purified populations were performed as described previously using the panel of fluorochrome-conjugated MoAbs listed in Table 1.13 Cell numbers were assessed using an improved Neubauer hemocytometer.
Total RNA and proteins were isolated from purified cell populations using TRIzol (Invitrogen, Paisley, United Kingdom) according to the manufacturer. To inhibit cellular protease diisopropyl fluorophosphate (DFP; Sigma, St Louis, MO) was added to the purified populations immediately before TRIzol isolation of RNA and proteins.
Microarray analysis
For gene expression analysis, total RNA was biotinylated and hybridized to HG-U133A and HG-U133B GeneChips (Affymetrix, Santa Clara, CA) according to the manufacturer's recommendations (www.affymetrix.com/pdf/expression_manual.pdf/).
Microarray fluorescence signals were normalized using the dCHIP v1.3 software.14 Expression indices for each gene were calculated using the perfect match/mismatch (PM/MM) difference model by Li and Wong.14 Negative values obtained by PM/MM calculations were converted into a defined background value of 20. All probe set lists were annotated with locus link identifications (IDs) provided by the NetAffx database (www.affymetrix.com/analysis/netaffx/index.affx) and converged into gene/EST (expressed sequence tag) lists by exclusion of redundant probe sets with identical locus link IDs. Genes were defined as expressed in cell populations if all replicates were assigned a present call by the MicroArray Suite 5 software (Affymetrix).
Statistical analysis was performed using the GeneSpring software (GeneSpring version 6.1; Silicon Genetics, Redwood City, CA). A Welsh 2-sample t test was applied to identify differentially expressed genes in PM, MY, bm-PMN, pb-PMN populations. The P values calculated by the Welsh t test were corrected for multiple testing (Benjamini-Hochberg)15 to estimate the false discovery rate for differentially regulated genes. A list of differentially expressed genes was generated at a false discovery rate of 0.05, and included genes with a 2-fold or more change of expression, and a minimum absolute difference of 75 between the highest and the lowest expression values.
Differentially expressed genes were annotated to gene categories according to their molecular functions using a simplified ontology tool provided by the GeneSpring 6.1 software and hierarchically clustered using the dChip v1.3 software.
Real-time RT-PCR
Changes of gene expression assessed by microarray analysis were confirmed by real-time reverse transcription-polymerase chain reaction (RT-PCR) for selected marker genes of granulocytic differentiation. Briefly, first-strand cDNA was generated by reverse transcription of 1 μg total RNA at 42°C for 1 hour using a T7-oligo(dT)24 primer and Superscript II according to the manufacturer's instructions (InVitrogen). First strand cDNA was then subjected to real-time PCR as described previously using forward primers, reverse primers, and probes listed in Table 2.16 The constitutively expressed housekeeping gene β-actin was used to normalize gene expression of marker genes.
List of primers and probes used for real-time PCR
. | Forward primer: 5′-3′ . | Reverse primer: 5′-3′ . | probe: 5′-3′ . |
|---|---|---|---|
| β-actin | CCT TTT TGT CCC CCA ACT TGA | TGG CTG CCT CCA CCC A | ATG TAT GAA GGC TTT TGG TCT CCC TGG GA |
| G-CSFR | CCC AGG CGA TCT GCA TAC TT | CAC AAA AGG CCA TCG GGT | AAG GAC CAG ATC ATG CTC CAT CCA TCC AGC |
| MPO | GTC CTG GTT AGC AGA GCT GGA | CAC ACA GCC CAG CTG CAG | CAG TGG TGC CTT GGA GGG ATA AGC C |
| Lactoferrin | CTC CCC AGG TGT GTT GGG | TAA GCA GAT GGA TGG GCA ATC | CCT TGG CTC CCC TGC TGA AGG TG |
| C/EBPα | GTT TTC TCG GAT ACT TGC CAA | ATT GGT CCC CCA GGA TCA A | AGA CTC TCC GTC GGC AGC TGG G |
| C/EBPϵ | GCT CCC TGG CAC GGT CTA A | ATC CAT GGT CTA TGT CTC AGG GTT | TCT GCG GAC CCC CAT CCT GCT |
. | Forward primer: 5′-3′ . | Reverse primer: 5′-3′ . | probe: 5′-3′ . |
|---|---|---|---|
| β-actin | CCT TTT TGT CCC CCA ACT TGA | TGG CTG CCT CCA CCC A | ATG TAT GAA GGC TTT TGG TCT CCC TGG GA |
| G-CSFR | CCC AGG CGA TCT GCA TAC TT | CAC AAA AGG CCA TCG GGT | AAG GAC CAG ATC ATG CTC CAT CCA TCC AGC |
| MPO | GTC CTG GTT AGC AGA GCT GGA | CAC ACA GCC CAG CTG CAG | CAG TGG TGC CTT GGA GGG ATA AGC C |
| Lactoferrin | CTC CCC AGG TGT GTT GGG | TAA GCA GAT GGA TGG GCA ATC | CCT TGG CTC CCC TGC TGA AGG TG |
| C/EBPα | GTT TTC TCG GAT ACT TGC CAA | ATT GGT CCC CCA GGA TCA A | AGA CTC TCC GTC GGC AGC TGG G |
| C/EBPϵ | GCT CCC TGG CAC GGT CTA A | ATC CAT GGT CTA TGT CTC AGG GTT | TCT GCG GAC CCC CAT CCT GCT |
Western blot analysis
Proteins copurifed with RNA from PM, MY, bm-PMN, and pb-PMN populations were electrophoresed on 7% and 10% sodium dodecyl sulfate (SDS) polyacrylamide gels (BDH Laboratory Supplies, Poole, United Kingdom) and transferred to nitrocellulose membranes (Amersham Bioscience, Uppsala, Sweden) by electroblotting. Subsequently, the membranes were incubated with primary antibodies raised against MPO (A0398; DAKO, Glostrup, Denmark), LF (A0186; DAKO), C/EBPϵ (C-22; Santa Cruz, Biotechnology, Santa Cruz, CA), C/EBPα (C-18; Santa Cruz) followed by a secondary horseradish peroxidase-conjugated swine antirabbit antibody (DAKO). Binding of antibodies was visualized by enhanced chemiluminescence (ECL; Amersham Bioscience). Loading of equal amounts of protein was assessed by reprobing membranes with a monoclonal primary anti-β-actin antibody (AC-15; Abcam, Cambridge, United Kingdom).
Results
Isolation of peripheral blood neutrophils and bone marrow populations highly enriched in promyelocytes, myelocytes, and neutrophils
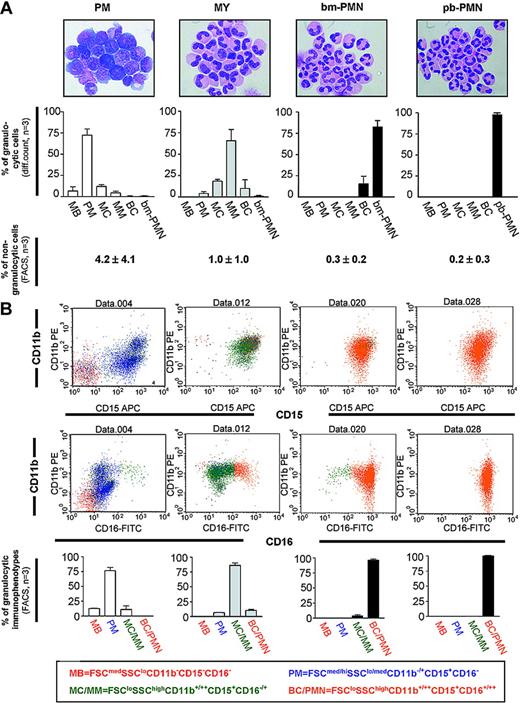
Highly enriched cell populations of PMs, MYs (the term MY refers to cells with both myelocytic and metamyelocytic morphology), and PMNs from human BM were isolated by combined density gradient centrifugation and immunomagnetic depletion of nongranulocytic cells including T and B cells, monocytes, eosinophils, megakaryocytes, and erythroid cells (Figure 1). Differential counts of the 3 purified populations clearly demonstrate a significant but not exclusive enrichment of either PMs, MYs, or bm-PMNs (Figure 1). The PM population contained mainly PMs (72% ± 6%, mean ± SD, n = 3), few MBs (myeloblasts; 7% ± 4%), and some MYs (12% ± 2%), resulting in a 6-fold higher frequency of PMs compared with MYs. In contrast, the MY population contained no MBs and a 21-fold lower frequency of PMs (4% ± 2%) compared with MYs (84% ± 13%). The most mature population contained mainly bm-PMNs (bone marrow neutrophils; 82% ± 8%), some BCs (band cells; 16% ± 9%), and almost no cell types representing earlier stages of differentiation (1% ± 1%).
Characterization of BM populations highly enriched in PMs, MYs, bm-PMNs, and pb-PMNs. (A, top row) Wright-Giemsa-stained cytospin preparations demonstrate the shape (× 1000 magnification) and the differential count of the purified BM and PB populations (mean ± SD, n = 3), including myeloblasts (MBs), promyelocytes (PMs), myelocytes (MCs), metamyelocytes (MMs), band cells (BCs), and neutrophils from bone marrow (bm-PMNs) and peripheral blood (pb-PMNs). (Bottom row) Two-color flow cytometry analysis was applied to determine the frequency of residual nongranulocytic cells (CD3, CD19, CD14, glycophorin-A, CD56, CD61) in cell populations following purification (mean ± SD, n = 3). (B, top and middle rows) Example of a typical 3-color flow cytometry analysis of BM and PB populations. (Bottom row) The purified populations were subcategorized as MBs, PMs, MCs/MMs, and BCs/PMNs based on cellular size, granularity, and expression profile of the CD15, CD11b, and CD16 surface markers (mean ± SD, n = 3).
Characterization of BM populations highly enriched in PMs, MYs, bm-PMNs, and pb-PMNs. (A, top row) Wright-Giemsa-stained cytospin preparations demonstrate the shape (× 1000 magnification) and the differential count of the purified BM and PB populations (mean ± SD, n = 3), including myeloblasts (MBs), promyelocytes (PMs), myelocytes (MCs), metamyelocytes (MMs), band cells (BCs), and neutrophils from bone marrow (bm-PMNs) and peripheral blood (pb-PMNs). (Bottom row) Two-color flow cytometry analysis was applied to determine the frequency of residual nongranulocytic cells (CD3, CD19, CD14, glycophorin-A, CD56, CD61) in cell populations following purification (mean ± SD, n = 3). (B, top and middle rows) Example of a typical 3-color flow cytometry analysis of BM and PB populations. (Bottom row) The purified populations were subcategorized as MBs, PMs, MCs/MMs, and BCs/PMNs based on cellular size, granularity, and expression profile of the CD15, CD11b, and CD16 surface markers (mean ± SD, n = 3).
The presence of MYs in both the PM population and the MY population probably reflects the continuous increase of density concomitant with differentiation among cells with myelocytic morphology. Thus, the MYs localized to the PM population have a lower density and are less differentiated compared with the major part of MYs, which constitute the MY population. Morphologic evaluation of pb-PMN (peripheral blood neutrophil) populations demonstrated a mean purity of 99.8% (range, 99.7%-99.9%; n = 3).
In a previous study, Terstappen et al17 applied multiparameter flow cytometry analysis to define stages of granulocytic differentiation based on cellular size, granularity, and the myeloid surface markers CD15, CD11b, and CD16. In accordance with that study, immunophenotypes representing MBs (FSCmedSSCloCD11b- CD15-CD16-) and PMs (FSCmed/hiSSClo/medCD11b+/-CD15+CD16-) were mainly detected in the PM population. However, the MY immunophenotype (FSCloSSChiCD11b+/++CD15+CD16-/+) was mainly detected in the MY population, whereas the PMN immunophenotype (FSCloSSChiCD11b-/+CD15+D16+/++) was detected almost exclusively in the bm-PMN population. Analysis of pb-PMNs revealed an immunophenotype similar to that of bm-PMNs (Figure 1).
These findings demonstrated that the isolated populations represented successive stages of granulocytic differentiation both by morphologic and immunophenotypical criteria. Additional flow cytometry analysis was applied to assess the frequency of residual nongranulocytic cells in the 4 isolated populations. As shown in Figure 1, the PM populations were contaminated with nongranulocytic cells by a mean of 4.2% (range, 1.8%-6%; n = 3), the MY populations by a mean of 1.0% (range, 0.5%-1.5%; n = 3), and the bm-PMN populations by a mean of 0.3% (range, 0.2%-0.4%; n = 3). The frequencies of contaminating nongranulocytic cells in pb-PMN populations were similar to those of bm-PMN populations (mean, 0.3%; range, 0.2%-0.4%; n = 3).
Microarray analysis further confirmed the low frequency of nongranulocytic cells in the isolated populations by failing to detect transcripts encoding lineage-specific genes highly expressed in T- and B-cells (CD3, CD19), monocytes (M-CSFR, detected at very low levels compared with G-CSFR), eosinophils (IL-5R), megakaryocytes (CD41/CD61), and erythroid cells (glycophorin-A). Hence, nongranulocytic cells in the isolated populations were unlikely to contribute to the gene expression profiles assessed by microarray analysis.
General characterization of granulocytic differentiation by microarray analysis
To investigate the molecular mechanisms that govern the transition from promyelocytes to mature neutrophils, we determined the gene expression profiles in highly purified human BM populations representing early (PMs), intermediate (MYs), and late (bm-PMNs) stages of terminal granulopoiesis. Among the 17 020 genes (44 760 probe sets) detected by the microarray, 11 310 genes (23 512 probe sets) were expressed in at least 1 of the BM populations (Affymetrix present/absent call). Moreover, 6700 genes (10 932 probe sets) were differentially expressed (Table S1, available on the Blood website; see the Supplemental Tables link at the top of the online article) as defined by a 2-fold or more change in gene expression and a false discovery rate 0.05 or less (assessed by Welch t test and Benjamini-Hochberg multiple correction). Differentially expressed genes were annotated to gene categories according to their molecular functions using a simplified ontology tool provided by the GeneSpring software package. This gene annotation revealed that differentially expressed genes were assigned to all of the major gene categories, including apoptosis, cell cycle, chaperones, enzymes, immunity proteins, kinases, motor proteins, signal transducers, structural proteins, and transcription factors.
In an attempt to define the “developmental distance” between PM, MY, and bm-PMN populations, we assessed the Pearson correlation coefficient (γ) by correlating the transcriptomes (all 44 760 probe sets; Affymetrix) of BM populations (Figure 2). The correlation coefficients among replicates of BM populations were in the range of 0.97 to 0.99. However, the correlation coefficients between BM populations were 0.81 (γPM - MY), 0.79 (γMY - bm-PMN), and 0.52 (γPM - bm-PMN) and, thus, provided a useful quantitative measure reflecting the hierarchical relationship between the 3 BM populations.
Developmental distance between PM, MY, bm-PMN, and pb-PMN populations. Schematic illustration of terminal granulocytic differentiation. The Pearson correlation coefficients (γ) were calculated by correlating the transcriptomes (all 44 760 probe sets on microarrays) of populations to define a quantitative measure reflecting the developmental distance between the 4 populations.
Developmental distance between PM, MY, bm-PMN, and pb-PMN populations. Schematic illustration of terminal granulocytic differentiation. The Pearson correlation coefficients (γ) were calculated by correlating the transcriptomes (all 44 760 probe sets on microarrays) of populations to define a quantitative measure reflecting the developmental distance between the 4 populations.
To explore at what point neutrophils complete differentiation, we compared gene expression patterns of bm-PMNs and pb-PMNs. The correlation coefficient between bm-PMN and pb-PMN transcriptomes revealed an almost identical gene expression pattern in the 2 populations (γbm-PMN - pb-PMN = 0.95, Figure 2), which strongly indicates that neutrophils complete their differentiation program in the BM microenvironment during steady-state hematopoiesis.
Overall, these findings indicate that the differentiation of promyelocytes into mature neutrophils is a highly complex process, which is governed by a massive change of gene expression involving all major functional gene categories.
Receptors and receptor ligands
To assess the cellular responsiveness toward immunoregulatory mediators and growth factors during granulocytic differentiation, we applied hierarchical clustering to identify expression patterns for receptors and receptor ligands annotated to the gene categories of signal transducers and immunity proteins (Figure 3).
Receptor and receptor ligands. Hierarchical clustering of differentially expressed receptors and receptor ligands in PM, MY, and bm-PMN populations annotated to the gene categories of signal transducers and immunity proteins. The mean relative expression levels for genes in BM populations are presented by a color scale, whereby red reflects high expression (3 × SD), white reflects mean expression, and blue reflects low expression (-3 × SD).
Receptor and receptor ligands. Hierarchical clustering of differentially expressed receptors and receptor ligands in PM, MY, and bm-PMN populations annotated to the gene categories of signal transducers and immunity proteins. The mean relative expression levels for genes in BM populations are presented by a color scale, whereby red reflects high expression (3 × SD), white reflects mean expression, and blue reflects low expression (-3 × SD).
Transcripts encoding the growth factor receptors FMS-like tyrosine kinase 3 (FLT-3) and c-kit, which sustain early hematopoiesis, were markedly down-regulated concomitant with cell cycle exit at the PM to MY transition (Figure 3). In contrast, growth factor receptors supporting terminal granulocytic differentiation, including the G-CSFR, GM-CSFR, and IL-6R were markedly up-regulated throughout differentiation.
Receptors for immunoregulatory mediators were generally expressed at low levels in PM and MY populations and markedly up-regulated in the bm-PMN population (Figure 3). Up-regulated receptors for cytokines and chemokines included interferon receptors, interleukin receptors (IL-R1, IL-R4, IL-R6, IL-R10, IL-R13, IL-R17, IL-R18), tumor necrosis factor receptors, tumor growth factor receptors, CXC chemokine receptors (IL8RA, IL8RB, CXCR4), and CC chemokine receptors (CCR1, CCR2, CCR3) (Figure 3). In addition, several members of the leukocyte immunoglobulin-like receptor family (LIR1, LIR2, LIR3, LIR4, LIR6, LIR7, LIR9) were found to be highly up-regulated in the bm-PMN population (Figure 3), which is the first documentation of LIR expression in neutrophils. This is of major interest as LIRs have been reported to bind major histocompatibility complex (MHC) class I molecules and to regulate the immune response of monocytes.18 In this context, one might speculate that various members of the LIR family might regulate the immune response of neutrophils. Other up-regulated molecules were critical for the recognition of microorganisms (Toll-like receptor 1 [TLR1], TLR2, TLR4, TLR6, TLR8; CD14; MYD88; MD-2; FPR1; TREM-1) and immunoglobulins (Fc receptors for IgA, IgE, IgG) (Figure 3). Finally, various death receptors were found to be highly up-regulated in the bm-PMN population (for details see “Apoptosis-related genes”).
Notably, some cytokines and chemokines were up-regulated in parallel with their receptors, including interleukin 1α/β (IL-1α/β), tumor necrosis factor (TNF), transforming growth factor-β (TGFβ), and ligands for the IL8RA and IL8RB (IL-8, growth-related oncogene-α [GROα], granulocyte chemotactic protein-2 [GCP-2]) (Figure 3). This is of particular interest, as autocrine stimulation of mature neutrophils in BM might prime cells for migration and immune response.
Taken together, the observed expression patterns for receptors and receptor ligands indicate a marked increase of cellular responsiveness toward immunoregulatory and apoptotic mediators concomitant with the termination of granulocytic differentiation.
Granule protein candidates
Neutrophils contain effector proteins of the innate immune system, which are stored in granules and transferred to phagosomes or the extracellular environment in response to various immunoregulatory mediators.1,2 Granulocytic differentiation is characterized by the sequential formation of primary, secondary, and tertiary granules and their constituting granule proteins (GPs).1,2 These findings have fostered the targeting-by-timing hypothesis, which states that targeting of GPs to a distinct subset of granules is determined by the time of their synthesis.19,20 Thus, primary, secondary, and tertiary GPs are readily identified by their unique gene expression profiles during granulocytic differentiation.
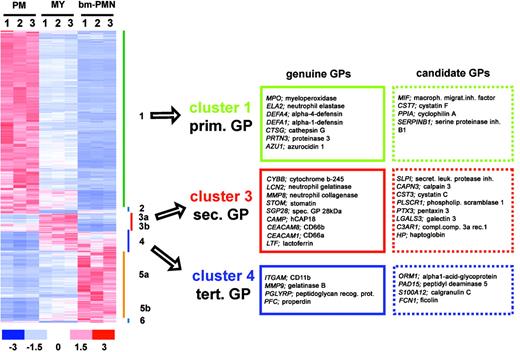
To identify new GPs, we searched for genes having expression profiles similar to those of genuine GPs. By hierarchical clustering we identified 3 gene clusters containing marker genes for primary (cluster 1), secondary (cluster 3), and tertiary (cluster 4) GPs (Figure 4). To further increase the probability of finding new GPs, we searched each cluster for genes having biologic functions similar to those of genuine GPs. By these criteria we identified 16 candidate GPs, including protease inhibitors, proteases, signaling molecules, and acute-phase proteins (Figure 4).
Identification of new GP candidates. (Left) Hierarchical clustering of 6700 differentially expressed genes in PM, MY, and bm-PMN populations. The relative expression levels for genes in each BM population (n = 9) are presented by a color scale, whereby red reflects high expression (3 × SD), white reflects mean expression, and blue reflects low expression (-3 × SD). (Right) Lists of genuine GPs and new GP candidates. Genes were defined as GP candidates if they had expression profiles similar to genuine GPs (ie, assigned to cluster 1, 3, 4) and had reported functions critical for immune response.
Identification of new GP candidates. (Left) Hierarchical clustering of 6700 differentially expressed genes in PM, MY, and bm-PMN populations. The relative expression levels for genes in each BM population (n = 9) are presented by a color scale, whereby red reflects high expression (3 × SD), white reflects mean expression, and blue reflects low expression (-3 × SD). (Right) Lists of genuine GPs and new GP candidates. Genes were defined as GP candidates if they had expression profiles similar to genuine GPs (ie, assigned to cluster 1, 3, 4) and had reported functions critical for immune response.
Among the primary GP candidates were the protease inhibitors cystatin F and serpin B1. Whereas cystatin F is a reported inhibitor of cathepsin proteases,21 serpin B1 is a well-characterized inhibitor of the primary granule proteases neutrophil elastase, cathepsin G, and proteinase-3.22 In addition, 2 other protease inhibitors exhibited expression profiles similar to secondary GPs. These included the secretory leukocyte protease inhibitor (SLPI), which is critical for proper wound healing by inhibiting elastase-dependent conversion of proepithelin to epithelin23 and cystatin C, which has been reported to inhibit autolytic cleavage of gelatinase, a protease stored in granules itself.24
These findings are in agreement with the concept that proteases and their cognate inhibitors are coexpressed in neutrophils to protect cells against proteolysis or to be exocytosed collectively upon activation and, thus, to promote a finely tuned proteolytic activity at sites of infection.
Another secondary GP candidate is galectin-3, a β-galactosidase-binding lectin, which facilitates bacterial phagocytosis by opsonization, and can induce a respiratory burst in neutrophils by binding to its receptors CD66a and CD66b.25 Notably, CD66a and CD66b are reported genuine GPs, which are mobilized to the cell surface by degranulation of neutrophils stimulated with bacterial lipopolysaccharide (LPS).25,26 Hence, to respond to galectin-3, neutrophils need to be primed by inflammatory mediators stimulating the exocytosis of granules. At present, galectin-3 is thought of as being released only by macrophages, epithelial cells, and endothelial cells in response to bacterial infection.25 However, our finding suggests that activated neutrophils can exocytose the GP candidate galectin-3 and its receptors CD66a and CD66b collectively, resulting in an autoregulatory enhancement of neutrophilic phagocytosis and respiratory burst.
The expression profiles of haptoglobin (Hp) and orosomucoid/α1-acid glycoprotein (AGP) were identical to those of secondary and tertiary GPs, respectively. Hp and AGP are both acute-phase proteins produced by hepatocytes in response to infections. The main task of Hp is to form a complex with free hemoglobin, which binds to the CD163 scavenger receptor expressed by macrophages, resulting in internalization and degradation of free hemoglobin.27 In contrast, AGP has a broad spectrum of immunoregulatory functions.28 Within minutes of injury, neutrophils are attracted to sites of infections, where they get activated and exocytose secondary and tertiary granules. In this context, our findings suggest that neutrophils are likely to serve as the primary local source of Hp and AGP at sites of infections. Indeed, this hypothesis is supported by preliminary data showing that Hp and AGP, like other secondary/tertiary GPs, are highly expressed at the protein level from the MY stage throughout granulocytic differentiation and are exocytosed by neutrophils following stimulation by phorbol myristate acetate (K.T.M. et al, unpublished data, 2004).
Overall, our cluster analysis implicates that neutrophils contain several yet unidentified effector proteins representing GP candidates.
Cell cycle-related genes
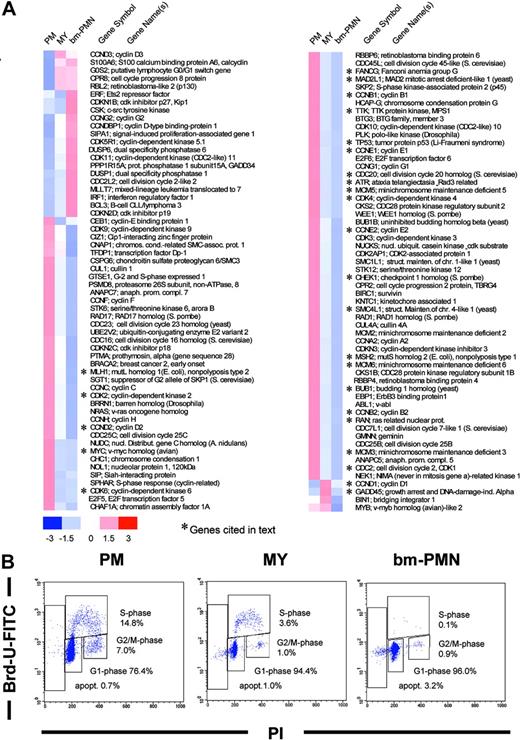
Cell cycle analysis demonstrated a high frequency of proliferating cells in the PM population (21.8% of cells in S/G2/M phase), a low frequency in the MY population (4.6% in S/G2/M phase), and a complete absence of proliferating cells among bm-PMNs (0.9% S/G2/M phase cells) (Figure 5). Hence, the abrupt decrease of proliferation in the MY population as compared with the PM population in our differentiation model prompted us to identify the changes of gene expression associated with cell cycle exit during terminal granulocytic differentiation. Cell cycle exit at the PM to MY transition was accompanied by a general down-regulation of transcripts encoding pro-proliferative genes (Figure 5). Potential mediators of this coordinated transcriptional repression are the E2 promoter binding factor (E2F) transcription factors, which have been shown to control transcription of several genes pivotal for G1/S phase progression, DNA replication, DNA repair, and mitosis.29-32 A detailed review of our cell cycle list revealed that all assigned E2F target genes30,31 were substantially down-regulated concomitant with cell cycle exit (Figure 5). Among the down-regulated E2F target genes were genes critical for G1/S phase progression and DNA replication (c-myc, cyclin E1 and E2, cyclin A, minichromosome maintenance [MCM] 3, 5, 6), DNA repair (MSH2, MLH1, Fanconi anemia/FANCG), DNA damage check-point (p53, CHEK1), and finally mitosis (cyclin B1 and B2, CDK1/CDC2, BUB1, CDC20, RAN, SMC4, MAD2L1, TTK) (Figure 5). E2F activity is primarily controlled by phosphorylation of the retinoblastoma protein (pRB) through cyclin-dependent kinases (CDKs) and their regulatory cyclins.33 In late G1/S phase CDK2, CDK4, and CDK6 phosphorylate pRB, resulting in the dissociation of inhibitory pRB/E2F complexes and subsequent transcription of E2F target genes. Microarray analysis demonstrated a concerted down-regulation of transcripts encoding the G1-phase CDKs (CDK2, CDK4, CDK6) at the PM to MY transition (Figure 5). These findings suggest a decrease of CDK activity at the PM to MY transition, which might contribute to the down-regulation of E2F target gene expression associated with cell cycle exit.
Cell cycle-related genes. (A) Hierarchical clustering of differentially expressed cell cycle-related genes in PM, MY, and bm-PMN populations. The mean relative expression levels for genes in BM populations are presented by a color scale, whereby red reflects a high expression (3 × SD), white reflects mean expression, and blue reflects low expression level (-3 × SD). (B) Representative flow cytometry analysis of G1, S, G2/M phases in PM, MY, bm-PMN populations. Cells to the left of the G1 population with less than 2n DNA content are undergoing apoptosis.
Cell cycle-related genes. (A) Hierarchical clustering of differentially expressed cell cycle-related genes in PM, MY, and bm-PMN populations. The mean relative expression levels for genes in BM populations are presented by a color scale, whereby red reflects a high expression (3 × SD), white reflects mean expression, and blue reflects low expression level (-3 × SD). (B) Representative flow cytometry analysis of G1, S, G2/M phases in PM, MY, bm-PMN populations. Cells to the left of the G1 population with less than 2n DNA content are undergoing apoptosis.
More recently, the transcription factors C/EBPα and C/EBPϵ have been shown to interact with E2F and to repress E2F-mediated transcription.34,35 Of interest, we found C/EBPα expression both at the mRNA and protein level in all BM populations, whereas C/EBPϵ mRNA and protein expression was up-regulated at the PM to MY transition when cell proliferation ceased (Figure 6). Thus, both C/EBPα and C/EBPϵ might contribute to cell cycle exit during granulocytic differentiation by inhibition of E2F target gene expression.
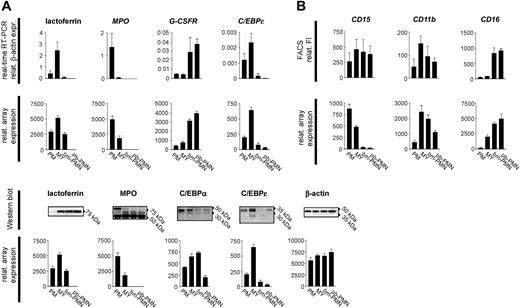
Validation of microarray data by real-time RT-PCR, Western blotting, and flow cytometry analysis. (A) Total RNA was purified from BM and PB populations and subjected to real-time RT-PCR and microarray analysis to compare gene expression profiles for selected marker genes of granulocytic differentiation (mean ± SD, n = 3). (B) Comparison of protein and mRNA profiles in BM and PB populations assessed by Western blotting, flow cytometry, and microarray analysis (mean ± SD, n = 3).
Validation of microarray data by real-time RT-PCR, Western blotting, and flow cytometry analysis. (A) Total RNA was purified from BM and PB populations and subjected to real-time RT-PCR and microarray analysis to compare gene expression profiles for selected marker genes of granulocytic differentiation (mean ± SD, n = 3). (B) Comparison of protein and mRNA profiles in BM and PB populations assessed by Western blotting, flow cytometry, and microarray analysis (mean ± SD, n = 3).
Apoptosis-related genes
The control of cell death is essential for the maintenance of cellular homeostasis in organisms. Apoptosis, the programmed form of cell death, operates by integrating multiple physiologic and pathologic death signals, which in turn trigger the release of apoptotic effectors from mitochondria (intrinsic pathway) and/or the activation of cytosolic effectors through death receptor signaling pathways (extrinsic pathway).36
We observed a marked differential expression of apoptosis-related genes, including members of the p53 pathway and B-cell lymphoma-2 (Bcl-2) family, caspases, and, in particular, death receptors and their corresponding signaling molecules (Figure 7).
Apoptosis-related genes. Hierarchical clustering of differentially expressed apoptosis-related genes in pm, MY, and bm-PMN populations. The mean relative expression levels for genes in BM populations are presented by a color scale, whereby red reflects high expression (3 × SD), white reflects mean expression level, and blue reflects low expression (-3 × SD).
Apoptosis-related genes. Hierarchical clustering of differentially expressed apoptosis-related genes in pm, MY, and bm-PMN populations. The mean relative expression levels for genes in BM populations are presented by a color scale, whereby red reflects high expression (3 × SD), white reflects mean expression level, and blue reflects low expression (-3 × SD).
The tumor suppressor p53 guards against genomic instability by its ability to induce apoptosis upon DNA damage.37 Transcripts for p53 (Figure 7) as well as the DNA damage check-point proteins (ATR, CHEK1, CHEK2), which stabilize and activate p53, were all down-regulated at the PM to MY transition (Figure 7), indicating that cell cycle exit is accompanied by a marked reduction of p53-dependent death control. More recently, the H1 histone isoform H1.2 (H1F2) was demonstrated to trigger apoptosis. Like all other H1 histones, H1.2 is released from the nucleus upon DNA damage, but exclusively activates the intrinsic apoptotic pathway by mediating cytochrome c release from mitochondria.38 Hence, H1.2 represents a direct link between DNA damage and induction of apoptosis through the intrinsic pathway. Transcripts encoding the H1.2 isoform but not other H1 isoforms increased as promyelocytes differentiated into mature neutrophils (Figure 7), suggesting that p53-independent apoptotic responses to DNA damage, affected by mediators such as H1.2, might be enhanced once cellular proliferation ceases during granulocytic differentiation.
Proapoptotic as well as antiapoptotic regulators of the extrinsic pathway were concomitantly up-regulated at the MY to bm-PMN transition (Figure 7). Among the proapoptotic proteins were death receptor ligands (TNF, tumor necrosis factor-related apoptosis-inducing ligand [TRAIL]), death receptors (TNFR1, TRAILR), and proteins propagating death receptor signals downstream (FADD, TRADD, initiator caspases 8 and 10), resulting in the activation of effector caspases and subsequent cell death.36 Up-regulated anti-apoptotic proteins of the extrinsic pathway included DcR1, a TRAIL decoy receptor, which cannot signal downstream, and FLIP, which inhibits death receptor signaling by blocking caspase 8 activation (Figure 7).36
The release of proapoptotic proteins from mitochondria constitutes a critical checkpoint in the intrinsic pathway, which is tightly regulated by the Bcl-2 family.36 The Bcl-2 proteins share 1 or more Bcl-2 homology domains (BH) and are divided into 3 main subclasses, the antiapoptotic BH1-4 proteins, and the proapoptotic BH1-3 and BH3-only proteins. Upon death signaling, the proapoptotic BH1-3 proteins BAX and BAK oligomerize within the mitochondrial membrane, an event which is thought to form pores releasing proapoptotic proteins.36,39 Transcripts encoding BAX and BAK were detected throughout granulocytic differentiation but not differentially expressed (data not shown). In contrast, BH1-4 and BH3-only family members that inhibit or promote BAX/BAK activity, respectively, were highly differentially regulated during granulocytic differentiation. Among the antiapoptotic BH1-4 proteins, only Bcl-2 was down-regulated, whereas Bcl-xL, Bfl-1/A1, and MCL-1 were up-regulated (Figure 7). Moreover, the proapoptotic BH3-only proteins BIM, NOXA, PUMA, and BID were all up-regulated throughout differentiation (Figure 7). The up-regulation of BID is of particular interest, as BID is activated through death receptor signaling (cleavage by caspase 8) and, thus, establishes a link between the extrinsic and intrinsic pathways in mature neutrophils (bm-PMN stage).40
The marked up-regulation of proapoptotic proteins suggests that granulocytic differentiation encompasses a priming of cells for apoptosis and particularly for apoptosis mediated by death receptor ligands. Apparently, the simultaneous up-regulation of antiapoptotic proteins might constitute a rheostat, which defines the threshold of responsiveness for apoptotic signals.41
Members of the NF-κB family have emerged as transcriptional regulators of proapoptotic as well as antiapoptotic proteins.42 Nuclear translocation of NF-κB and subsequent target gene expression is mediated by signaling through various receptors, including the IL-1R and TNFR.43 Notably, granulocytic differentiation was accompanied by a marked up-regulation of transcripts encoding IL-1R, TNFR1, and their corresponding ligands, suggesting an enhancement of NF-κB transcriptional activity. Accordingly, several apoptosis-related NF-κB target genes42 were up-regulated in mature neutrophils. These include the proapoptotic target genes FAS, the TRAILR, TNF, and TRAIL. Among the up-regulated antiapoptotic target genes were Bcl-xL, Bfl-1/A1, DcR1, FLIP, IEX1, GAAD45β, and MnSOD (Figure 7). Thus, our findings indicate that NF-κB is an important regulator of the apoptotic machinery emerging during terminal granulocytic differentiation.
In conclusion, these data indicate that the control of cell death during granulocytic differentiation is partially regulated through the p53-dependent pathway in proliferating cells (ie, at the PM stage). Cell cycle exit at the PM to MY transition is accompanied by a decline of p53-dependent death control and subsequent implementation of death control by H1.2 and death receptor pathways in mature neutrophils.
Validation of microarray data by real-time RT-PCR and Western blotting
To validate the microarray data, changes of gene expression in the PM, MY, bm-PMN, and pb-PMN populations were assessed by real-time RT-PCR and Western blot analysis (Figure 6). Real-time RT-PCR for selected markers of granulocytic differentiation, including MPO, LF, C/EBPϵ, C/EBPα, and G-CSFR, demonstrated gene expression profiles identical to the microarray expression profiles (Figure 6). Additional comparison of microarray data with previously reported Northern blot analysis of PM, MY, bm-PMN, and pb-PMN populations revealed identical expression profiles for 21 genes, including transcription factors and granule proteins.12,19 Given the conformity of microarray data and real-time RT-PCR/Northern blot data, the microarray analysis of the PM, MY, bm-PMN, and pb-PMN populations reflects a highly valid transcriptional profile of terminal granulopoiesis.
Changes of protein and mRNA expression were identical for some but not all investigated markers. Whereas protein and transcript profiles correlated well for C/EBPϵ, G-CSFR, CD16, and CD11b, they did not correlate for the early marker CD15, and the granule proteins MPO and LF (Figure 6). The latter is in accordance with previous studies demonstrating that primary granule proteins (MPO) and secondary granule proteins (LF) are synthesized transiently at the PM and MY stages, respectively, and remain stored in granules throughout terminal granulocytic differentiation.5,19
Discussion
Previously, no genomic approach has been used to assess the transcriptional program of terminal granulocytic differentiation in vivo. In the present study, we applied microarray technology to systematically profile the gene expression patterns in human BM populations highly enriched in PMs, MYs and bm-PMNs. The high number of differentially regulated genes (6700 genes/10 932 probe sets) reported here clearly exceeds that reported in other studies.44-47 However, those studies differ markedly from ours in several ways. First, gene expression was profiled using microarrays containing lower numbers of probe sets (12 600) compared with the microarrays used in the present study (44 760). Second, leukemic as well as genetically modified cell lines rather than primary cells were used. And finally, differentiation was induced in vitro by chemicals or a limited number of cytokines. As a consequence, transcriptional profiles for selected marker genes expressed in cell lines during terminal granulocytic differentiation differed partially from those observed in our in vivo study of human BM populations. For example, the human leukemic cell lines NB4 and HL60, which differentiate into morphologic mature neutrophils in response to retinoic acid, fail to express secondary/tertiary GPs.48,49 In contrast, the murine EML and MPRO cell lines, which were generated by transduction of a dominant-negative retinoic acid receptor (RAR) into normal murine progenitors, express secondary/tertiary GPs when differentiating into mature neutrophils in response to retinoic acid.50 However, these cell lines demonstrate expression profiles for transcription factors that differ partially from our in vivo profiles. Whereas, transcription factors like C/EBPα, C/EBPβ, c-myc, and c-myb demonstrated comparable expression profiles during MPRO and in vivo differentiation, others like p53, GATA2, NF-κB1, signal transducer and activator of transcription 3 (STAT3), C/EBPγ, and most importantly C/EBPϵ did not (Table S2).44,51 Overall, these findings indicate that expression profiles reported for in vitro differentiation of human and murine cell lines differ partially from those observed during in vivo differentiation.
Hence, at present our study provides the most complete and physiologic reliable data set representing the transcriptional program of terminal granulocytic differentiation.
To describe the differentiation process at the global level, we performed functional clustering of differentially regulated genes and defined the developmental distances among the BM populations. Those analyses demonstrate the involvement of all major gene categories in granulopoiesis and a striking developmental distance between the PM and bm-PMN populations as measured by a poor correlation of transcriptomes (γPM - bm-PMN = 0.52). Additional correlation of bm-PMN and pb-PMN transcriptomes revealed a high similarity of both populations (γbm-PMN - pb-PMN = 0.95), indicating a termination of granulocytic differentiation in the BM microenvironment.
More recently, Akashi et al52 reported a developmental distance of 0.87 (γHSC - CMP) between highly purified multipotential HSCs and lineage-restricted CMPs. In this context, our data suggest a less significant change of gene expression during early as compared with terminal hematopoietic differentiation. This observation might be explained by the fact that early differentiation of HSCs toward lineage-restricted progenitors requires a transcriptional program, which progressively down-regulates genes affiliated with multiple lineages (loss of function program). In contrast, terminal differentiation requires a transcriptional program, which directs the acquisition of new and highly specific functions critical for immune response concomitant with cell cycle exit (gain of function program).
In an attempt to define general mechanisms governing terminal differentiation, we reviewed differentially regulated genes assigned to the functional categories of cell cycle, apoptosis, receptors, and receptor ligands.
Flow cytometry analysis demonstrated the abrupt decrease of proliferation at the pm to MY transition, which allowed us to define the changes of gene expression associated with cell cycle exit during terminal differentiation. Cessation of proliferation was accompanied by a marked down-regulation of various metabolic pathways, mitochondrial genes, and virtually all proteasome-constituting genes (data not shown). In addition, transcripts encoding pro-proliferative genes, including 20 E2F target genes and the E2F-activity promoting kinases CDK2, CDK4, and CDK6, were markedly down-regulated concomitant with growth arrest. Hence, cell cycle exit during differentiation was characterized by a highly coordinated decline of cellular activity and the shutdown of proliferative activity, which to some extent was governed by inhibition of E2F target gene expression.
The transcription factors C/EBPα and C/EBPϵ have been shown to repress E2F-mediated transcription34,35 and were both expressed in PMs and MYs. Thus, C/EBPα and C/EBPϵ might contribute to cell cycle exit during granulocytic differentiation by inhibition of E2F target gene expression. This scenario is of particular interest as it directly links transcription factors critical for lineage commitment (C/EBPα) and terminal differentiation (C/EBPϵ) to cell cycle exit.8,11 Likewise, cell cycle exit might be linked to lineage-specific growth factors directing terminal differentiation, as G-CSF has been shown to promote transcription of C/EBPϵ through the G-CSFR signaling pathway.10
Primary GPs are early markers of granulocytic differentiation and are apparently transactivated by C/EBPα.5 Hierarchical clustering revealed identical expression profiles for primary GPs and pro-proliferative genes (ie, expressed in PMs and down-regulated at pm to MY transition when proliferation ceased). These findings suggest a general mechanism where key regulators of differentiation such as C/EBPα initiate differentiation (ie, transactivation of primary GPs), and successively promote cell cycle exit by repression of E2F-mediated transcription.
The expression patterns for apoptosis-related genes indicated that cell death is partially controlled through the p53-dependent pathway in proliferating PMs and through H1.2 and death receptor pathways in mature bm-PMNs.
This points to a general mechanism in which cell cycle exit and subsequent completion of differentiation define a switch of death control, which is accompanied by decreased responsiveness toward cancer chemotherapy concomitant with an increase of responsiveness toward death receptor ligands. These findings might have both biologic and clinical implications. First, high local or systemic levels of death receptor ligands induced by infection will primarily target terminally differentiated cells (bm-PMN) and not their progeny (pm, MY) and, thus, not interfere essentially with differentiation or tissue regeneration. Second, differentiation therapies for leukemia, such as the treatment of acute promyelocytic leukemia (APL) by retinoic acid, might reduce p53-dependent death control and consequently the responsiveness toward cancer chemotherapy. Indeed, the latter is substantiated by a prospectively randomized study showing that treatment of APL by retinoic acid and subsequent chemotherapy resulted in a significantly higher relapse rate as compared with the parallel treatment by retinoic acid and chemotherapy (16% versus 6%, P = .04).53
The immune response of neutrophils is orchestrated by various immunoregulatory mediators through receptor signaling. Microarray analysis revealed that receptors for chemokines, cytokines, and microorganisms were markedly up-regulated at the MY to bm-PMN transition, including several receptors not yet identified in neutrophils. In addition, some chemokines and cytokines were up-regulated in parallel with their receptors, suggesting an autocrine stimulation of bm-PMNs. Two of the up-regulated ligand-receptor pairs in bm-PMNs were IL-1/IL-1R and TNF/TNFR, which are potent inducers of NF-κB activity. This is of particular interest, since IL-1 and TNF are NF-κB target genes themselves.43 Hence, NF-κB might be activated constitutively in bm-PMNs by autocrine loops involving the ligand-receptor pairs IL-1/IL-1R and TNF/TNFR. In support of this notion, NF-κB activity was markedly increased at the MY to bm-PMN transition as defined by the up-regulation of a high number of NF-κB target genes related to immune response and apoptosis. Additional support is provided by Lu et al,54 who recently reported a constitutive NF-κB activation by autocrine loops in various cell lines.
These findings indicate that gain of responsiveness for immunoregulatory/apoptotic mediators is a most terminal event in granulocytic differentiation, which to some extent might be regulated by autocrine priming through the NF-κB signaling pathway. These findings further suggest a general mechanism whereby terminal differentiation is directed by a fail-safe program, which promotes completion of differentiation by avoiding premature responsiveness to activating stimuli.
In summary, the present study demonstrates that terminal granulocytic differentiation is a most complex process governed by a highly coordinated regulation of distinct genetic programs. In addition, this study identifies several new proteins in neutrophils, including effector proteins and surface receptors critical for cellular immune response. Finally, the present study provides the first human data set of terminal granulocytic differentiation, which might be used in comparative genomic studies to define dysregulated genes in myeloid leukemias, myelodysplastic syndromes, and disorders of neutrophils.
Prepublished online as Blood First Edition Paper, October 28, 2004; DOI 10.1182/blood-2004-08-3346.
Supported in part by The Novo Nordisk Foundation, the Amalie Jørgensens Memorial Foundation, the Danish Cancer Research Foundation, the Danish Medical Research Council, the Gangsted Foundation, the Toyota Foundation, the Carlsberg Foundation, and the Lundbeck Foundation. K.T.M. is the recipient of a scholarship from the IMK Foundation and Rigshospitalet.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank our colleagues Lene Udby, Pia Klausen, Carsten Niemann, Mikkel Schuster, and Bo Porse for critical review of the manuscript. The expert technical assistance of Marianne Lodahl is highly appreciated.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal