Abstract
Constitutively activating internal tandem duplication (ITD) mutations of the receptor tyrosine kinase FLT3 (Fms-like tyrosine kinase 3) play an important role in leukemogenesis, and their presence is associated with poor prognosis in acute myeloid leukemia (AML). To better understand FLT3 signaling in leukemogenesis, we have examined the changes in gene expression induced by FLT3/ITD or constitutively activated wild-type FLT3 expression. Microarrays were used with RNA harvested before and after inhibition of FLT3 signaling. Pim-1 was found to be one of the most significantly down-regulated genes upon FLT3 inhibition. Pim-1 is a proto-oncogene and is known to be up-regulated by signal transducer and activator of transcription 5 (STAT5), which itself is a downstream target of FLT3 signaling. Quantitative polymerase chain reaction (QPCR) confirmed the microarray results and demonstrated approximately 10-fold decreases in Pim-1 expression in response to FLT3 inhibition. Pim-1 protein also decreased rapidly in parallel with decreasing autophosphorylation activity of FLT3. Enforced expression of either the 44-kDa or 33-kDa Pim-1 isotypes resulted in increased resistance to FLT3 inhibition-mediated cytotoxicity and apoptosis. In contrast, expression of a dominant-negative Pim-1 construct accelerated cytotoxicity in response to FLT3 inhibition and inhibited colony growth of FLT3/ITD-transformed BaF3 cells. These findings demonstrate that constitutively activated FLT3 signaling up-regulates Pim-1 expression in leukemia cells. This up-regulation contributes to the proliferative and antiapoptotic pathways induced by FLT3 signaling. (Blood. 2005;105: 1759-1767)
Introduction
FLT3 (Fms-like tyrosine kinase 3) is a member of the class III receptor tyrosine kinase family, which includes KIT (c-kit receptor tyrosine kinase), FMS (macrophage-stimulating factor receptor), and 2 genes for the platelet-derived growth factor (PDGF) receptors. Both KIT and FMS play important roles in hematopoiesis.1 FLT3 is expressed by a number of human and murine cell lines of both myeloid and B-lymphoid lineage.2 FLT3 is also highly expressed on the malignant cells in most cases of acute myeloid leukemia (AML) and acute B-lineage leukemia (ALL).3,4 Internal tandem duplication (ITD) mutations of the juxtamembrane domain-coding sequence of the FLT3 gene have been identified in 17% to 34% of patients with AML and 5% of patients with myelodysplastic syndrome (MDS).5,6 In vitro studies have shown that FLT3/ITD receptors dimerize in a ligand-independent manner, leading to autophosphorylation of the receptor through constitutive activation of the tyrosine kinase domain.7 Recently, constitutive activation of FLT3 has also been shown to occur through autocrine mechanisms.8
Constitutive activation of FLT3 leads to autonomous, cytokine-independent growth with subsequent transformation of cells.9,10 The signaling through constitutively activated FLT3 is mediated at least partially by the Ras and signal transducer and activator of transcription 5 (STAT5) pathways.10-12 STAT5 appears to be an important target molecule in constitutively activated FLT3-mediated leukemogenesis, and its phosphorylation is inhibited by selective FLT3 tyrosine kinase inhibitors.13-17
CEP-701 is a potent and selective inhibitor for autophosphorylation of both wild-type and constitutively activated mutant FLT3 forms.13 CEP-701 improves survival in a murine leukemia model of FLT3/ITD-transformed cells and induces cytotoxicity in primary AML cells containing FLT3/ITD mutants.13
The proto-oncogene Pim-1 was first identified by its induction by proviral insertion in murine leukemia virus-induced T-cell lymphomas.18 It is a serine threonine kinase with increased expression in a variety of murine and human acute leukemias.19,20 Expression of Pim-1 increases cell mitogenesis and survival independent of growth factors.21 Pim-1 also synergizes with c-Myc in leukemogenesis and enhances transcriptional activity of c-Myb.22,23 Pim-1 may also protect hematopoietic cells from apoptosis induced by genotoxic stress or growth factor withdrawal, perhaps by directly targeting nuclear effectors.24
Expression of Pim-1 RNA and protein are normally induced by a number of cytokines, including granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte colony-stimulating factor (G-CSF), interleukin 3 (IL-3), IL-6, and interferon-α (IFN-α). This suggests that Pim-1 may partially mediate signal transduction through these receptors.25,26 Pim-1 is induced through Janus kinase 2 (Jak2)/STAT5-mediated growth factor signaling, implying it may play a role in the proliferation and/or differentiation of hematopoietic cells.20,25,27,28 Expression of Pim-1 protein specifically occurs in the G1/S-phase of the cell cycle, where it may also be important.29 Pim-1 is up-regulated in BaF3 cells expressing constitutively active mutant STAT5.30
Several binding and phosphorylation targets of Pim-1 have been identified. These include p100, which is an activator of the c-Myb transcription factor23 ; cdc25A, a protein phosphatase which activates Cdk2, leading to cell-cycle progression31 ; HP-1, a heterochromatin-binding protein that participates in gene silencing32 ; PAP-1 (phosphatidate phosphohydrolase-1), a protein functioning in transcription repression and splicing regulation33 ; TFAF2/SNX6 (tumor necrosis factor receptor-associated factor 4 and associated factor 2/sorting nexin 6)34 ; nuclear mitotic apparatus protein35 ; and p21, which is a cyclin-dependent kinase inhibitor.36 Pim-1 colocalizes with each of these proteins in the nucleus. Pim-1 expression alone is sufficient to induce cytokine independence in murine hematopoietic cells.37 Pim-1 was reported to cooperate with the antiapoptotic protein A1 in breakpoint cluster region/abelson murine leukemia (BCR/ABL)-mediated leukemogenesis.38 In this study, we have found that Pim-1 is overexpressed in cell lines expressing constitutively activated FLT3, and its expression dramatically decreases in response to FLT3 inhibitors. Pim-1 induction may be important in FLT3-mediated leukemogenesis through effects on cell proliferation and antiapoptosis.
Materials and methods
Reagents
CEP-701 and CEP-5214 were kindly provided by Cephalon (West Chester, PA) and stored as a 4-mM stock solution in dimethyl sulfoxide (DMSO) at -70 °C. They were diluted into RPMI-1640 media just before use. Rabbit anti-human FLT3 antibody (S18) and mouse anti-human Pim-1 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal mouse antiphosphotyrosine antibody (4G10) and recombinant protein A-agarose were purchased from Upstate Biotechnology (Lake Placid, NY). Horseradish peroxidase-conjugated secondary antibody and the enhanced chemiluminescence (ECL) detection system were from Amersham (Arlington Heights, IL). Recombinant murine IL-3, human flt3 ligand (FL), and GM-CSF were purchased from PeproTech (Rocky Hill, NJ).
Cells and plasmids
Cells were routinely maintained in RPMI-1640 medium with 10% heat-inactivated fetal bovine serum (FBS; Gemini Bio-Products, Woodland, CA) at 37°C in 5% CO2. MV4-11 cells were maintained in Iscoves modified Dulbecco medium (IMDM) with 20% FBS. Retroviral expression pLXSN plasmids for wild-type human 33-kDa and murine 44-kDa Pim-1 proteins and the dominant-negative PimNT81 mutant were used.39 The retroviral packaging line AmphoPack-293 was used to produce retroviruses (Clontech, Palo Alto, CA).
Patient samples
Bone marrow samples were collected with informed consent from patients with de novo AML or from healthy donors as part of a protocol approved by the Johns Hopkins Institutional Review Board. Mononuclear cells were separated and stored frozen as described previously.16
Gene expression profiling: RNA labeling, hybridization, and analysis
RNA labeling and hybridization was performed using the previously described techniques used by the Cancer Genetics Branch of the National Human Genome Research Institute (CGB/NHGRI).40,41 Approximately 50 to 100 μg treated and untreated control RNA were oligo (dT)-primed and labeled using reverse transcription, incorporating fluorescently labeled cyanine-3 (Cy-3) and Cy-5 deoxyuridine triphosphates (dUTPs; Amersham Pharmacia Biotech, Piscataway, NJ). Microarray slides (12 601 clones) derived from sequence-verified IMAGE consortium clones processed at the CGB/NHGRI and spotted by Agilent Technologies (Palo Alto, CA) were used for hybridization in the MOLM-14 and MV4-11 experiments. Microarray slides containing 15 360 sequence-verified cDNA clones were obtained from the National Human Genome Research Institute/National Institute of Neurological Disorders and Stroke/National Institute of Mental Health microarray core facility were used for the EOL-1 experiments. Gene names were assigned according to build 141 of the UniGene human sequence collection (www.ncbi.nlm.nih.gov/UniGene/). Microarray slides were scanned on an Agilent DNA Microarray Scanner. Image analyses were performed as previously described using DeArray software (Scanalytics, Fairfax, VA).42
RNA preparation and quantitative real-time polymerase chain reaction (QPCR)
Total RNA for cDNA microarray and QPCR experiments were extracted by Trizol reagent (Invitrogen, Carlsbad, CA) and purified by RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Quantitative real-time reverse transcription (RT)-PCR was performed using an iCycler iQ multicolor real-time PCR system (Bio-Rad, Richmond, CA). cDNA was synthesized and then amplified from 200 ng RNA using a “One-step” QuantiTect SYBR Green PCR kit (Qiagen). PCR cycles were 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. Cloned β-actin cDNA was used as a standard for quantification using specific sense (5-tgcgtgacattaaggagaag-3) and antisense (5-gctcgtagctcttctcca-3) primers. As an internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ribosomal protein S16 RNA were also detected by quantitative RT-PCR using specific primers (5′-CACGAACCACGGCACTGATT-3′ and 5′-TTTTCTTGCTGCCAGTCTGGAC-3′ for GAPDH, 5′-ATATTCGGGTCCGTGTGAAG-3′ and 5′-CTTGGAGGCTTCATCCACAT-3′ for S16). Each of the PCR assays was run in triplicate, and the gene copy numbers were estimated from the threshold amplification cycle numbers using software supplied with the iCycler IQ Thermal Cycler. Pim-1 expressions from human and mouse were amplified with the following primers: human Pim-1, 5′-CGAGCATGACGAAGAGATCAT-3′ and 5′-TCGAAGGTTGGCCTATCTGA-3′; mouse Pim-1, 5′-GATCATCAAGGGCCAAGTGT-3′ and 5′-GATGGTTCCGGATTTCTTCA-3′.
Proliferation/apoptosis assay
Cell proliferation assays using 3-4,5-dimethylthiazol-2,5-diphenyltetrazolium (MTT) and apoptosis assays using annexin V binding were performed as described previously.13 Briefly, for the MTT assay, 50 μL media was divided into aliquots into triplicate wells of 96-well plates for each CEP-701 concentration. All wells contained the same concentration of DMSO. Cell suspension in media (50 μL) was added to each well (50 000 cells/well). Plates were incubated for 24 or 48 hours at 37°C in 5% CO2, then the MTT assay was performed according to the manufacturer's instructions (Roche, Indianapolis, IN). For the annexin V binding apoptosis assay, cells were incubated for 24 hours in media with increasing or fixed CEP-701 concentration, then annexin V and 7-aminoactinomycin D (7-AAD) binding was performed and assessed by flow cytometry according to the manufacturer's instructions (Clontech).
Colony-forming assay
FLT3/ITD-expressing BaF3 cells (BaF3/ITD) were electroporated with 15 μg of the N-terminal deletion dominant-negative Pim-1 expression vector (Pim-1/NT81) or control vector (pLXSN), both of which confer G418 resistance. One day after electroporation, cells were seeded at concentrations of 100, 500, and 2500 cells per dish in 1 mL culture mix using MethoCult (StemCell Technologies, Vancouver, BC, Canada) containing 1% methylcellulose in Iscoves MDM, 30% FBS, 1% bovine serum albumin (BSA), 2 mM l-glutamine, 0.1 mM 2-mercaptoethanol, and 0.6 mg/mL G418. The colonies were grown at 37°C in 5% CO2 and counted on day 7.
Generation of retrovirus and infection of cells
High-titer retrovirus was generated by transfection of the AmphoPack-293 retrovirus packaging line. Briefly, 5 × 106 AmphoPack-293 cells were seeded 1 day before transfection with 24 μg plasmid DNA (pLXSN constructs) by the lipofectamine method (Invitrogen). The virus-containing medium was harvested every 48 hours after transfection and filtered through a 0.45-μm filter. After addition of 8 μg/mL Polybrene (Sigma, St Louis, MO), the medium was used to infect BaF3 cells for 24 hours.
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting were performed as described previously.13 Briefly, cells were incubated in culture medium with CEP-701 or CEP-5214 for various times at 37°C, then washed with ice-cold phosphate-buffered saline (PBS), and lysed with 1% NP-40 lysis buffer. Clarified lysate (500 μg) per sample was incubated at 4°C overnight with anti-FLT3 antibody or anti-Pim-1 monoclonal antibody, followed by addition of protein A agarose (Upstate Biotechnology) for an additional 2 hours of incubation. After electrophoresis and transfer to Immobilon membranes (Millipore, Bedford, MA), immunoblotting was performed using antiphosphotyrosine antibody 4G10 to assess phosphorylated FLT3, or the appropriate protein-specific antibodies. Protein bands were visualized using chemiluminescence and scanned with an AGFAArcus 1200 laser scanner.
Results
Pim-1 RNA expression decreases in response to FLT3 inhibition and recovers when inhibition is released
To better understand how FLT3 contributes to leukemogenesis, we examined the global changes in gene expression mediated by inhibition of constitutively activated FLT3. A number of potent, selective inhibitors of FLT3 have been developed, and several are currently in clinical trials. The FLT3 inhibitor CEP-701 inhibits autophosphorylation of wild-type and constitutively activated mutant FLT3 in vitro in human FLT3-expressing myeloid leukemia-derived and murine FLT3-transformed model cell lines.13 A second inhibitor, CEP-5214, also inhibits FLT3 phosphorylation with approximately the same potency as CEP-701 (D.S., unpublished data, October 2000). Because these inhibitors can reversibly turn off FLT3 signaling, we made RNA from several cell lines which express activated FLT3, before and after incubation with CEP-701, and following increasing periods of inhibitor washout. This included the EOL-1 cell line, which expresses wild-type FLT3 which is constitutively activated by autocrine expression of FL,8 the FLT3/ITD mutant MOLM-14 and MV4-11 cell lines, and FLT3/ITD-transformed murine BaF3 cells. The effect of these treatments on cytotoxicity and on FLT3 phosphorylation and expression in the EOL-1 cell line is shown in Figure 1. CEP-701 inhibits FLT3 phosphorylation in a dose-dependent manner with close to complete inhibition occurring by 50 nM (Figure 1A, inset). The inhibition is reversible in that FLT3 phosphorylation returns after inhibitor is washed away (Figure 1B, lanes 4,5 versus 2,3). CEP-701 induced the same changes in FLT3 phosphorylation in the other cell lines (data not shown).
CEP-701 inhibits FLT3 autophosphorylation reversibly and induces cytotoxicity in EOL-1 cells. (A) EOL-1 cells were incubated with increasing concentrations of CEP-701. The graph shows cytotoxicity assessed by MTT assay after 48 hours. The inset shows whole-cell extracts (500 μg/sample) that were prepared after 1 hour of exposure to CEP-701 and immunoprecipitated with anti-human FLT3 antibody, followed by separation in 8% polyacrylamide gel electrophoresis and immunoblotting with antiphosphotyrosine antibody. The membrane was stripped and reprobed with anti-FLT3 antibody to demonstrate equal loading of FLT3 in each lane. (B) Aliquots of EOL-1 cells were incubated with 50 nM CEP-701 for 4 hours and 8 hours. Additional cell aliquots were then washed by PBS to remove the inhibitor and incubated for an additional 4 and 8 hours. Most of the cells from each of these time points were harvested for RNA preparation for microarray analysis. A small fraction was harvested for analysis of FLT3 phosphorylation status as shown.
CEP-701 inhibits FLT3 autophosphorylation reversibly and induces cytotoxicity in EOL-1 cells. (A) EOL-1 cells were incubated with increasing concentrations of CEP-701. The graph shows cytotoxicity assessed by MTT assay after 48 hours. The inset shows whole-cell extracts (500 μg/sample) that were prepared after 1 hour of exposure to CEP-701 and immunoprecipitated with anti-human FLT3 antibody, followed by separation in 8% polyacrylamide gel electrophoresis and immunoblotting with antiphosphotyrosine antibody. The membrane was stripped and reprobed with anti-FLT3 antibody to demonstrate equal loading of FLT3 in each lane. (B) Aliquots of EOL-1 cells were incubated with 50 nM CEP-701 for 4 hours and 8 hours. Additional cell aliquots were then washed by PBS to remove the inhibitor and incubated for an additional 4 and 8 hours. Most of the cells from each of these time points were harvested for RNA preparation for microarray analysis. A small fraction was harvested for analysis of FLT3 phosphorylation status as shown.
RNA from each of the cell lines from the same time points of CEP-701 treatment was then hybridized to cDNA microarrays. Changes in gene expression were then analyzed, looking for genes whose expression consistently changed in each of the cell lines in response to FLT3 inhibition and then returned toward baseline after inhibition was released. Several genes showed changes in response to FLT3 inhibition by 50 nM CEP-701 for 4 hours and 8 hours (Figure 2). Among the genes most consistently affected was Pim-1, which was down-regulated upon FLT3 inhibition and then returns toward increased levels after inhibition was released by washing away the inhibitor (Table 1). Because of its known role as a proto-oncogene and its consistent expression changes in response to FLT3 inhibition, Pim-1 was chosen for further study.
Gene expression changes in EOL-1, MV4-11, and MOLM-14 cells treated with CEP-701. RNAs harvested from cell cultures treated as shown in Figure 1B were hybridized to cDNA microarray slides to examine changes in gene expression. RNA from uninhibited cells was labeled with Cy-5 dUTPs (green fluorescence signal). RNAs from inhibited cells or cells after inhibitor washout were labeled with Cy-3 dUTPs (red fluorescence signal). The uninhibited cell RNA was used as the control to examine changes in inhibited cell RNAs after 4 and 8 hours of inhibition or 4 hours and 8 hours release after inhibition. The inhibited RNA after 4 and 8 hours of inhibition or 4 hours and 8 hours of release after inhibition were represented as 4 hours inhibited, 8 hours inhibited, 4 hours released, and 8 hours released, respectively. The figure shows a pseudocolor representation of the intensity ratios of the raw data from the 2-channel fluorescent images. These data are quality filtered, and the top 15 genes ordered by degree of decreased expression in the EOL-1 cells inhibited by CEP-701 for 8 hours are shown.
Gene expression changes in EOL-1, MV4-11, and MOLM-14 cells treated with CEP-701. RNAs harvested from cell cultures treated as shown in Figure 1B were hybridized to cDNA microarray slides to examine changes in gene expression. RNA from uninhibited cells was labeled with Cy-5 dUTPs (green fluorescence signal). RNAs from inhibited cells or cells after inhibitor washout were labeled with Cy-3 dUTPs (red fluorescence signal). The uninhibited cell RNA was used as the control to examine changes in inhibited cell RNAs after 4 and 8 hours of inhibition or 4 hours and 8 hours release after inhibition. The inhibited RNA after 4 and 8 hours of inhibition or 4 hours and 8 hours of release after inhibition were represented as 4 hours inhibited, 8 hours inhibited, 4 hours released, and 8 hours released, respectively. The figure shows a pseudocolor representation of the intensity ratios of the raw data from the 2-channel fluorescent images. These data are quality filtered, and the top 15 genes ordered by degree of decreased expression in the EOL-1 cells inhibited by CEP-701 for 8 hours are shown.
cDNA microarray results of Pim-1 expression from several human myeloid leukemia-derived cell lines in response to FLT3 inhibition
. | 4-h inhibition with CEP-701 . | 4-h release after 4-h inhibition . | 8-h inhibition with CEP-701 . | 8-h release after 8-h inhibition . |
|---|---|---|---|---|
| EOL-1, fold change | 0.16 | 0.34 | 0.17 | 0.40 |
| MOLM-14, fold change | 0.31 | 0.48 | 0.45 | 0.70 |
| MV4-11, fold change | 0.27 | 0.41 | 0.26 | 0.31 |
. | 4-h inhibition with CEP-701 . | 4-h release after 4-h inhibition . | 8-h inhibition with CEP-701 . | 8-h release after 8-h inhibition . |
|---|---|---|---|---|
| EOL-1, fold change | 0.16 | 0.34 | 0.17 | 0.40 |
| MOLM-14, fold change | 0.31 | 0.48 | 0.45 | 0.70 |
| MV4-11, fold change | 0.27 | 0.41 | 0.26 | 0.31 |
The table shows the fold difference of raw data of Pim-1 signals in CEP-701-treated cells relative to untreated controls.
P < .02, derived from 1-sided pairwise t test from 6 independent experiments (4-h inhibition versus 4-h release after 4-h inhibition and 8-h inhibition versus 8-h release after 8-h inhibition from EOL-1, MOLM-14, and MV4-11). The P value was calculated from log-transformed data.
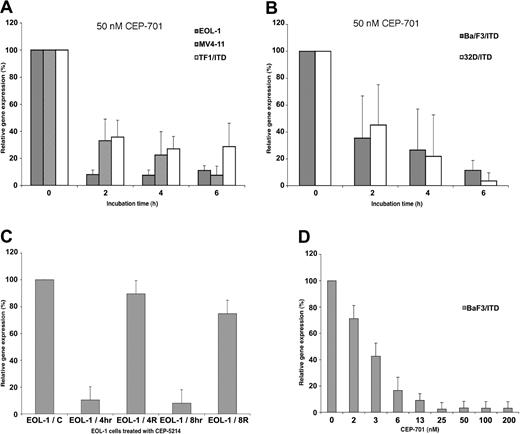
To confirm that Pim-1 expression was a consistent downstream target of FLT3 signaling, we performed an independent time-course of FLT3 inhibition using several human FLT3-expressing myeloid leukemia-derived (EOL-1, MV4-11) and human (TF1/ITD) and murine (BaF3/ITD, 32D/ITD) FLT3-transformed model cell lines using 2 different FLT3 inhibitors, CEP-701 and CEP-5214. The resulting RNA was then used in a series of QPCR experiments to examine changes in Pim-1 expression in response to FLT3 inhibition with 50 nM CEP-701 (Figure 3A-B). The RNA levels of Pim-1 exhibited at least 3-fold decrease in each of the human leukemic-derived cell lines with FLT3 inhibition within 2 hours. Levels in some cell lines decreased to as little as 10% the levels observed in the absence of FLT3 inhibition. The same changes were also observed in both of the murine FLT3/ITD-transformed cell lines. A second FLT3 inhibitor (CEP-5214) was also used to confirm that the observed down-regulation of Pim-1 expression did not result from effects specific to CEP-701. Like CEP-701, FLT3 inhibition with CEP-5214 also resulted in a down-regulation of Pim-1 expression, and expression returned toward baseline after washing out the inhibitor to allow FLT3 signaling to resume (Figure 3C). A dose-response experiment with CEP-701 was also carried out with the BaF3/ITD cell line for 2 hours. A dose dependency was observed for the BaF3/ITD cells with increasing down-regulation observed in the 2 to 13 nM range (Figure 3D). Maximal inhibition of Pim-1 expression in EOL-1 cells was obtained by 10 nM CEP-701 treatment (data not shown).
Pim-1 decreases in response to FLT3 inhibition in EOL-1, MV4-11, TF1/ITD, BaF3/ITD, and 32D/ITD cells. Cell lines were incubated for increasing times or with increasing concentrations of CEP-701 or CEP-5214. RNA was then harvested and used for Pim-1 expression analysis by QPCR. GAPDH or ribosomal S16 gene expression were used as internal controls to normalize the level of Pim-1 expression. Inhibition of Pim-1 expression by 50 nM CEP-701 with increasing time of treatment in (A) the constitutively activated FLT3-expressing human leukemia-derived cell lines, EOL-1, MV4-11, TF1/ITD; and (B) the constitutively activated FLT3/ITD-transformed murine cell lines, BaF3/ITD and 32D/ITD. (C) Changes in Pim-1 expression in EOL-1 cells after treatment and release from 50 nM CEP-5214 inhibition. EOL-1/C indicates control; EOL-1/4h, 4-hour inhibition; EOL-1/4R, 4-hour release after 4 hours of inhibition; EOL-1/8h, 8-hour inhibition; EOL-1/8R, 8-hour release after 8 hours of inhibition. (D) Pim-1 response in BaF3/ITD cells to increasing concentrations of CEP-701 treatment for 2 hours.
Pim-1 decreases in response to FLT3 inhibition in EOL-1, MV4-11, TF1/ITD, BaF3/ITD, and 32D/ITD cells. Cell lines were incubated for increasing times or with increasing concentrations of CEP-701 or CEP-5214. RNA was then harvested and used for Pim-1 expression analysis by QPCR. GAPDH or ribosomal S16 gene expression were used as internal controls to normalize the level of Pim-1 expression. Inhibition of Pim-1 expression by 50 nM CEP-701 with increasing time of treatment in (A) the constitutively activated FLT3-expressing human leukemia-derived cell lines, EOL-1, MV4-11, TF1/ITD; and (B) the constitutively activated FLT3/ITD-transformed murine cell lines, BaF3/ITD and 32D/ITD. (C) Changes in Pim-1 expression in EOL-1 cells after treatment and release from 50 nM CEP-5214 inhibition. EOL-1/C indicates control; EOL-1/4h, 4-hour inhibition; EOL-1/4R, 4-hour release after 4 hours of inhibition; EOL-1/8h, 8-hour inhibition; EOL-1/8R, 8-hour release after 8 hours of inhibition. (D) Pim-1 response in BaF3/ITD cells to increasing concentrations of CEP-701 treatment for 2 hours.
Thus, the QPCR results correlate with the microarray data shown previously. These findings suggest that FLT3 up-regulates Pim-1 expression in leukemic cells and may be part of the pathway by which FLT3 transforms cells. Therefore, Pim-1 expression appears to be highly dependent on constitutively activated FLT3 signaling in several cell lines, and its expression appears to be regulated at the level of RNA.
Pim-1 protein decreases in response to FLT3 inhibition
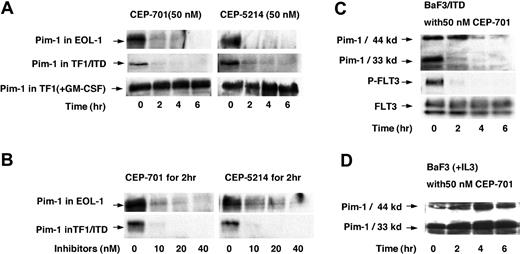
RNA levels do not always correlate with changes in protein expression. To determine whether the changes in mRNA result in changes to protein expression, cell lysates of EOL-1 and TF1/ITD cells were immunoprecipitated with anti-Pim-1 antibody followed by immunoblotting for Pim-1 (Figure 4). Pim-1 protein levels were high in the absence of FLT3 inhibition and greatly decreased by 2 hours of treatment with 50 nM CEP-701 or CEP-5214, the earliest time point assayed (Figure 4A). Thus, Pim-1 protein levels parallel the observed decreases in Pim-1 RNA expression. Pim-1 protein expression is known to be induced by GM-CSF in TF1 cells, which are dependent on GM-CSF for growth, and lack FLT3 expression. Pim-1 protein levels were unaffected by CEP-701 or CEP-5214 treatment of GM-CSF-stimulated TF1 cells (Figure 4A). This lack of effect on the GM-CSF-mediated signaling pathway leading to Pim-1 expression confirms that the Pim-1 response to CEP-701 is not a direct effect on Pim-1 but only occurs when FLT3 signaling is responsible for inducing expression of Pim-1. The inhibition of Pim-1 expression was very sensitive to FLT3 inhibition as most of the changes in expression occurred by 10 nM, a level only 3-fold the IC50 (inhibitory concentration 50%) (Figure 4B).
Pim-1 protein expression decreases in response to FLT3 inhibition. EOL-1, TF1/ITD, and BaF3/ITD cells were incubated with 50 nM CEP-701 or CEP-5214 for 0 to 6 hours or incubated with an increasing concentration of the inhibitors for 2 hours. Cell extracts were immunoprecipitated with anti-Pim-1 antibody followed by immunoblotting with the same antibody. (A) Time course of treatment with 50 nM CEP-701 or CEP-5214 in EOL-1, TF1/ITD, and TF1 (treated with GM-CSF) cells. (B) Dose response to increasing concentrations of CEP-701 or CEP-5214 treatment of EOL-1 and TF1/ITD cells for 2 hours. (C) Changes in Pim-1 expression, FLT3 phosphorylation, and expression in BaF3/ITD cells in response to treatment with 50 nM CEP-701 for increasing times. Note that mouse Pim-1 also expresses a 44-kDa isoform. (D) Pim-1 expression in BaF3 cells treated with IL-3 plus 50 nM CEP-701 for increasing times.
Pim-1 protein expression decreases in response to FLT3 inhibition. EOL-1, TF1/ITD, and BaF3/ITD cells were incubated with 50 nM CEP-701 or CEP-5214 for 0 to 6 hours or incubated with an increasing concentration of the inhibitors for 2 hours. Cell extracts were immunoprecipitated with anti-Pim-1 antibody followed by immunoblotting with the same antibody. (A) Time course of treatment with 50 nM CEP-701 or CEP-5214 in EOL-1, TF1/ITD, and TF1 (treated with GM-CSF) cells. (B) Dose response to increasing concentrations of CEP-701 or CEP-5214 treatment of EOL-1 and TF1/ITD cells for 2 hours. (C) Changes in Pim-1 expression, FLT3 phosphorylation, and expression in BaF3/ITD cells in response to treatment with 50 nM CEP-701 for increasing times. Note that mouse Pim-1 also expresses a 44-kDa isoform. (D) Pim-1 expression in BaF3 cells treated with IL-3 plus 50 nM CEP-701 for increasing times.
We also examined the dependence of Pim-1 expression on FLT3/ITD signaling in BaF3/ITD cells (Figure 4C-D). Murine cells express both a 44-kDa and 33-kDa form of Pim-1. The 44-kDa protein results from use of an upstream alternative translation initiation site.43 As shown in Figure 4C, the level of both isoforms of Pim-1 decrease in response to inhibition of FLT3 by 50 nM CEP-701. CEP-701 does not interfere with the Pim-1 induction pathway mediated by IL-3 in BaF3 cells (Figure 4D). This is further evidence that the observed suppression of Pim-1 expression in BaF3/ITD cells occurs in response to FLT3 inhibition.
Pim-1 is highly expressed in FLT3/ITD-transformed cells but is little induced by FL stimulation of wild-type FLT3-expressing cells
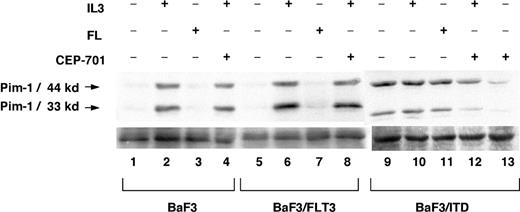
Pim-1 is known to be induced by a variety of cytokines, including GM-CSF, G-CSF, IL-3, IL-6, and IFN-α. It is possible that “normal” FLT3 signaling induced by FL stimulation of wild-type FLT3 also stimulates Pim-1 expression. To investigate this possibility, parental BaF3, BaF3/FLT3, and BaF3/ITD cells were examined under conditions of cytokine deprivation or stimulation for 2 hours with IL-3 or FL (Figure 5). While IL-3 was able to induce Pim-1 expression in BaF3 parental and BaF3/FLT3 cells (lane 1 versus 2 and 5 versus 6), FL was unable to do so (lane 7), despite increasing FLT3 phosphorylation (data not shown). Treatment with CEP-701 did not block IL-3-mediated Pim-1 expression (lanes 4, 8, and 12). It did, however, interfere with FLT3/ITD-mediated Pim-1 expression (lane 13 versus 9). The same results were seen using TF-1, TF-1/FLT3, and TF-1/ITD cells (data not shown). To confirm the Western blot results, we measured Pim-1 RNA levels by QPCR. Pim-1 RNA levels in BaF3/ITD cells were highly elevated with or without FL, but Pim-1 RNA was barely increased by FL stimulation in BaF3/FLT3 cells (data not shown). Thus, signaling through transient FL-activated wild-type FLT3 is not the same as signaling through ITD mutant FLT3. This FLT3/ITD-dependent pattern may explain why the presence of ITDs have bad prognostic implications in patients with AML.44
Pim-1 is expressed in BaF3/ITD cells but not by FL stimulation of BaF3/FLT3 cells. Comparison of Pim-1 expression in BaF3 cells, FLT3-transformed BaF3 cells, and FLT3/ITD-transformed BaF3 cells treated with or without IL-3, FL, or CEP-701. Cell extracts (100 μg/sample) were resolved by 12% polyacrylamide gel electrophoresis and immunoblotted with anti-Pim-1 antibody. Lanes 1, 5, and 9 have no cytokines or CEP-701 treatment; lanes 2, 6, and 10, IL-3 (1 ng/mL) for 2 hours; lanes 3, 7, 11, FL (10 ng/mL) for 2 hours; lanes 4, 8, 12, IL-3 (1 ng/mL) and 50 nM CEP-701 for 2 hours; and lane 13, 50 nM CEP-701 for 2 hours.
Pim-1 is expressed in BaF3/ITD cells but not by FL stimulation of BaF3/FLT3 cells. Comparison of Pim-1 expression in BaF3 cells, FLT3-transformed BaF3 cells, and FLT3/ITD-transformed BaF3 cells treated with or without IL-3, FL, or CEP-701. Cell extracts (100 μg/sample) were resolved by 12% polyacrylamide gel electrophoresis and immunoblotted with anti-Pim-1 antibody. Lanes 1, 5, and 9 have no cytokines or CEP-701 treatment; lanes 2, 6, and 10, IL-3 (1 ng/mL) for 2 hours; lanes 3, 7, 11, FL (10 ng/mL) for 2 hours; lanes 4, 8, 12, IL-3 (1 ng/mL) and 50 nM CEP-701 for 2 hours; and lane 13, 50 nM CEP-701 for 2 hours.
Pim-1 plays an important role in FLT3-mediated cell survival
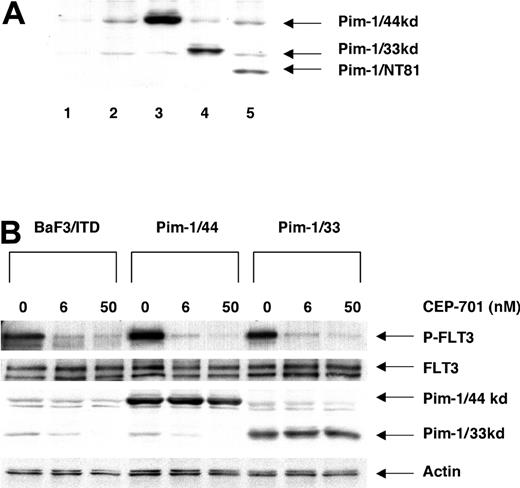
Pim-1 might be an important part of the process by which FLT3/ITD signaling contributes to leukemogenesis. To investigate the biologic function of Pim-1 in the FLT3/ITD signaling pathway, we transfected BaF3/ITD cells with wild-type 44-kDa Pim-1 (Pim-1/44), wild-type 33-kDa Pim-1 (Pim-1/33), and a dominant-negative N-terminal deletion mutant of Pim-1 (Pim-1/NT81). Expression of the Pim-1 constructs was confirmed by Western blotting of cell lines that were cloned by limiting dilution (Figure 6A). FLT3 signaling is turned off by treatment with CEP-701 for 1 hour, and endogenous Pim-1 expression was inhibited in turn (Figure 6B). In contrast, expression of the transfected Pim-1 isotypes was not affected by FLT3 inhibition (Figure 6B).
Expression of Pim-1 constructs in stably transfected BaF3 cells. (A) Expression of Pim-1 constructs was detected by monoclonal anti-Pim-1 antibody from abstracts of transfected cell lines. Extracts (100 μg/sample) were resolved by separation in 12% polyacrylamide gels and immunoblotted with anti-Pim-1 antibody. Lane 1 shows BaF3 cells deprived of IL-3 for 8 hours; lane 2, BaF3/ITD; lane 3, Pim-1/44 (44 kDa); lane 4, Pim-1/33 (33 kDa); lane 5, Pim-1/NT81 (27.5 kDa). (B) Expression of Pim-1, FLT3, and Actin after treatment with or without CEP-701 for 1 hour. Pim-1 and Actin were directly assessed by immunoblotting with the anti-Pim-1 and anti-Actin antibodies. FLT/ITD expression and phosphorylation were detected by immunoprecipitation with FLT3 antibody (S-18) followed by immunoblotting with 4G10.
Expression of Pim-1 constructs in stably transfected BaF3 cells. (A) Expression of Pim-1 constructs was detected by monoclonal anti-Pim-1 antibody from abstracts of transfected cell lines. Extracts (100 μg/sample) were resolved by separation in 12% polyacrylamide gels and immunoblotted with anti-Pim-1 antibody. Lane 1 shows BaF3 cells deprived of IL-3 for 8 hours; lane 2, BaF3/ITD; lane 3, Pim-1/44 (44 kDa); lane 4, Pim-1/33 (33 kDa); lane 5, Pim-1/NT81 (27.5 kDa). (B) Expression of Pim-1, FLT3, and Actin after treatment with or without CEP-701 for 1 hour. Pim-1 and Actin were directly assessed by immunoblotting with the anti-Pim-1 and anti-Actin antibodies. FLT/ITD expression and phosphorylation were detected by immunoprecipitation with FLT3 antibody (S-18) followed by immunoblotting with 4G10.
These same cell lines were then subjected to the MTT assay in the presence of increasing concentrations of CEP-701. Cytotoxicity was induced potently by CEP-701 in BaF3/ITD cells with an IC50 of 5 nM and complete inhibition induced by 50 nM (Figure 7A). Forced expression of the 44-kDa or 33-kDa isotypes of Pim-1 each shifted the dose-response curves to CEP-701 to the right (ie, more resistant to the effects of the drug; approximately 5-fold increase in the IC50), and cytotoxicity was not induced as completely at higher levels of inhibitor. In contrast, the expression of the dominant-negative construct (NT81) further sensitized the cells to the effects of FLT3 inhibition as noted by the shift of the dose-response curve to the left. The enhanced resistance to FLT3 inhibition by forced expression of wild-type Pim-1 isotypes provides support that the cytotoxic effect of FLT3 inhibition might be mediated, at least partially, through blocking the expression of Pim-1.
Enforced Pim-1 expression renders BaF3/ITD cells more resistant to cytotoxicity induced by CEP-701 while dominant-negative Pim-1 expression inhibits colony growth. (A) Triplicate samples of 50 000 cells were incubated with increasing concentration of CEP-701 for 24 hours, and cytotoxicity was assessed by MTT assay. The graph displays the MTT results for each cell line normalized to the untreated controls. Error bars represent standard deviation from 3 independent experiments. (B) Pim-1/NT81 inhibits colony formation by FLT3/ITD. BaF3/ITD cells were electroporated with 15 μg of the expression vectors for the N-terminal deletion form of Pim-1 (Pim-1/NT81) or control vector (pLXSN). One day after electroporation, 100, 500, and 2500 cells were seeded on triplicate dishes containing fetal calf serum (FCS), IMDM, 1% methylcellulose, and G418. The colonies were grown at 37°C in 5% CO2 and counted on day 7. The mean colony numbers are counted from triplicate dishes of 100, 500, and 2500 plated cells. The graph shows the relative mean colony number difference and standard deviation of Pim-1/NT81 transfectants compared with control vector transfectants from each group from 3 independent experiments. The inset shows one each of the 10-mm culture dishes from control vector and Pim-1/NT81 electroporations. (C) Cells were incubated with increasing concentrations of CEP-701 for 24 hours and assayed for viability by annexin V and 7-AAD binding by fluorescence activated cell sorting (FACS) analysis. The graph displays the percentage of cell death induced in each cell line by the treatment normalized to the untreated controls. Error bars represent standard deviation from 3 independent experiments.
Enforced Pim-1 expression renders BaF3/ITD cells more resistant to cytotoxicity induced by CEP-701 while dominant-negative Pim-1 expression inhibits colony growth. (A) Triplicate samples of 50 000 cells were incubated with increasing concentration of CEP-701 for 24 hours, and cytotoxicity was assessed by MTT assay. The graph displays the MTT results for each cell line normalized to the untreated controls. Error bars represent standard deviation from 3 independent experiments. (B) Pim-1/NT81 inhibits colony formation by FLT3/ITD. BaF3/ITD cells were electroporated with 15 μg of the expression vectors for the N-terminal deletion form of Pim-1 (Pim-1/NT81) or control vector (pLXSN). One day after electroporation, 100, 500, and 2500 cells were seeded on triplicate dishes containing fetal calf serum (FCS), IMDM, 1% methylcellulose, and G418. The colonies were grown at 37°C in 5% CO2 and counted on day 7. The mean colony numbers are counted from triplicate dishes of 100, 500, and 2500 plated cells. The graph shows the relative mean colony number difference and standard deviation of Pim-1/NT81 transfectants compared with control vector transfectants from each group from 3 independent experiments. The inset shows one each of the 10-mm culture dishes from control vector and Pim-1/NT81 electroporations. (C) Cells were incubated with increasing concentrations of CEP-701 for 24 hours and assayed for viability by annexin V and 7-AAD binding by fluorescence activated cell sorting (FACS) analysis. The graph displays the percentage of cell death induced in each cell line by the treatment normalized to the untreated controls. Error bars represent standard deviation from 3 independent experiments.
BaF3/ITD cells are completely dependent on FLT3 signaling for their growth and survival in the absence of IL-3. To further determine the role of Pim-1 in FLT3 signaling, we transfected those cells with the dominant-negative form of Pim-1 (Pim-1/NT81) and examined the effect of colony growth. Colony growth was decreased by approximately 50% (average of 3 different experiments performed in triplicate), further supporting the role of Pim-1 in FLT3/ITD-mediated transformation (Figure 7B).
Enforced Pim-1 expression increases the resistance of BaF3/ITD cells to apoptosis induced by CEP-701
While forced expression of Pim-1 isotypes renders cells more resistant to CEP-7010induced cytotoxicity observed in the MTT assay, this can occur by several mechanisms, including increased proliferation, increased metabolism, and increased survival. We have previously shown that CEP-701 induces apoptosis in BaF3/ITD cells.13 Thus, we examined whether the effects on cytotoxicity mediated by forced expression of Pim-1 protein might result from decreased apoptosis. Apoptosis was measured by the annexin V binding assay of the BaF3/ITD, Pim-1/44, Pim-1/33, and Pim-1/NT81 cell lines following CEP-701 treatment (Figure 7C). BaF3/ITD cells showed a dose-dependent increase in the proportions that were annexin V and 7-AAD positive. Approximately 60% of these cells were apoptotic after treatment with 50 nM CEP-701 for 24 hours. Forced expression of Pim-1/44 or Pim-1/33 proteins decreased CEP-701-induced apoptosis to 40% and 30%, respectively, at the same level of CEP-701 treatment. Dominant-negative Pim-1 protein expression (NT81) caused a slight increase in the proportion of apoptotic cells. These results indicate that Pim-1 plays an antiapoptotic role in FLT3/ITD-mediated signaling.
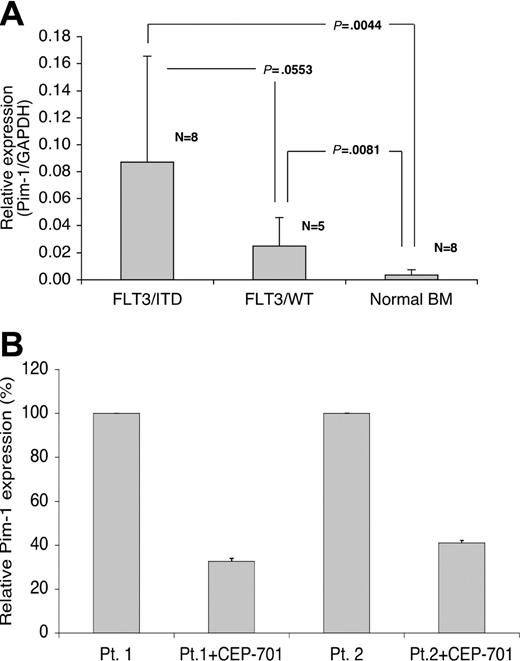
Pim-1 is overexpressed in FLT3/ITD-positive samples from patients with AML and its expression decreases in response to FLT3 inhibition
To determine whether these observations had clinical relevance, we determined by QPCR the expression levels of Pim-1 in FLT3/ITD-positive and nonmutant FLT3 AML samples, as well as in normal bone marrow. The highest levels of Pim-1 expression occurred in FLT3/ITD samples. Pim-1 expression was more than 25-fold higher in FLT3/ITD-positive AML samples compared with normal bone marrow (Figure 8A). Interestingly, one of the FLT3/ITD samples was homozygous for the mutation (ie, no wild-type FLT3 detectable), and this sample showed the highest level of Pim-1 expression. Pim-1 levels in nonmutant FLT3 AML samples were also elevated relative to normal bone marrow.
Pim-1 is overexpressed in FLT3/ITD-positive samples from patients with AML, and its expression is decreased after treatment with CEP-701. (A) Samples from patients with AML were analyzed for Pim-1 expression by QPCR and compared with expression in normal bone marrow. The graph shows the relative expression levels of Pim-1 normalized to GAPDH in FLT3/ITD, FLT3/WT (wild-type) AML samples and normal bone marrow samples. The results show the mean and standard deviation for duplicate QPCR results from 3 independent experiments. The data were analyzed by Student t test and P values are shown. (B) Two FLT3/ITD-positive AML patient blast samples (patient 1 and patient 2) were incubated with 50 nM CEP-701 for 6 hours, and total RNA was harvested for QPCR. The results show the mean and standard deviation for triplicate QPCR results from 3 independent experiments.
Pim-1 is overexpressed in FLT3/ITD-positive samples from patients with AML, and its expression is decreased after treatment with CEP-701. (A) Samples from patients with AML were analyzed for Pim-1 expression by QPCR and compared with expression in normal bone marrow. The graph shows the relative expression levels of Pim-1 normalized to GAPDH in FLT3/ITD, FLT3/WT (wild-type) AML samples and normal bone marrow samples. The results show the mean and standard deviation for duplicate QPCR results from 3 independent experiments. The data were analyzed by Student t test and P values are shown. (B) Two FLT3/ITD-positive AML patient blast samples (patient 1 and patient 2) were incubated with 50 nM CEP-701 for 6 hours, and total RNA was harvested for QPCR. The results show the mean and standard deviation for triplicate QPCR results from 3 independent experiments.
We also investigated the relation between FLT3 inhibition and Pim-1 expression in FLT3/ITD-positive samples from patients with AML. The RNA levels of Pim-1 decreased by more than 2-fold in both primary FLT3/ITD-positive AML samples after FLT3 inhibition (Figure 8B). Thus, Pim-1 expression in FLT3/ITD-positive primary AML samples is consistent with the results found in constitutively activated FLT3-expressing cell lines.
Discussion
Pim-1 was originally isolated as a frequent site of retroviral integration that cooperated to generate leukemia/lymphoma in the Moloney murine leukemia virus (MoMuLV)-infected mouse model. Pim-1 is normally induced by a variety of cytokines through the Jak/Stat pathway and contributes to cytokine-dependent cell survival signaling in normal hematopoietic cells. Pim-1 fits the definition of a true oncogene, in that its enforced expression in transgenic mice leads to an increased incidence of tumors.45 Increased expression of Pim-1 is found in prostate cancer cell lines and primary samples examined by cDNA microarray and tissue microarray.46 In this report, we found Pim-1 is one of several genes whose expression consistently decreased in microarray experiments in several myeloid leukemia cell lines when constitutively activated FLT3 was inhibited by small molecule tyrosine kinase inhibitors. We found Pim-1 expression to be highly dependent on FLT3/ITD signaling in a variety of cell lines in which FLT3 is constitutively activated. Pim-1 is strongly induced by FLT3-ITD mutations in the absence of added cytokines, and its level of expression when FLT3 signaling is inhibited with CEP-701 closely parallels the level of activated FLT3. FLT3/ITD affects Pim-1 at the level of RNA, and this translates directly into changes in the level of protein expression. Importantly, these changes occur rapidly, lessening the chances that they represent a response to far downstream events (eg, apoptosis). Moreover, Pim-1 expression is still induced by IL-3 and GM-CSF in BaF3 and TF1 cells, treated with CEP-701. Thus, CEP-701 does not interfere with the IL-3- or GM-CSF-mediated Pim-1 induction pathways, but only affects Pim-1 induction dependent on the FLT3/ITD signaling pathway. In addition, inhibition of Pim-1 expression is not CEP-701 specific because another FLT3-selective inhibitor, CEP-5214, causes the same changes. In support of our findings, Pim-1 was recently identified by gene expression profile using microarray as one of the discriminative genes in samples from patients with AML with FLT3/ITD mutations.47
FLT3 mutations activate the tyrosine kinase activity of FLT3 in the absence of FL binding. Mutationally activated FLT3 appears to suppress apoptosis and increases cell division by activating multiple intracellular pathways, but the exact signaling pathways important for transformation are not thoroughly understood. A number of proteins, including STAT5 and extracellular signal-regulated kinase 1 and 2 (ERK1/2), have been identified as downstream components of constitutively activated FLT3 signaling pathways. The strong phosphorylation/activation of STAT5 by constitutively activated FLT3, either by ITD mutation (MV4-11, MOLM-14, BaF3/ITD cells) or autocrine stimulation (EOL-1), contrasts with its limited activation by FL-stimulated “normal” FLT3 signaling. Pim-1 may be one of the components of constitutively activated FLT3 signaling pathways downstream of STAT5. In fact, STAT5 is strongly and constitutively phosphorylated/activated by FLT3/ITD mutation but only weakly stimulated by wild-type FLT3.12 This points to one of the known differences in signaling between wild-type FLT3 and FLT3/ITD mutations.44 Therefore, Pim-1 may be important in FLT3/ITD-mediated leukemogenesis and may explain why the presence of FLT3/ITD mutations indicate poor prognosis in patients with AML. The highest levels of Pim-1 expression were seen in FLT3/ITD-positive samples from patients with AML. Elevated Pim-1 levels, relative to normal bone marrow, were also seen in nonmutant FLT3 AML samples where it may be induced by other signaling pathways.
The findings that forced Pim-1 expression in BaF3/ITD cells makes the cells more resistant to CEP-701-induced cytotoxicity and that dominant-negative Pim-1 expression inhibited colony growth support the idea that induction of Pim-1 may be an important part of FLT3/ITD signaling. Pim-1 may have this effect by blocking proapoptotic signaling molecules such as Bad (Bcl-2-associated death promoter), which has been reported to be a substrate of Pim-1 and Pim-2 phosphorylation.45,48 Bad is a proapoptotic protein, and Bad-induced cell death can be reversed with phosphorylation by Pim-1 or Pim-2.48 Expression of either of the isoforms of Pim-1 inhibited cytotoxicity and apoptosis induced by CEP-701. The 2 isoforms of murine Pim-1 have similar functions but work through different Bcl-2 family members.39 Pim-1/33 protein expression decreased more quickly than Pim-1/44 after CEP-701 treatment, which may reflect differences in the half-lives of the 2 isoforms.43
The reason for the incomplete inhibition of the Pim-1 dominant-negative form (NT81) in affecting the growth and survival of BaF3/ITD cells may result from its inability to block all forms of the Pim family, which also includes Pim-2 and Pim-3. Pim-1/NT81 expression might be compensated by other Pim family members or by Akt activation in BaF3/ITD cells. Pim-2 is structurally and functionally similar to Pim-1, which is also important to cell survival in factor-dependent cell lines. In support of this idea, Pim-1 knock-out mice do not show significant disorders.49 However, nearly all Pim-1 transgenic mice, but only 15% of nontransgenic mice, develop T-cell lymphomas within 200 days with a single low dose of ENU (N-ethyl-N-nitrosourea).50 Recently, Pim-2 expression was also found to be dependent on FLT3/ITD signaling in 32D/ITD cells.51 Therefore, further study of the role that the Pim family plays in FLT3/ITD-involved hematologic malignancies is clearly warranted.
FLT3 inhibition results in not only induction of apoptosis but also in G1/S cell-cycle arrest in FLT3/ITD-expressing cells.17,52 Pim-1 might also be involved in promotion of cell-cycle progression in STAT5-induced Pim-1-expressing cells because Pim-1 expression is sufficient to induce IL-3-independent growth of Ba/F3 cells.30,53 Pim-1 is known to phosphorylate CdC25A, which is a key molecule in cell-cycle progression. CdC25A is required for the G1/S transition, as microinjection of anti-CdC25A antibodies effectively block cell-cycle progression from G1 into S phase. However, enforced expression of Pim-1 isotypes in the cell lines studied here did not rescue the block of G1/S cell-cycle transition induced by CEP-701 (data not shown). Thus, other cell-cycle regulatory molecules such as CDKs, cyclins, c-Myc, and/or cell-cycle inhibitors likely also play important roles in CEP-701-mediated cell-cycle arrest. This may explain why enforced Pim-1 expression does not completely rescue BaF3/ITD cells from CEP-701 inhibition. For example, simultaneous up-regulation of both the antiapoptotic protein A1 and Pim-1 is required for in vitro transformation and in vivo leukemogenesis mediated by BCR/ABL.38 Also, constitutive expression of Pim-1 together with c-Myc fully compensates for loss of STAT3-mediated cell-cycle progression, antiapoptotic signaling, and Bcl-2 expression.54 Thus, the discovery of proteins that cooperate with Pim-1 in FLT3-mediated cell-cycle and antiapoptotic signaling will improve our understanding of how constitutively activated FLT3 signaling participate in leukemogenesis.
Prepublished online as Blood First Edition Paper, October 21, 2004; DOI 10.1182/blood-2004-05-2006.
Supported by grants for National Cancer Institute (NCI; CA90668, CA70970, CA91177, CA100632), Leukemia and Lymphoma Society, and Burroughs Wellcome Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Patrick Brown, Rui Zheng, Peili Chen, Li Li, Obdulio Piloto, and Allen Williams for helpful discussions; Susan Jones-Bolin and Bruce Ruggeri of Cephalon, Inc for providing CEP-701 and CEP-5214; and William Matsui for providing normal human bone marrow samples. D.S. is the Douglas Kroll Research Foundation Translational Researcher of the Leukemia and Lymphoma Society and the recipient of the Kyle Haydock Professorship in Oncology.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal