Abstract
We studied immunoglobulin variable heavy-chain (IGHV) repertoire and mutational status in 553 patients with chronic lymphocytic leukemia (CLL) from the Mediterranean area to gain insight into the potential pathogenetic role of antigenic stimulation. The most commonly represented IGHV genes mirrored the usage of normal B cells, with the exception of IGHV1-18, IGHV3-30.3, and IGHV4-59 that were underrepresented. The IGHV3-21 gene, frequently expressed in Northern European CLL, was present only in 16 cases (2.9%). Based on HCDR3 cluster analysis, cases using IGHV3-21 could be grouped in 2 subsets of similar frequency. The first one (7 of 16 cases) carried a similar HCDR3 amino acid sequence (common-HCDR3 subset), virtually identical to the Scandinavian IGHV3-21 CLL. These cases used the IGHJ6 gene; 4 of 7 were unmutated; 6 of 7 carried the Vλ2-14 (IGLV3-21) light-chain gene with a similar LCDR3. All expressed CD38 and had a progressive disease. The second subset (9 of 16) was characterized by heterogeneous HCDR3 rearrangements (nonhomogeneous-HCDR3 subset), diverse IGHJ and IGV light-chain gene usage, variable IGHV mutational status (5 of 9 unmutated), variable CD38 expression, and variable clinical course (4 of 9 progressed). The first subset suggests a potential antigenic element rarely encountered in the Mediterranean area, possibly responsible for a negative outcome. The second subset may reflect the physiologic heterogeneity of expression of IGHV3-21 rearrangements in the normal repertoire and is characterized by a variable clinical outcome. (Blood. 2005;105:1678-1685)
Introduction
The study of immunoglobulin variable heavy-chain (IGHV) genes in B-lymphoproliferative disorders has helped in tracing the developmental stage of neoplastic transformation by assigning a normal B-cell counterpart to individual B-cell malignancies and has provided a new prognostic tool. This progress is well illustrated by chronic lymphocytic leukemia (CLL)1 where the analysis of the IGHV repertoire and mutation pattern has helped to define at least 2 disease subsets, one with unmutated IGHV genes and one with somatically mutated genes, that is, less than 98% homology with the closest germline gene.2,3 The mutational status translates into a different clinical outcome, with patients with unmutated IGHV CLL having a considerably poorer survival and an overall worse prognosis.
IGHV repertoire analysis in CLL has demonstrated biases both at the IGH subgroup level, with IGHV1, IGHV3, and IGHV4 being the most frequently used subgroup, and at the specific gene level, with IGHV1-69, IGHV3-07, IGHV3-23, and IGHV4-34 being the most frequently used IGHV genes.3-6 Importantly, somatic mutations do not appear to occur uniformly among IGHV gene subgroups; rather they display a hierarchy of mutations (IGHV3>IGHV4>IGHV1).4 Differences appear even more striking when considering individual IGHV genes, with IGHV1-69 carrying very few mutations as opposed to IGHV3-07, IGHV3-23, and IGHV4-34, which show a high load of mutations.
Even if IGH subgroup frequencies are similar in different cohorts, relative frequencies of individual genes vary among series. Possible reasons for these differences include size of cohorts, heterogeneity of the patient's ancestry, and the type of medical facility (referral versus primary centers) where the patients are evaluated, which may bring in unwanted selection biases.
Recently an overrepresentation of the IGHV3-21 gene, accounting for more than 11% of cases, has been described in a Scandinavian cohort of patients with CLL.7,8 Most cases (68%) were mutated, albeit with a low mutation “load” (between 2% and 5%). Interestingly, these mutated IGHV3-21 cases had a worse overall survival than the remaining patients with mutated CLL. Similar results were shown in a study of 83 British patients where IGHV3-21 was used in 9.6% of cases (most having 2% to 5% mutation) and was associated with poor survival9 although this series might carry a particular bias due to its focus on p53 dysfunction. A similar frequency was also reported in a Belgian series of 91 patients with 8 cases (8.8%) expressing the IGHV3-21 gene, mainly mutated.10
It remains to be elucidated whether the IGHV3-21 gene by itself escapes the general rule of mutated versus unmutated IGHV in predicting clinical prognosis in CLL11 and thus represents a so far unique example of a mutated gene that is associated with reduced survival.
In this present study, we retrospectively analyzed and compared the IGHV repertoire and mutation status in a series of 553 patients with CLL followed in different institutions from the Mediterranean area (France, Greece, Italy, and Spain). Specifically, we defined the commonly used IGHV repertoire in CLL patients of Mediterranean descent and focused our attention on the frequency, molecular features, and clinical significance of IGHV3-21 gene usage in this large series of patients. Our analysis provides evidence that IGHV3-21-expressing CLL cases in Southern Europe can be divided into 2 groups: one with similar molecular and clinical features to the Northern European cases, though much less frequent, and another one, more heterogeneous at both the biologic and clinical levels.
Patients, materials, and methods
Patient group
A total of 553 unselected patients with CLL from different institutions, located in Paris (France, 150 cases), Thessaloniki and Athens (Greece, 191 cases), Torino (Italy, 97 cases), and Barcelona (Spain, 115 cases) were studied for IGHV gene usage and mutational status (Table 1). All patients met the diagnostic criteria of the National Cancer Institute-Working Group (NCI-WG).12 The mean age of the 337 men and 216 women was 64.1 years (range, 24-91 years; Table 1). Written informed consent was obtained at study entry. The study was approved by the local Ethics Review Committee of each institution.
Characteristics of all CLL patients analyzed
. | France . | Greece . | Italy . | Spain . | Total . |
|---|---|---|---|---|---|
| No. of cases | 150 | 191 | 97 | 115 | 553 |
| Mean age, y | 61.1 | 67.2 | 69.8 | 58.0 | 64.1 |
| Sex, no. M/no. F | 89/61 | 116/75 | 60/37 | 72/43 | 337/216 |
| No. mutated/no. unmutated | 91/59 | 113/78 | 59/38 | 57/58 | 320/233 |
. | France . | Greece . | Italy . | Spain . | Total . |
|---|---|---|---|---|---|
| No. of cases | 150 | 191 | 97 | 115 | 553 |
| Mean age, y | 61.1 | 67.2 | 69.8 | 58.0 | 64.1 |
| Sex, no. M/no. F | 89/61 | 116/75 | 60/37 | 72/43 | 337/216 |
| No. mutated/no. unmutated | 91/59 | 113/78 | 59/38 | 57/58 | 320/233 |
Within this cohort, 16 patients carrying the IGHV3-21 gene in the expressed immunoglobulin (8 men, 8 women; mean age, 70.7 years) were studied in detail by analyzing the following parameters measured at diagnosis or during the follow-up: CD38 expression, immunoglobulin light-chain (LC) rearrangement, disease stage according to Binet13 or Rai modified criteria,14 history of treatment, and progressive or stable disease as defined by the NCI-WG.12
Determination of IGHV, IGKV, and IGLV gene mutational status
In all cases, the analysis of IGHV-D-J genes was done on leukemic cells obtained from peripheral blood samples after isolation by Ficoll gradient. Total RNA was extracted using the guanidinium thiocyanate method and reverse-transcribed into cDNA using Moloney murine leukemia virus (MMLV) reverse transcriptase and random hexamers as primers. gDNA was isolated by standard phenol chloroform extraction followed by ethanol precipitation.
The IGHV-D-J gene sequences were determined as previously described,15-18 by amplifying with polymerase chain reaction (PCR) either cDNA, using the appropriate sense IGHV leader primer and the antisense IGHC primer,4 or DNA with consensus primers for the framework 1 and IGHJ genes, as previously described19 or following the BIOMED-2 protocol.20 Amplification of the immunoglobulin κ and λ LC gene rearrangements (IGKV-J and IGLV-J) was performed according to BIOMED-2 protocols20 and as previously described.21
Purified PCR products were sequenced either directly or after a cloning procedure using an automated DNA sequencer. Sequence data were analyzed using the international Immunogenetics database (IMGT, http://imgt.cines.fr; initiator and coordinator: Marie-Paule Lefranc, Montpellier, France) and more particularly, the IMGT/V-QUEST22 and IMGT/Junction Analysis23 to determine the respective homology of each gene with the closest germline counterpart. The sequences with a germline homology 98% or higher were considered unmutated, and those with a homology less than 98% as mutated.2,3 The mutational status of 242 of 553 sequences was published previously.17,18,24
To facilitate direct reference and comparison with previous studies (mainly those by Tobin et al3,4 ), in the only case of the IGLV3-21 gene we have followed the Kawasaki nomenclature, after which this gene is named Vλ2-14. Assessment of the distribution of mutations and possible impact of antigen selection on the hypermutation process (for cases with > 2% deviation from the closest germline V gene) was performed by the multinomial model proposed by Lossos et al.25 Each codon was assessed separately for the presence of somatic mutations. When more than one nucleotide substitution was encountered within a 3-bp codon sequence, it was postulated that mutations were introduced sequentially from the first through the third nucleotide in order to be assigned as being of the replacement (R mutation; leading to amino acid substitution in the encoded polypeptide) or silent (S mutation; leaving the encoded polypeptide unaltered) type.
Immunophenotypic analysis
Immunophenotypic analysis was performed on fresh blood samples or cells cryopreserved at diagnosis to define the isotype and the LC clonal restriction and analyze the CD38 expression of the leukemic population.
Statistical analysis
Differences between median cumulative survivals and median times to undergo progressive disease were analyzed by the Kaplan-Meier method. The Fisher exact test was performed to determine the significance of differences among frequencies of events between patient subsets.
Results
IGHV gene usage in CLL is similar in 4 Mediterranean countries
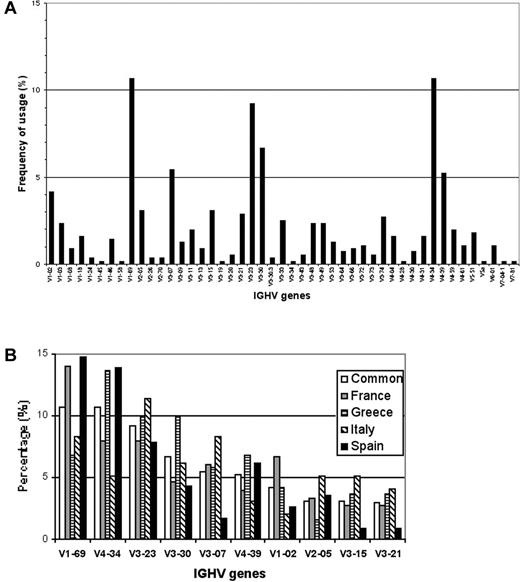
Productive IGHV-D-J rearrangements were sequenced in 553 patients with CLL followed in different clinical centers from 4 different Mediterranean countries (Table 1). The IGHV genes used by these leukemic cell populations were derived from all the 7 human IGHV gene families. As shown in Figure 1A-B, among the 47 functional IGHV genes used, the most frequently encountered were IGHV1-69 (59; 10.7%), IGHV4-34 (59; 10.7%), IGHV3-23 (51; 9.2%), IGHV3-30 (37; 6.7%), IGHV3-07 (30; 5.4%), IGHV4-39 (29; 5.2%), and IGHV1-02 (22; 4.2%); collectively, these genes accounted for more than 50% of cases. As a whole, this large and geographically diverse series confirmed previously published results obtained in smaller series.3-6 Considering the percentage of somatic mutations present in the sequences and using the 98% cut-off value,2,3 320 (57.9%) cases were considered mutated and the remaining 233 (42.1%) had a germline configuration. The most represented IGHV gene within the mutated subset was IGHV4-34, which was used in 59 cases overall (50 mutated), whereas IGHV1-69 was the most frequently used in the unmutated group, being expressed in 59 patients overall (52 unmutated) as shown in Table 2.
Frequency of specific IGHV genes used in Mediterranean CLL. (A) Frequencies of IGHV genes among 553 CLL cases of Mediterranean descent. (B) Relative frequencies of the 10 most frequent IGHV genes in the whole series (□), in the French (▦), Greek (▤), Italian (▧) and Spanish (▪) cohorts.
Frequency of specific IGHV genes used in Mediterranean CLL. (A) Frequencies of IGHV genes among 553 CLL cases of Mediterranean descent. (B) Relative frequencies of the 10 most frequent IGHV genes in the whole series (□), in the French (▦), Greek (▤), Italian (▧) and Spanish (▪) cohorts.
IGHV gene usage in all mutated and unmutated cases
Rank . | Mutated IGHV gene . | % . | Unmutated IGHV gene . | % . |
|---|---|---|---|---|
| 1 | IGHV4-34 | 15.6 | IGHV1-69 | 22.7 |
| 2 | IGHV3-23 | 13.8 | IGHV4-39 | 8.6 |
| 3 | IGHV3-07 | 9.1 | IGHV3-30 | 7.3 |
| 4 | IGHV3-30 | 6.3 | IGHV1-02 | 6.9 |
| 5 | IGHV3-15 | 4.1 | IGHV4.34 | 3.9 |
| 6 | IGHV3-74 | 4.1 | IGHV3-21 | 3.9 |
| 7 | IGHV2-05 | 3.4 | IGHV3-48 | 3.9 |
| 8 | IGHV4-39 | 2.8 | IGHV3-49 | 3.9 |
| 9 | IGHV4-59 | 2.8 | IGHV3-23 | 3.0 |
| 10 | IGHV4-31 | 2.5 | IGHV3-33 | 3.0 |
Rank . | Mutated IGHV gene . | % . | Unmutated IGHV gene . | % . |
|---|---|---|---|---|
| 1 | IGHV4-34 | 15.6 | IGHV1-69 | 22.7 |
| 2 | IGHV3-23 | 13.8 | IGHV4-39 | 8.6 |
| 3 | IGHV3-07 | 9.1 | IGHV3-30 | 7.3 |
| 4 | IGHV3-30 | 6.3 | IGHV1-02 | 6.9 |
| 5 | IGHV3-15 | 4.1 | IGHV4.34 | 3.9 |
| 6 | IGHV3-74 | 4.1 | IGHV3-21 | 3.9 |
| 7 | IGHV2-05 | 3.4 | IGHV3-48 | 3.9 |
| 8 | IGHV4-39 | 2.8 | IGHV3-49 | 3.9 |
| 9 | IGHV4-59 | 2.8 | IGHV3-23 | 3.0 |
| 10 | IGHV4-31 | 2.5 | IGHV3-33 | 3.0 |
Some IGHV genes appeared to be suppressed when their frequency was compared to the normal B-cell repertoire.26 Specifically, the IGHV3-30.3 gene was practically absent in our patients' repertoire with only 2 cases (1 French and 1 Italian) among the 553 cases examined. This finding is in keeping with the previously reported decrease in the use of this gene by Fais et al4 and clearly contrasts with the situation both in normal B-cell subsets and in other previously published CLL series.3 An undescribed4 suppression of IGHV1-18 and of IGHV4-59 genes was also evident in comparison with normal B-cell subsets4 because these 2 genes were productively rearranged in only 9 (1.6%) and 11 (2.0%) cases, respectively. Of interest, the IGHV1-18 gene was never detected in the Italian cohort.
The IGHV3-21 gene is used in a minority of Mediterranean CLL cases
Among 553 sequenced patients, the IGHV3-21 gene was used in only 16 cases (2.9%), being overall the 10th most frequently used IGHV segment (Figure 1A; Table 3). Importantly, in 2 national cohorts this gene barely scored among the top 10 most used IGHV genes with the interesting case of the Spanish series where only 1 of 115 cases productively rearranged the IGHV3-21 gene (Figure 1B; Table 3). Seven of the 16 cases (43.8%) carried somatic mutations, but only 3 of 7 had a low level of IGHV mutations (between 2% and 4%), which has been indicated as a typical feature of this rearranged gene in CLL.8,27 The overall mutations ranged between 2.4% and 7.1%, with a mean value of 4.8%. The IGHJ6 gene was used in 9 cases (56%), the IGHJ4 in 6 cases, and the IGHJ5 segments in one case.
Characteristics of all CLL patients using IGHV3-21 gene
. | Total . | France . | Greece . | Italy . | Spain . |
|---|---|---|---|---|---|
| No. IGHV3-21 cases | 16 | 4 | 7 | 4 | 1 |
| Frequency, % | 2.9 | 2.7 | 3.7 | 4.1 | 0.9 |
| Mean age, y | 70.7 | 63.5 | 67 | 79.5 | 90* |
| Sex, no. F/no. M | 8/8 | 2/2 | 4/3 | 1/3 | 1/0 |
| No. mutated/no. unmutated | 7/9 | 2/2 | 2/5 | 2/2 | 1/0 |
| IGHV homology, no. | |||||
| At least 98% | 9 | 2 | 5 | 2 | 0 |
| 98%-96% | 3 | 0 | 1 | 1 | 1 |
| Less than 96% | 4 | 2 | 1 | 1 | 0 |
| LC, no. κ/no. λ | 7/9 | 2/2 | 4/3 | 1/3 | 0/1 |
| IGV LC homology, no. | |||||
| At least 98% | 14 | 3 | 7 | 3 | 1 |
| 98%-96% | 2 | 1 | 0 | 1 | 0 |
| Less than 96% | 0 | 0 | 0 | 0 | 0 |
| CD38, % | |||||
| Neg | 5 | 0 | 4 | 1 | 0 |
| Pos | 10 | 3 | 3 | 3 | 1 |
| Stage at diagnosis | |||||
| A | 8 | 1 | 5 | 2 | 0 |
| B | 5 | 3 | 1 | 1 | 0 |
| C | 3 | 0 | 1 | 1 | 1 |
| Clinical course | |||||
| Stable | 5 | 0 | 4 | 1 | 0 |
| Progressive | 11 | 4 | 3 | 3 | 1 |
| Need for treatment | 11 | 4 | 3 | 3 | 1 |
| CLL-related death | 4 | 2 | 0 | 1 | 1 |
. | Total . | France . | Greece . | Italy . | Spain . |
|---|---|---|---|---|---|
| No. IGHV3-21 cases | 16 | 4 | 7 | 4 | 1 |
| Frequency, % | 2.9 | 2.7 | 3.7 | 4.1 | 0.9 |
| Mean age, y | 70.7 | 63.5 | 67 | 79.5 | 90* |
| Sex, no. F/no. M | 8/8 | 2/2 | 4/3 | 1/3 | 1/0 |
| No. mutated/no. unmutated | 7/9 | 2/2 | 2/5 | 2/2 | 1/0 |
| IGHV homology, no. | |||||
| At least 98% | 9 | 2 | 5 | 2 | 0 |
| 98%-96% | 3 | 0 | 1 | 1 | 1 |
| Less than 96% | 4 | 2 | 1 | 1 | 0 |
| LC, no. κ/no. λ | 7/9 | 2/2 | 4/3 | 1/3 | 0/1 |
| IGV LC homology, no. | |||||
| At least 98% | 14 | 3 | 7 | 3 | 1 |
| 98%-96% | 2 | 1 | 0 | 1 | 0 |
| Less than 96% | 0 | 0 | 0 | 0 | 0 |
| CD38, % | |||||
| Neg | 5 | 0 | 4 | 1 | 0 |
| Pos | 10 | 3 | 3 | 3 | 1 |
| Stage at diagnosis | |||||
| A | 8 | 1 | 5 | 2 | 0 |
| B | 5 | 3 | 1 | 1 | 0 |
| C | 3 | 0 | 1 | 1 | 1 |
| Clinical course | |||||
| Stable | 5 | 0 | 4 | 1 | 0 |
| Progressive | 11 | 4 | 3 | 3 | 1 |
| Need for treatment | 11 | 4 | 3 | 3 | 1 |
| CLL-related death | 4 | 2 | 0 | 1 | 1 |
Actual age of the single patient of the Spanish series.
Evidence of 2 CLL IGHV3-21-expressing subsets based on the immunoglobulin amino acid sequence
Cluster analysis of the number and sequence of the amino acids present in the complementary-determining region 3 of the heavy-chain (HCDR3) rearrangements allowed grouping of IGHV3-21 cases in 2 subsets. One subset comprised 7 of 16 cases (Table 4) with a very short and strikingly similar HCDR3 sequence (all with 9 codons; common-HCDR3 subset). The conserved sequence was ARDANAMDV in 2 cases, whereas 1 case differed by one amino acid, and the remaining 4 cases differed by either 2 or 3 amino acids in 2 cases in different positions (Table 5). This sequence is virtually identical to the one published earlier by Tobin et al3,4 (ARDANGMDV). Interestingly, despite carrying a short HCDR3 sequence, all cases used the IGHJ6 gene that is the longest IGHJ gene in the human genome. The first 2 amino acids in the conserved HCDR3 sequence (AR) derive from the IGHV3-21 gene, whereas the last 4 (AMDV) from the IGHJ6 gene. The sixth residue was either a glycine (G) in 4 of 7 of our cases as in most of Scandinavian sequences, and is germline encoded by the IGHJ6 gene, or an alanine (A; 3 of 7 cases), both being amino acids with an aliphatic side chain.
Characteristics of the IGHV3-21-expressing CLL patients belonging to the common-HCDR3 subset
. | Fra1 . | Fra3 . | Gre1 . | Ita1 . | Ita3 . | Ita4 . | Spa1 . |
|---|---|---|---|---|---|---|---|
| No. of cases | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Age, y | 69 | 46 | 56 | 72 | 69 | 91 | 90 |
| Sex | F | M | M | M | M | F | F |
| IGHJ gene | IGHJ6 | IGHJ6 | IGHJ6 | IGHJ6 | IGHJ6 | IGHJ6 | IGHJ6 |
| IGHV homology, % | 94.9 | 98.3 | 98.0 | 96.3 | 98.6 | 98.3 | 96.9 |
| LC | Vλ2-14 | IGKV3-15 | Vλ2-14 | Vλ2-14 | Vλ2-14 | Vλ2-14 | Vλ2-14 |
| IGJ LC gene | IGLJ3 | IGKJ1 | IGLJ3 | IGLJ3 | IGLJ3 | IGLJ3 | IGLJ3 |
| IGV LC homology, % | 99.7 | 100 | 100 | 99 | 98.6 | 98.3 | 99.7 |
| CD38, % | 7 | 47 | 7.7 | 99 | 53 | 99 | 46 |
| Stage at diagnosis | B | B | B | B | C | A | C |
| Progression | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Need for treatment | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CLL-related death | No | Yes | No | Yes | No | No | Yes |
. | Fra1 . | Fra3 . | Gre1 . | Ita1 . | Ita3 . | Ita4 . | Spa1 . |
|---|---|---|---|---|---|---|---|
| No. of cases | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Age, y | 69 | 46 | 56 | 72 | 69 | 91 | 90 |
| Sex | F | M | M | M | M | F | F |
| IGHJ gene | IGHJ6 | IGHJ6 | IGHJ6 | IGHJ6 | IGHJ6 | IGHJ6 | IGHJ6 |
| IGHV homology, % | 94.9 | 98.3 | 98.0 | 96.3 | 98.6 | 98.3 | 96.9 |
| LC | Vλ2-14 | IGKV3-15 | Vλ2-14 | Vλ2-14 | Vλ2-14 | Vλ2-14 | Vλ2-14 |
| IGJ LC gene | IGLJ3 | IGKJ1 | IGLJ3 | IGLJ3 | IGLJ3 | IGLJ3 | IGLJ3 |
| IGV LC homology, % | 99.7 | 100 | 100 | 99 | 98.6 | 98.3 | 99.7 |
| CD38, % | 7 | 47 | 7.7 | 99 | 53 | 99 | 46 |
| Stage at diagnosis | B | B | B | B | C | A | C |
| Progression | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Need for treatment | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CLL-related death | No | Yes | No | Yes | No | No | Yes |
Fra indicates France; Gre, Greece; Ita, Italy; Spa, Spain.
Heavy-and light-chain CDR3 amino acid sequences of the IGHV3-21-using CLL cases
Case . | HCDR3 AA sequence . | IG light chain gene . | LCDR3 AA sequence . |
|---|---|---|---|
| Cases with consensus sequence* | |||
| Ita3 | ARDANAMDV | Vλ2-14 (IGLV3-21) | QVWDSGSDHPWV |
| Ita4 | ...G.G... | Vλ2-14 (IGLV3-21) | ............ |
| Ita1 | ...M..... | Vλ2-14 (IGLV3-21) | ............ |
| Gre1 | .I.R.G... | Vλ2-14 (IGLV3-21) | .....S...... |
| Spa1 | .T.R.G... | Vλ2-14 (IGLV3-21) | ............ |
| Fra1 | ....DG... | Vλ2-14 (IGLV3-21) | .....S...... |
| Fra3 | ......... | IGKV3-15 | QQYNNWPPET |
| Cases with completely unrelated sequence† | |||
| Fra4 | ARDRTYPDAYFDF | IGKV3D-15 | QQYYNVWPPGR |
| Gre3 | AKDYDSSGANFQH | IGLV1-51 | GTWDTSLSAWL |
| Ita2 | ARVGNYDRYFFDF | IGKV1-12 | QQANSFPYS |
| Gre5 | ARIWDIVVVPDAMIWA | IGKV1-27 | QKYNSAQFT |
| Gre4 | ARDRRSIVVVVAATPVY | IGKV4-1 | QQSYSTLVEA |
| Fra2 | ATSGKDTTMVQGGHFDY | Vλ2-14 (IGLV3-21) | QVWDSSSDHPFV |
| Gre7 | ASQLWFDRVNNYYYMDV | IGKV1-33/1D-33 | QQYDNLPRYT |
| Gre2 | AREPLSVLLWFGESFGGGFDY | Vλ2-14 (IGLV3-21) | QVWDSGSDHPWV |
| Gre6 | ARDRLLGYCSSTSCWDYYYYYGMDV | IGKV1-17 | LQHNSYPWYT |
Case . | HCDR3 AA sequence . | IG light chain gene . | LCDR3 AA sequence . |
|---|---|---|---|
| Cases with consensus sequence* | |||
| Ita3 | ARDANAMDV | Vλ2-14 (IGLV3-21) | QVWDSGSDHPWV |
| Ita4 | ...G.G... | Vλ2-14 (IGLV3-21) | ............ |
| Ita1 | ...M..... | Vλ2-14 (IGLV3-21) | ............ |
| Gre1 | .I.R.G... | Vλ2-14 (IGLV3-21) | .....S...... |
| Spa1 | .T.R.G... | Vλ2-14 (IGLV3-21) | ............ |
| Fra1 | ....DG... | Vλ2-14 (IGLV3-21) | .....S...... |
| Fra3 | ......... | IGKV3-15 | QQYNNWPPET |
| Cases with completely unrelated sequence† | |||
| Fra4 | ARDRTYPDAYFDF | IGKV3D-15 | QQYYNVWPPGR |
| Gre3 | AKDYDSSGANFQH | IGLV1-51 | GTWDTSLSAWL |
| Ita2 | ARVGNYDRYFFDF | IGKV1-12 | QQANSFPYS |
| Gre5 | ARIWDIVVVPDAMIWA | IGKV1-27 | QKYNSAQFT |
| Gre4 | ARDRRSIVVVVAATPVY | IGKV4-1 | QQSYSTLVEA |
| Fra2 | ATSGKDTTMVQGGHFDY | Vλ2-14 (IGLV3-21) | QVWDSSSDHPFV |
| Gre7 | ASQLWFDRVNNYYYMDV | IGKV1-33/1D-33 | QQYDNLPRYT |
| Gre2 | AREPLSVLLWFGESFGGGFDY | Vλ2-14 (IGLV3-21) | QVWDSGSDHPWV |
| Gre6 | ARDRLLGYCSSTSCWDYYYYYGMDV | IGKV1-17 | LQHNSYPWYT |
Seven cases showed a consensus HCDR3 sequence or a high homology to it, differing by 1, 2, or 3 amino acids at different positions (common-HCDR3 subset), whereas 6 of 7 had a consensus LCDR3 sequence, differing by 1 amino acid in 2 cases. The remaining 9 patients expressed a completely unrelated sequence (nonhomogeneous-HCDR3 subset). Dots indicate homology.
Common-HCDR3 subset.
Nonhomogeneous HCDR3 subset.
Four of these sequences were unmutated (≥ 98% homology), whereas the remaining 3 were mutated: 2 cases were, respectively, 96.3% and 96.9% homologous with the closest germline gene, whereas one was 94.9% homologous (Table 4). The impact of antigen selection on the hypermutation process for these 3 mutated cases was assessed by determining replacement (R) as opposed to silent (S) mutations in the CDRs and S as opposed to R mutations in framework regions (FRs). The probability that excess (for CDR) or scarcity (for FR) of R mutations occurred by chance was calculated by the multinomial distribution model described by Lossos et al25 and following IMGT definitions for FR and CDR lengths. Statistically significant evidence for positive selection by antigen was obtained in all 3 mutated cases; there were 2 cases with higher R/S mutation ratios in CDRs and one case with higher and lower R/S mutation ratios in CDRs and FRs, respectively, than those theoretically expected given the inherent germline RfCDRs and RfFRs values.
The second subset of IGHV3-21 cases (9 of 16; Table 6) carried HCDR3 rearrangements that were totally unrelated to each other and to the consensus sequence (nonhomogeneous-HCDR3 subset). Their mean amino acid length was almost twice longer (16.9 codons) than the consensus (Table 5). The use of IGHJ genes was also heterogeneous with 6 cases using IGHJ4, 1 using IGHJ5, and 2 using IGHJ6. Five cases were unmutated and the remaining 4 were mutated, with 3 of 4 exhibiting a high mutation “load” (< 95%).
Characteristics of the IGHV3-21—expressing CLL patients belonging to the nonhomogeneous-HCDR3 subset
. | Fra2 . | Fra4 . | Gre2 . | Gre3 . | Gre4 . | Gre5 . | Gre6 . | Gre7 . | Ita2 . |
|---|---|---|---|---|---|---|---|---|---|
| No. of cases | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Age, y | 54 | 85 | 65 | 67 | 70 | 60 | 81 | 70 | 86 |
| Sex | M | F | F | F | F | M | F | M | M |
| IGHJ gene | IGHJ4 | IGHJ4 | IGHJ4 | IGHJ5 | IGHJ4 | IGHJ4 | IGHJ6 | IGHJ6 | IGHJ4 |
| IGHV homology, % | 98.3 | 93.2 | 99.0 | 94.6 | 97.6 | 98.0 | 99.0 | 98.6 | 92.9 |
| LC | Vλ2-14 | IGKV3D-15 | Vλ2-14 | IGLV1-51 | IGKV4-1 | IGKV1-27 | IGKV1-1 | IGKV1-33 | IGKV1-12 |
| IGJ LC gene | IGLJ1 | IGKJ1 | IGLJ3 | IGLJ3 | IGKJ3 | IGKJ3 | IGKJ2 | IGKJ2 | IGKJ2 |
| IGV LC homology, % | 100 | 97.0 | 100 | 98.3 | 100 | 99.0 | 100 | 100 | 96.5 |
| CD38, % | 35 | ND | 37 | 2.5 | 1.4 | 12 | 27 | 3.2 | 0.5 |
| Stage at diagnosis | B | A | A | A | A | A | C | A | A |
| Progression | Yes | Yes | No | No | Yes | No | Yes | No | No |
| Need for treatment | Yes | Yes | No | No | Yes | No | Yes | No | No |
| CLL-related death | No | Yes | No | No | No | No | No | No | No |
. | Fra2 . | Fra4 . | Gre2 . | Gre3 . | Gre4 . | Gre5 . | Gre6 . | Gre7 . | Ita2 . |
|---|---|---|---|---|---|---|---|---|---|
| No. of cases | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Age, y | 54 | 85 | 65 | 67 | 70 | 60 | 81 | 70 | 86 |
| Sex | M | F | F | F | F | M | F | M | M |
| IGHJ gene | IGHJ4 | IGHJ4 | IGHJ4 | IGHJ5 | IGHJ4 | IGHJ4 | IGHJ6 | IGHJ6 | IGHJ4 |
| IGHV homology, % | 98.3 | 93.2 | 99.0 | 94.6 | 97.6 | 98.0 | 99.0 | 98.6 | 92.9 |
| LC | Vλ2-14 | IGKV3D-15 | Vλ2-14 | IGLV1-51 | IGKV4-1 | IGKV1-27 | IGKV1-1 | IGKV1-33 | IGKV1-12 |
| IGJ LC gene | IGLJ1 | IGKJ1 | IGLJ3 | IGLJ3 | IGKJ3 | IGKJ3 | IGKJ2 | IGKJ2 | IGKJ2 |
| IGV LC homology, % | 100 | 97.0 | 100 | 98.3 | 100 | 99.0 | 100 | 100 | 96.5 |
| CD38, % | 35 | ND | 37 | 2.5 | 1.4 | 12 | 27 | 3.2 | 0.5 |
| Stage at diagnosis | B | A | A | A | A | A | C | A | A |
| Progression | Yes | Yes | No | No | Yes | No | Yes | No | No |
| Need for treatment | Yes | Yes | No | No | Yes | No | Yes | No | No |
| CLL-related death | No | Yes | No | No | No | No | No | No | No |
LC usage is different in the 2 IGHV3-21 subsets
As defined by cytofluorographic analysis, 9 of the 16 IGHV3-21 cases expressed monoclonal λ LCs on the cell surface and the remaining 7 cases expressed κ LCs. Considering only mutated IGHV3-21 cases (7 of 16), regardless of their HCDR3 composition, 3 expressed a clonal κ LC rearrangement and 4 cases expressed a λ LC rearrangement.
To facilitate direct reference and comparison with previous studies (mainly Tobin et al3,4 ), we have followed the Kawasaki nomenclature, after which this gene is named as Vλ2-14, only in the case of the IGLV3-21 gene. The sequence of LC rearrangements of all IGHV3-21 showed that among the 9 λ-expressing cases, 8 cases expressed Vλ2-14 (IGLV3-21), and the remaining one expressed IGLV1-51 (Vλ1-19). All λ-expressing cases used the IGLJ3 gene except one using IGLJ1. In contrast, the 7 κ-expressing cases used heterogeneous IGKV genes: IGKV3-15 (L2), IGKV3D-15 (L16), IGKV4-1 (B3), IGKV1-27 (A20), IGKV1-17 (A30), IGKV1-33/1D-33 (O8/O18), and IGKV1-12 (L5). In these cases, IGKJ usage was also quite heterogeneous: IGKJ1 was used in 2 cases, IGKJ2 in 3 cases, and IGKJ3 in 2 cases.
Taken together, 14 immunoglobulin LC sequences showed more than 98% homology to their respective germline gene. In particular, all Vλ2-14 (IGLV3-21)-using cases were unmutated.
Considering only the cases belonging to the common-HCDR3 subset, 6 of 7 used a Vλ2-14 (IGLV3-21) rearrangement (Table 4), and the remaining case expressed the IGKV3-15 gene. In these 6 Vλ2-14 (IGLV3-21)-expressing cases cluster analysis of the CDR3 of the LC (LCDR3) rearrangements showed a conserved sequence (QVWDSGSDHP) that was present in 4 cases, whereas 2 cases differed by only one amino acid (QVWDSSSDHP; Table 5). This sequence is virtually identical to the consensus published earlier by Tobin et al3,4 (QVWDGSSDHP), differing only by 2 amino acids at the fifth and sixth positions.
Correlation between clinical outcome and IGHV3-21 gene expression
When considering all IGHV3-21 patients together (Table 3), 4 of 16 died of CLL-related causes, leading to a median survival time of 98 months, which overlaps the median survival of unmutated CLL patients.3 Given the low number of IGHV3-21 cases in our series, the comparative analysis of median survival curves of mutated versus unmutated cases did not reach statistical significance.
Five patients had stable disease, not requiring any treatment; the remaining 11 had clinical progression during follow-up and needed therapy. Among patients who progressed, 6 had a germline configuration in both heavy-chain and LC IGV genes; 3 showed a low level of mutations on their IGHV genes (homologies ranging between 97.6% and 96.3%) but expressed unmutated IGV LC genes. Finally, the remaining 2 patients had highly mutated IGHV genes (94.9% and 93.2% homology), which were associated in one with unmutated IGV LC and in the other with low levels of somatic mutations (97.0%) of the LC variable gene. Interestingly, 9 of 10 tested patients with progressive disease were CD38+.
If patients are grouped according to HCDR3 amino acids sequence, all those belonging to the common-HCDR3 subgroup (Table 4) had a progressive disease (100%) as compared to 4 of the 9 cases of the nonhomogeneous subset (44.4%; P = .03; Table 6). In addition, the patients with a common-HCDR3 IGHV3-21 seemed to experience also a shorter median time to progression (3.5 months versus 60.4 months) though statistical significance was not completely reached (P = .06), due to the low number of progressions in the nonhomogeneous subset.
All 7 cases with a common-HCDR3 expressed CD38 on the cell surface, though only 4 cases had unmutated IGHV genes.
Discussion
In the present multicenter study, we analyzed the immunoglobulin gene repertoire of 553 patients who had CLL and were of Mediterranean origin (Table 5). Our analysis shows that several of the most common IGHV genes in CLL are used at a frequency that is not disease-related, but mirrors the repertoire of normal peripheral blood IgM+ B cells (both CD5+ and CD5-)26 with the exception of the well-known overrepresentation of the IGHV1-69 gene,3-6 especially among unmutated cases. Similar to normal peripheral blood IgM+ B cells, the CLL cases studied were found to use frequently the IGHV3-23, IGHV3-07, and IGHV4-34 genes (as previously reported in smaller series of patients3,4 ) as well as the IGHV4-39 and IGHV1-02 genes. The frequency of the IGHV4-39 gene resembled that of normal CD5- B cells, whereas the frequency of the IGHV1-02 gene was comparable to that of normal IgM+ CD5+ peripheral blood B cells. In addition, in our series the frequency of expression of other genes previously indicated as suppressed in the CLL repertoire in 2 distinct but smaller series of patients3,4 was found similar to that of normal B lymphocytes (both CD5+ and CD5-).26 In particular, IGHV3-30 gene frequency (6.7%) was similar to that of both CD5+ and CD5- B cells, whereas the IGHV3-33 gene was present in 2.5% of the cases showing a usage similar to normal CD5+ B cells.
In contrast, the IGHV3-49 gene, which is rarely expressed in normal B cells, was rather frequently expressed in our cohort (2.4% of cases), especially among Italian patients where it was present in 5.1% of the cases. Conversely, the IGHV1-18, IGHV3-30.3, and IGHV4-59 genes that are common in normal B cells were rarely represented in our series. This lack of representation has not been previously reported in CLL.
The differences in IGHV gene usage between our study and previously reported series may be explained, at least in part, by the small size of the patient populations previously analyzed. However, considering that the expressed immunoglobulin repertoire of both normal and leukemic B cells is shaped by molecular processes as well as by antigenic selection and affinity maturation mechanisms. Such discrepancies may be real and reflect a geographic difference that could allow peculiar genetic or environmental elements to differently shape both the normal and the “leukemic” repertoire. This possibility is highlighted by the different usage of the IGHV3-21 gene among the CLL patients in our multicenter, Mediterranean-based study, as compared to the previously published study originating from Scandinavia.8 In the latter series, this gene was second in frequency (> 11%) only surpassed by IGHV1-69. Furthermore, it was the most frequently used gene in mutated rearrangements. Similar results were reported in a second larger study from the same group (265 cases),7 thereby excluding a bias attributed to small sampling. In addition, a high frequency was also reported in British (9.6%) and Belgian studies (8.8%), though on smaller series of patients.9,10 In contrast, the IGHV3-21 gene barely ranked among the top 10 used IGHV genes in our cohort, being the 10th overall (2.9% of the cases) and the 5th among the unmutated cases. This frequency is at least 3 times lower than the frequency observed in patients from Northern Europe. To further underscore this finding in the Spanish series there was only one IGHV3-21 case among the 115 cases analyzed. Similar to our series, a low frequency for IGHV3-21 usage was suggested in a previous study that combined CLL IGHV sequences from unrelated and geographically diverse cohorts.3,4 The report included CLL cases from heterogeneous sources and referred to IGVH sequences present in databases, not allowing to draw any conclusion regarding the actual gene frequency in terms of population or geographic distribution. Therefore, more studies are needed to finally elucidate whether the reported differences in terms of frequencies may be more wide-spread than so far reported.
The apparent geographic bias shown by our own data may theoretically be due to specific and peculiar genetic background, depending on variations in germline composition of the IGHV locus28 or to the effect of a potential environmental variable less frequently encountered in different regions. Because normal IgM+ peripheral blood B cells and in particular CD5+ B cells have been described to express the IGHV3-21 gene at a low frequency,4,26 it would be interesting to analyze the normal usage of this gene among the Scandinavian population to evaluate whether a bias also exists in the normal repertoire or if it is restricted to CLL cells.
A detailed analysis of the Scandinavian series7,8 revealed that, besides a higher frequency of usage, several peculiarities characterize Scandinavian IGHV3-21-expressing CLL.28 The authors show that an individual IGHV gene can have by itself a prognostic impact in CLL. In their cohort the patients using IGHV3-21 gene, although usually mutated, showed a poor overall prognosis and were therefore an exception to the rule of mutated CLL having good prognosis. In addition IGHV3-21 genes are mainly expressed in combination with a λ LC (28 of 31 cases), which is almost exclusively the Vλ2-14 (IGLV3-21) gene (24 of 27) and demonstrates remarkable similarities in their CDR3s. In our Mediterranean cohort, collectively IGHV3-21 cases (Table 3) express κ and λ LC chain in similar frequencies and usually have IGHV sequences in germline configuration, which explains the occurrence of a therapy-requiring progressive disease in several cases. Interestingly, the few patients with mutations who had progressive disease were mainly CD38+.2,17 These discrepancies may be reconciled by considering that, based on the HCDR3 sequences, we here describe the existence of 2 distinct IGHV3-21 subsets of similar frequency. Less than 50% of the cases do carry an almost identical amino acid consensus sequence (common-HCDR3 subset), together with a similar LCDR3 on the IGLV gene. Both are virtually identical to those commonly expressed in Scandinavian CLL.8 In this respect it is interesting to note a repetitive glycine (G) to alanine (A) variation at HCDR3 position 6 in both Scandinavian and Mediterranean sequences. Glycine is the germline configuration of the IGHJ6 gene. However, the G>A substitution does not seem to bring significant charge or conformation differences, as both G and A amino acids have nonpolar side chains and similar size.
The common-HCDR3 subset shows a strong bias in the expression of λ LCs and in particular of the Vλ2-14 (IGLV3-21) gene. In addition, a worse prognosis due to progressive disease is also more frequent in this subset as compared to the remaining cases (P = .03). Nevertheless, most cases are actually unmutated and those that are not express some levels of CD38 on the cell surface. This makes it challenging to define the actual role of the IGHV3-21 rearrangement by itself in predicting the outcome as compared to the other biologic prognostic factors.
More than 50% of our IGHV3-21-expressing cases show HCDR3 rearrangements that are totally unrelated to each other and to the consensus sequence (nonhomogeneous-HCDR3 subset). They use diverse IGHJ and IGV LC genes and 5 of 9 cases are unmutated. They also show variable CD38 expression and have a heterogeneous clinical course with only 4 of 9 patients experiencing progressive disease. This subset may mirror the heterogeneity and low frequency of expression of IGHV3-21 rearrangements in the normal repertoire of peripheral blood B cells, as shaped by the germline composition, molecular mechanisms, and distinct selective influences.
The first subset, with the evidence of a similar antigen-binding site in so many different CLL cohorts, indicates that a potential environmental variable may be the selecting element for the clonal founder of this leukemic progeny in all European regions. We obtained statistically significant evidence for positive selection by antigen for all 3 mutated cases with a short HCDR3. This subset may be differently selected and expanded in different regions, according to the prevalence of the selective element in the specific geographic environment. Given the usually bad prognosis of this subset of patients it becomes interesting to hypothesize that a potential antigenic stimulation may be eventually responsible for an aggressive clinical course.
Interestingly, no consensus HCDR3 or LCDR3 sequences were identified among 5 multiple myeloma cases, one acute lymphoblastic leukemia case, and one follicular lymphoma case using the IGHV3-21 gene (K.S. and C.B., unpublished observations, 2004) supporting the idea of a selective pressure acting preferentially in CLL. According to this hypothesis, it is notable that among 89 IGHV3-21 (+) sequences from the IMGT database, mainly obtained from libraries of normal B cells as well as from antibodies against known pathogens, there were only 3 IGHV3-21 sequences with a 9 codons-long HCDR3 similar to the common-HCDR3 subset sequence, all using the IGHJ6 gene and all derived from CLL cases.
In contrast, the second subset includes cases heterogeneous at both the biologic and clinical levels, carrying diverse HCDR3 rearrangements and showing variable clinical outcomes. This subset may mirror the low frequency of expression of IGHV3-21 rearrangements in the normal repertoire of peripheral blood B cells as well as their biologic heterogeneity, shaped by the germline composition, molecular mechanisms, and distinct selective influences, thereby likely favoring a variable clinical course.
These findings bring in important implications. It was recently reported that unrelated CLL patients from different cohorts using other IGHV genes may share common CDR3s.29-31 Consequently, it would not be unreasonable to speculate that important biologic and (perhaps) prognostic information might be gained by defining not only the usage of specific genes (eg, IGHV3-21) and their mutational load but also the configuration of HCDR3 and the associated light chain of each patient among a subset of cases using similar IGHV genes.
In conclusion, the striking difference in the overall frequency of IGHV3-21 usage between North and South European CLL may be explained by the overweight in terms of percentage of the common-HCDR3 subset as compared to the nonhomogeneous IGHV3-21-expressing subset in the Scandinavian series. The lower frequency of the common-HCDR3 CLL subset in the Mediterranean region suggests that the hypothetical antigenic element may be less frequently encountered around the Mediterranean Sea. Still, it is unequivocally present, as demonstrated by those patients from France, Greece, Italy, and Spain who share strikingly similar HCDR3 sequences with Scandinavian patients.
Prepublished online as Blood First Edition Paper, October 5, 2004; DOI 10.1182/blood-2004-07-2606.
Supported in part by Associazione Italiana per la Ricerca sul Cancro (AIRC), Progetto Oncologia CNR-MIUR, MURST 40%, Association Claude Bernard, the José Carreras International Leukemia Foundation (EM-03; CR-03), and Generalitat de Catalunya 2001/SGR/00381.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank to Prof Marie-Paule Lefranc and Dr Veronique Giudicelli (Laboratoire d'Immunogenetique Moleculaire, LIGM, Universite Montpellier II, UPR CNRS) for sharing with us a wealth of insight on IG genes, offering valuable help in data analysis, and assisting us in navigating through the IMGT database and resources.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal