Abstract
Adult T-cell leukemia (ATL) caused by human T-cell leukemia virus type 1 (HTLV-1) infection, occurs in 2% to 4% of the HTLV-1 carriers with a long latent period, suggesting that additional alterations participate in the development of ATL. To characterize and identify novel markers of ATL, we examined the expression profiles of more than 12 000 genes in 8 cases of acute-type ATL using microarray. One hundred ninety-two genes containing interleukin 2 (IL-2) receptor α were up-regulated more than 2-fold compared with CD4+ and CD4+CD45RO+ T cells, and tumor suppressor in lung cancer 1 (TSLC1), caveolin 1, and prostaglandin D2 synthase showed increased expression of more than 30-fold. TSLC1 is a cell adhesion molecule originally identified as a tumor suppressor in the lung but lacks its expression in normal or activated T cells. We confirmed ectopic expression of the TSLC1 in all acute-type ATL cells and in 7 of 10 ATL- or HTLV-1–infected T-cell lines. Introduction of TSLC1 into a human ATL cell line ED enhanced both self-aggregation and adhesion ability to vascular endothelial cells. These results suggested that the ectopic expression of TSLC1 could provide a novel marker for acute-type ATL and may participate in tissue invasion, a characteristic feature of the malignant ATL cells.
Introduction
Adult T-cell leukemia (ATL) is a malignant and fatal disorder of CD4+ T cells caused by an infection of human T-cell leukemia virus type I (HTLV-1). Carriers of HTLV-1 are observed in the southern part of Kyushu in Japan and the Caribbean basin.1 ATL develops among these carriers with a very long latency: the lifetime risk of the disease in HTLV-1 carriers is estimated to be 2.6% in women and 4.5% in men.1
HTLV-1 encodes a transcriptional activator, Tax, in its pX region, which activates various cellular genes and creates an autocrine loop involving interleukin-2 (IL2) and its cognate IL-2 receptor α (IL2RA). Tax alters many transcriptional pathways: it activates cyclic AMP (adenosine monophosphate) response element binding protein (CREB), activator protein-1 (AP-1), and nuclear factor-kB (NFKB); represses p53; and interferes with several cell cycle regulators, including cyclins and cdk inhibitors (p15 and p16).2 Thus, these multiple functions of Tax are believed to participate in the immortalization of HTLV-1–infected cells.
In contrast, the role of Tax in advanced ATL cells is unclear. A previous study has shown that the expression of Tax is undetectable in circulating ATL cells, while a genetically defective provirus was observed in more than half of patients with ATL examined.3 Moreover, a report indicated that the 5′ long terminal repeat (LTR) of the HTLV-1 provirus is hypermethylated.4 These findings suggest that the HTLV-1 provirus is inactivated in ATL cells either by methylation in its 5′ LTR or by deletion of the proviral DNA. When the long latent period of about 50 years in ATL is considered, at least 5 additional genetic or epigenetic events appear to be required for the development of the overt disease.5 However, no cytogenetic abnormalities specific to ATL have been identified,6 although human leukemias often show reciprocal chromosomal translocations and associated genetic alterations.7 It is reported that the tumor suppressor genes p53, p15, and p16 are altered in aggressive ATL through loss of the chromosomes or methylation of the promoter.8 However, a molecular view of the multistep process of leukemogenesis in ATL remains to be obtained.
In the present study, we established the expression profiles and identified highly expressed genes in acute-type ATL cells to be available for specific markers using a GeneChip microarray containing oligonucleotide hybridization probes for more than 12 000 genes. After making expression profiles from the panels of ATL patients, we identified 3 genes that were markedly up-regulated more than 30-fold in ATL cells, including tumor suppressor in lung cancer 1 (TSLC1), caveolin 1 (CAV1), and prostaglandin D2 synthase (PGDS). Then, we confirmed the results by semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) or Northern blot hybridization or both. Among them, TSLC1 was especially interesting because it was originally identified as a tumor suppressor gene in lung cancer and is involved in cell adhesion.9 We have confirmed that TSLC1 is highly and ectopically expressed in all primary ATL cells and most HTLV-1–infected T-cell or ATL cell lines, whereas no expression was detected in CD4+ T cells or many other leukemia cells not related to HTLV-1 infection. Introduction of TSLC1 into human ATL cell line, ED, enhanced both self-aggregation activity and adhesion ability to human vascular endothelial cells. The biologic significance of TSLC1 in the multistage leukemogenesis of ATL will be discussed, focusing on the possible involvement of TSLC1 in adhesion and invasion.
Patients, materials, and methods
Patient samples
Eight patients with ATL were examined. All samples were collected at the time of admission to our hospital before the patients started chemotherapy. Relevant clinical data, profiles, and the extent of organ involvement for each case are summarized in Table 1. The mean age at diagnosis was 65.7 years. Diagnosis of ATL was made on the basis of clinical features, hematologic characteristics, serum antibodies against HTLV-1 antigens, and insertion of the HTLV-1 viral genome into leukemia cells by Southern blot hybridization. Using Shimoyama criteria,10 the 8 patients were diagnosed as having acute-type ATL. The infiltration of the lymphoid organs, skin, and liver by ATL cells was radiographically and histochemically confirmed by examining biopsy samples. Mononuclear cells were obtained from heparinized blood or ascites by Histopaque density gradient centrifugation (Sigma, St Louis, MO). After separation, ATL cell enrichment of more than 90% was confirmed by using 2-color flow cytometric analysis. All samples were separated by Histopaque density gradient centrifugation, quickly frozen within 3 hours, and cryopreserved at –80°C. This study was approved by the Institutional Review Board (IRB) at Miyazaki Medical Collage, University of Miyazaki. Informed consent was obtained from all blood and tissue donors according to the Helsinki Declaration.
Clinical features of patients with acute type of ATL
. | . | . | . | . | . | . | . | Involved organs . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case no. . | Age, y/sex . | WBC count, × 103/mL . | Flower cells . | LDH, IU/L . | Ca, mg/dL . | CD4, % . | CD25, % . | Skin . | Lymph node . | Liver/spleen . | Others . | ||
| 1 | 82/M | 48.6 | + | 1189 | 10.6 | 94.1 | 47.4 | - | + | - | - | ||
| 2 | 63/M | 5.1 | + | 2281 | 8.8 | 68.7 | 74.3 | - | + | - | AS/PE | ||
| 3 | 67/F | 29.8 | + | 757 | 9.1 | 4.8 | 58.9 | - | + | + | - | ||
| 4 | 70/M | 60.9 | + | 1003 | 11.2 | 96.4 | 86.2 | + | - | + | - | ||
| 5 | 60/F | 30.9 | + | 3187 | 20.1 | ND | ND | - | + | + | PE | ||
| 6 | 66/M | 30.4 | + | 506 | 8.8 | 94.7 | 93.7 | + | + | ND | - | ||
| 7 | 35/M | 114.4 | + | 666 | 9.2 | 97.8 | 93.3 | + | + | + | - | ||
| 8 | 73/M | 16.6 | + | 378 | 15.1 | 63.2 | 52.4 | + | + | + | - | ||
. | . | . | . | . | . | . | . | Involved organs . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case no. . | Age, y/sex . | WBC count, × 103/mL . | Flower cells . | LDH, IU/L . | Ca, mg/dL . | CD4, % . | CD25, % . | Skin . | Lymph node . | Liver/spleen . | Others . | ||
| 1 | 82/M | 48.6 | + | 1189 | 10.6 | 94.1 | 47.4 | - | + | - | - | ||
| 2 | 63/M | 5.1 | + | 2281 | 8.8 | 68.7 | 74.3 | - | + | - | AS/PE | ||
| 3 | 67/F | 29.8 | + | 757 | 9.1 | 4.8 | 58.9 | - | + | + | - | ||
| 4 | 70/M | 60.9 | + | 1003 | 11.2 | 96.4 | 86.2 | + | - | + | - | ||
| 5 | 60/F | 30.9 | + | 3187 | 20.1 | ND | ND | - | + | + | PE | ||
| 6 | 66/M | 30.4 | + | 506 | 8.8 | 94.7 | 93.7 | + | + | ND | - | ||
| 7 | 35/M | 114.4 | + | 666 | 9.2 | 97.8 | 93.3 | + | + | + | - | ||
| 8 | 73/M | 16.6 | + | 378 | 15.1 | 63.2 | 52.4 | + | + | + | - | ||
+ indicates infiltration of ATL cells into each organ; - indicates no infiltration. WBC indicates white blood cells; LDH, lactate dehydrogenase; AS, ascites; PE, pleural effusion; ND, not done.
CD4+ and CD4+CD45RO+ T cells
CD4+ T cells were obtained from 5 healthy volunteers. All samples were quickly frozen within 3 hours and cryopreserved at –80°C after separation as follows. Highly purified CD4+ T cells were obtained through negative depletion from the peripheral blood cells by using a RosetteSep cocktail (CD8, CD16, CD19, CD36, CD56) as recommended by the manufacturer (StemCell Technologies, Vancouver, BC, Canada).11 After centrifugation, all hematopoietic cells except CD4+ T cells were precipitated, and the CD4+ T cells were enriched more than 90%, as confirmed by flow cytometric analysis. CD4+CD45RO+ T cells were obtained from 3 other volunteers and separated from CD4+ T cells with anti-CD45RA monoclonal antibodies (Becton Dickinson, Mountain View, CA) and immunobeads (Dynal AS, Oslo, Norway). Resting T cells were used as a reference, because ATL cells and T cells were freshly isolated and spared from unnecessary stimulation by cell culture or antibody selection. For the usage of activated T cells, CD4+ T cells were stimulated with 10 μg/mL phytohemagglutinin (PHA) for 48 hours or with 1 μg immobilized anti–human CD3 antibody (UCHT1; Becton Dickinson Biosciences) and 10 μg soluble anti–human CD28 (CD28.2; Becton Dickinson Biosciences) for 72 hours according to manufacturer's recommendation.
Hematopoietic cell fractions
A series of hematopoietic cell fractions were prepared as previously reported.12 Briefly, human bone marrow mononuclear cells were isolated by Ficoll-Hypaque centrifugation from healthy volunteers. CD34+ cells and megakaryocytes in bone marrow were isolated using DYNABEADS M-450 (Dynal) coupled with monoclonal antibodies against CD34 (QBWND10) and human platelet glycoprotein IIb/IIIa (GpIIb/IIIa; TP80; Seikagaku-Kogyo, Tokyo, Japan). Human monocytes, T cells, and B cells in peripheral blood were purified by sorting with Epics Elite (Coulter, Miami, FL) after labeling with monoclonal antibodies against CD14, CD2, and CD20. Erythrocyte and neutrophil fractions were separated from peripheral blood by Ficoll-Hypaque centrifugation.
Cell lines
Ten of the cell lines used (Jurkat, MOLT-4, KAWAI, MOLT3, MOLT15, MOLT16, SKW3, HUT78, St Luke, and Chiba) are HTLV-I–negative human T-cell acute lymphoblastic leukemia (T-ALL) cell lines. Four cell lines (KOB, SO-4, KK-1, and ST-1) are IL2-dependent ATL cell lines.13 ED, Su9T, and S1T are IL2-independent ATL lines.14,15 MT-2, HUT-102, and OMT are human T-cell lines transformed by HTLV-1 infection.16-18 KOB, SO-4, KK-1, ST-1, and OMT were kindly provided by Dr Y. Yamada (Nagasaki University), ED cells by Dr M. Maeda (Kyoto University), and Su9T and S1T cells by Dr N. Arima (Kagoshima University). IL2-dependent ATL cell lines and the murine IL2-dependent T-cell line CTLL219 were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), and 100 JRU/mL recombinant human IL2 (Takeda Chemical Industries, Osaka, Japan). HTLV-I–negative cell lines, cell lines transformed with HTLV-1, and IL2-independent ATL cell lines were maintained in the same medium without IL2. The HTLV-1–negative leukemia cell lines consisted of 17 myelocytic/monocytic (HL-60, HNT-34, Kasumi-2, Kasumi-3, Kasumi-4, Kumamoto, UCSD/AML1, KG-1, INV3, MO, MO7, Hiroshima-1, THP1, Hiroshima-2, LGC-2, U937, KH88), 3 megakayrocytic (UT7/GM, CMK, MOLM1), 2 erythrocytic (HEL, K562), and 4 B-lymphocytic (SCMC-L1, TAKEDA, SKW1, UTP-2) lines and were maintained in the RPMI 1640 medium without IL2.20
Flow cytometric analysis
Cells were washed with phosphate-buffered saline (PBS) containing 2% FBS and incubated for 30 minutes with monoclonal antibodies, phycoerythrin (PE)–labeled anti-CD4 and fluorescein isothiocyanate (FITC)–labeled anti-CD25 monoclonal antibodies (Becton Dickinson), or control isotype-matched murine immunoglobulin G (IgG) for double staining. Treated cells were analyzed on a FACScan (Becton Dickinson).
Oligonucleotide microarray
The protocol used for the sample preparation and microarray processing is available from Affymetrix (Santa Clara, CA). Briefly, at least 5 μg purified RNA was reverse transcribed by Superscript II reverse transcriptase (Life Technologies, Grand Island, NY) using the primer T7-dT24 containing a T7 RNA polymerase promoter. After a second strand of cDNA was synthesized by RNase H, Escherichia coli DNA polymerase, and E coli DNA ligase, in vitro transcription was performed on the cDNA to produce biotin-labeled cRNA with a MEGAscript High Yield Transcription Kit (Ambion, Austin, TX) as recommended by the manufacturer. After the cRNA was linearly amplified with T7 polymerase, the biotinylated cRNA was cleaned with an RNeasy Mini Column (Qiagen, Valencia, CA), fragmented to 50 to 200 nucleotides, and then hybridized to Affymetrix HU95A arrays. The stained microarray was scanned with a GeneArray Scanner (Affymetrix) and signal was calculated with Affymetrix software, Microarray Suite 5.0. All of the data were scaled with the global scaling method to adjust the target intensity to 300.
Data analysis
The expression value for each gene was determined by calculating the average of differences (perfect match intensity minus mismatch intensity) of the probe pairs in use for the gene. Fold change was calculated for each sample against the median of the controls. The small and negative expression levels were clipped-off to be equal to a cutoff value arbitrarily chosen as 100. Then we chose the genes showing an increase in expression of more than 2-fold in the ATL cells compared with CD4+ T cells or CD4+CD45RO+ T cells with statistical significance of P less than .01 by z test. A 2-fold cutoff was chosen, based on statistical information provided by Affymetrix.21
RT-PCR
Total cellular RNA from cryopreserved cells was extracted with Trizol (Life Technologies) according to the protocol provided by the manufacturer. First-strand cDNA was synthesized from 1 μg total cellular RNA in a 20-μL reaction volume using an RNA-PCR kit (Takara Shuzo, Kyoto, Japan) with oligo dT primers. Primers were 5′-ATCCCGTGGAGACTCCTCAA-3′ (forward) and 5′-AACACGTAGACTGGGTATCC-3′ (reverse) for HTLV-1 Tax, 5′-ATGATCGATATCCAGAAAGACACT-3′ (forward) and 5′-GTACTTCTAGATACCGCTGGG-3′ (reverse) for TSLC1, 5′-ATGATCGATATCCAGAAAGACACT-3′ (forward) and 5′-ATTTTGCAGACGCTCTCAGCA-3′ (reverse) for IL2RA, 5′-ATGCCTTACTCTTCTTGAAAT-3′ (forward) and 5′-TGATCAGCTGAAATGGGAA-3′ (reverse) for CAV1, 5′-CCTCTCACTGGCTCGTGATT-3′ (forward) and 5′-GGAGGGAGCATGTGGATTAT-3′ (reverse) for PGDS, 5′-ATCACTCTCTTTAATCACTACT-3′ (forward) and 5′-ACTTAATTATCAAGTTAGTGTTG-3′ (reverse) for IL2, 5′-TCCTTCTGCATCCTGTCGGCT-3′ (forward) and 5′-CCAGAGATGGCCACGGCTGCT-3′ (reverse) for β-actin. The length of the semiquantitative RT-PCR for each gene was as follows; 30 cycles for HTLV-1 Tax, IL2RA, CAV1, and PGDS; 32 cycles for TSLC1 and IL2; and 25 cycles for β-actin. The PCR products were fractionated on 2% agarose gels and visualized by ethidium bromide staining.
Northern blot analysis
Messenger RNA was extracted from the cells using FastTrack 2.0 as recommended by the manufacturer (Invitrogen, Carlsbad, CA). Messenger RNA (3 μg) was separated by electrophoresis on a denaturing 1% agarose gel containing formaldehyde in 4-morpholinopropanesulfonic acid buffer (MOPS) and blotted overnight onto nylon membranes (BIODYNE Pall BioSupport, East Hills, NY). The membranes were prehybridized in 50 mM sodium phosphate, 0.5% sodium dodecyl sulfate (SDS), and 100 μg/mL salmon sperm DNA for 2 hours at 42°C and then hybridized overnight with 32P-radiolabeled probes. A 961-base pair (bp) PCR-derived fragment (nt 411-1371) was used to detect TSLC1.9 Radiolabeled probes were generated using a Prime-lt II Random Primer Labeling Kit (Stratagene, La Jolla, CA).
Plasmids and transfection
We amplified the entire coding sequence of TSLC1 by PCR using the plasmid pcTSLC19 (primers, 5′-GAATTCCCCGACATGGCGTAGTGTA-3′ and 5′-CTCGAGCTAGATGAAGTACTCTTTCTT-3′). Amplified fragments were digested with restriction endonucleases, EcoRI and XhoI, and subcloned into pMSCVneo (Becton Dickinson) to obtain the plasmid MSCV-TSLC1. MSCV-TSLC1 and pMSCVneo were transfected into RetroPack PT 67 packaging cells (Becton Dickinson) with FuGENE 6 reagent treatment according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). Retroviral supernatant was harvested 48 hours after transfection and supplemented with 8 μg/mL polybrene to infect ED cells. Stable retrovirus-producing ED cells were selected using G418 (800 μg/mL). The construction of pTSLC1-GFP (green fluorescent protein) was described previously.9 To isolate CTLL2 cells stably expressing TSLC1-GFP, CTLL2 cells were transfected with pTSLC1-GFP using electroporation, according to the manufacturer's directions. Stable transformants were selected in the media containing 400 μg/mL Geneticin (G418; Sigma) and were maintained in the presence of 400 μg/mL G418.
Cell adhesion assay
ED transformants in serum-free RPMI 1640 medium were labeled with a Vybrant Cell Adhesion Assay Kit (Molecular Probes, Eugene, OR) for 60 minutes at 37°C. Human umbilical vein endothelial cells (HUVECs; 3.5 × 104) were plated onto 96-well plates and cultured until 80% to 90% confluent. A total of 1 × 105 labeled ED transformants in 100 μL RPMI 1640 medium with 10% FBS were then added to each well. After being left for 30 minutes at 37°C, the wells were washed 3 times with warmed RPMI medium. After further washing, the fluorescence of adherent cells was measured at an excitation wavelength of 485 nm and emission wavelength of 530 nm using a Fluoroscan fluorescence reader (Titertek, Helsinki, Finland). All experiments were performed in triplicate.
Promoter methylation analysis
Promoter methylation was analyzed using bisulfite-SSCP (single-strand conformation polymorphism) coupled with a sequencing method as described previously.9 After denaturing treatment with NaOH (0.3 M), genomic DNA (2 μg) was incubated with sodium bisulfite (3.1 M; Sigma) and hydroquinone (0.8 mM; Sigma), purified, and treated with NaOH (0.2 M). The modified DNA (100 ng) was subjected to PCR to amplify the promoter sequence of the TSLC1 gene with primers, one of which was end-labeled with Texas Red as described previously.9,22 The PCR product was diluted 10 times with a loading buffer (90% deionized formamide, 0.01% New Fuchsin, and 10 mM EDTA[ethylenediaminetetraacetic acid]), heat-denatured for 3 minutes at 95°C, cooled on ice for 3 minutes, and then loaded onto the gel (0.5 × MDE Gel Solution; BMA, Rockland, ME). Electrophoresis was carried out for 120 minutes at 20°C using SF5200 (Hitachi Electronics Engineering, Tokyo, Japan) with cooling systems (single-strand conformation polymorphism [SSCP] analysis). The results were analyzed using a DNA Fragment Analyzer (Hitachi Electronics Engineering Co.). Hypermethylation, partial methylation, and unmethylation were determined from the mobility of the fragments in comparison with that of clones with known sequences. The PCR products were also subcloned to confirm the sequences in at least 4 clones.
Results
Profile of gene expression in ATL cells compared with CD4+ and CD4+CD45RO+ T cells
To search for novel markers of acute-type ATL cells, we initially made gene expression profiles of primary ATL cells using an oligonucleotide microarray (HU95A; Affymetrix) that contains a total of 12 625 genes. Since it is reported that ATL cells belong to CD4+, CD8–, and CD45RO+ cells,23 we used CD4+ and CD4+CD45RO+ T cells in peripheral blood from healthy volunteers as controls. Although ATL is a relatively rare disease, we collected leukemia cells from 8 patients with acute ATL and compared them with 5 samples of CD4+ and 3 samples of CD4+CD45RO+ T cell subsets. A search for genes whose expression in acute ATL cells showed more than a 2-fold increase compared with the control cells yielded 422 and 610 genes from the comparison with CD4+ and CD4+CD45RO+ T cells, respectively (data not shown). Then, we detected 192 genes whose expression was up-regulated in acute ATL cells compared with both T-cell subsets (Table 2).
Highly expressed genes in ATL cells compared with normal CD4+ and CD4+CD45RO+ T cells
. | . | . | Fold change . | . | |
|---|---|---|---|---|---|
| Accession no. . | Symbol . | Gene . | CD4+ . | CD45RO+ . | |
| AF070648 | CAV1 | Caveolin 1* | 34.82 | 49.74 | |
| AF150241 | PGDS | Prostaglandin D2 synthase | 31.21 | 12.56 | |
| AL080181 | TSLC1 | Tumor suppressor in lung cancer 1 | 30.36 | 36.86 | |
| M36711 | TFAP2A | Transcription factor AP-2 alpha | 19.14 | 11.28 | |
| AF079529 | PDE8B | Phosphodiesterase 8B | 17.58 | 13.07 | |
| AB002305 | ARNT2 | Aryl-hydrocarbon receptor nuclear translocator 2 | 17.15 | 22.59 | |
| X58288 | PTPRM | Protein tyrosine phosphatase, receptor type, M | 15.89 | 5.12 | |
| Y00664 | NMYC | V-myc myelocytomatosis viral related oncogene, neuroblastoma | 15.30 | 12.11 | |
| J03802 | PTHLH | Parathyroid hormone-like hormone*† | 14.10 | 8.68 | |
| AF022797 | KCNN4 (hKCa4) | Potassium intermediate/small conductance calcium-activated | 12.78 | 6.05 | |
| X00948 | RLN2 | Relaxin 2 (H2) | 12.02 | 8.85 | |
| M25753 | CCNB1 | CyclinB1 | 11.53 | 5.33 | |
| W26466 | NA | 32f11 | 11.20 | 10.83 | |
| X65233 | ZNF80 | Zinc finger protein 80 (pT17) | 10.54 | 7.10 | |
| U49089 | DLG3 | Discs, large (Drosophila) homolog 3 | 9.60 | 6.79 | |
| AL080146 | CCNB2 | CyclinB2 | 9.56 | 7.18 | |
| AI760162 | NA | wg58e09.x1 | 9.19 | 4.34 | |
| D87119 | TRB2 | Tribbles homolog 2 | 9.18 | 4.24 | |
| AF030107 | RGS13 | Regulator of G-protein signaling 13 | 8.33 | 9.41 | |
| AF024714 | AIM2 | Interferon-inducible protein (AIM2) absent in melanoma | 8.26 | 5.74 | |
| D13633 | DLG7 | Discs large homolog 7 | 7.53 | 3.06 | |
| M37712 | CDC2L2 | Cell division cycle 2-like 2 | 7.16 | 7.26 | |
| X01057 | IL2RA | Interleukin 2 receptor, alpha*† | 6.78 | 3.73 | |
| AI375913 | NA | tc14c08.x1 | 6.62 | 4.23 | |
| AB020316 | UST | Uronyl-2-sulfotransferase | 6.58 | 3.72 | |
| L25876 | CDKN3 | CIP2 (cyclin-dependent interacting protein 2) cyclin-dependent kinase inhibitor 3 | 6.39 | 8.06 | |
| D38073 | MCM3 | Minichromosome maintenance deficient 3 | 6.34 | 2.71 | |
| AB014566 | DAAM1 | Disheveled associated activator of morphogenesis 1 | 6.27 | 5.55 | |
| AF007155 | LOC254531 | PISC domain containing hypothetical protein | 6.14 | 4.20 | |
| L47276 | HUMTOPATR | Homo sapiens alpha topoisomerase truncated-form mRNA | 6.12 | 4.03 | |
| U37426 | KIF11 | Kinesin family member 11 | 6.12 | 3.15 | |
| AB012911 | FZD6 | Frizzled homolog 6 | 6.12 | 5.48 | |
| AL080121 | DKFZp564O0823 | Highly similar to rat castration induced prostatic apoptosis related protein-1 | 6.10 | 6.96 | |
| X83490 | TNFRSF6 | Tumor necrosis factor receptor superfamily member 6* (Fas/APO-1) | 6.02 | 3.85 | |
| AL050107 | TAZ | Transcriptional co-activator with PDZ (PSD-95, DLG, and ZO-1)—binding motif | 5.81 | 5.67 | |
| J04088 | TOP2A | Topoisomerase (DNA) II alpha | 5.59 | 4.52 | |
| D42055 | NEDD4 | Neural precursor cell expressed, developmentally down-regulated 4 | 5.47 | 2.02 | |
| D38251 | POLR2E | Polymerase (RNA) II (DNA directed) polypeptide E | 5.40 | 8.51 | |
| M74089 | LOC91137 | Hypothetical protein BC017169 | 5.28 | 3.81 | |
| D13643 | DHCR24 | 24-dehydrocholesterol reductase | 5.17 | 5.52 | |
| D14657 | KIAA0101 | KIAA0101 gene product | 4.94 | 3.93 | |
| AA203476 | NA | zx55e01.rl | 4.92 | 2.31 | |
| M13755 | GIP2 | Interferon, alpha-inducible protein | 4.86 | 6.20 | |
| U63743 | KIF2C | Kinesin family member 2C | 4.69 | 3.78 | |
| J03473 | ADPRT | ADP-ribosyltransferase | 4.52 | 3.58 | |
| U43916 | EMP1 | Epithelial membrane protein 1 | 4.49 | 2.75 | |
| Z70519 | TNFRSF6 | FAS soluble protein* | 4.44 | 2.69 | |
| U73379 | UBE2C | Ubiquitin-conjugating enzyme E2C | 4.43 | 4.57 | |
| D86062 | C21orf33 | Chromosome 21 open reading frame 33 | 4.41 | 7.29 | |
| AI127424 | NA | qb75b02.x1 | 4.36 | 6.10 | |
| AC004528 | C19orf6 | Chromosome 19 open reading frame 6 | 4.31 | 4.16 | |
| AL050069 | DOK5 | Docking protein 5 | 4.27 | 5.45 | |
| U81561 | PTPRN2 | Protein tyrosine phosphatase, receptor type, N polypeptide 2 | 4.24 | 5.04 | |
| M15205 | TK1 | Thymidine kinase 1, soluble | 4.21 | 4.71 | |
| M84443 | GALK2 | Galactokinase 2 | 4.09 | 2.29 | |
| D63861 | PPID | Peptidylprolyl isomerase D (cyclophilin D) | 4.08 | 2.34 | |
| X02883 | HSTCRAC | Human gene for T-cell receptor alpha chain C region | 4.05 | 2.81 | |
| D00596 | TYMS | Thymidylate synthetase | 4.03 | 4.17 | |
| U03494 | TFCP2 | Transcription factor CP2 (CCAAT protein) | 3.99 | 3.15 | |
| U31601 | JAK3 | Janus kinase 3 (a protein tyrosine kinase, leukocyte) | 3.95 | 3.48 | |
| AF009426 | C18orf1 | Chromosome 18 open reading frame 1 | 3.93 | 2.87 | |
| AL022398 | ADORA2BP | Adenosine A2b receptor pseudogene | 3.90 | 2.80 | |
| M97935 | STAT1 | Signal transducer and activator of transcription 1 | 3.89 | 4.36 | |
| AA919102 | NA | o184h02.sl | 3.86 | 2.11 | |
| U88964 | ISG20 | Interferon-stimulated gene | 3.81 | 3.21 | |
| AJ010953 | ATP2CI | ATPase, Ca++ transporting, type 2C, member 1 | 3.73 | 2.87 | |
| U53003 | C21orf33 | Chromosome 21 open reading frame 33 | 3.69 | 2.76 | |
| M36711 | NA | DNA-binding protein Ap-2 | 3.67 | 3.09 | |
| M34641 | FGFR1 | Fibroblast growth factor receptor 1 | 3.62 | 2.37 | |
| AI652660 | wb30c10.x1 | 3.62 | 3.47 | ||
| AJ000534 | SGCE | Sarcoglycan, epsilon | 3.61 | 3.07 | |
| D50920 | THRAP4 | Thyroid hormone receptor associated protein 4 | 3.48 | 2.05 | |
| AB024704 | TPX2 | Microtubule-associated protein homolog | 3.45 | 3.20 | |
| X14798 | ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 | 3.44 | 3.36 | |
| Y11681 | Y11681 | Mitochondrial ribosomal protein S12 | 3.44 | 7.90 | |
| D26155 | SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | 3.42 | 3.10 | |
| AL031737 | NA | Human DNA sequence from clone 8B22 on chromosome 1p35.1-36.21 | 3.42 | 2.35 | |
| M86699 | TTK | TTK (threonine/tyrosine kinase) protein kinase | 3.40 | 2.38 | |
| U16799 | ATP1B1 | ATPase, Na+/K+ transporting, beta 1 polypeptide | 3.39 | 3.81 | |
| AB018270 | MYO1D | Myosin ID | 3.34 | 2.57 | |
| U76638 | BARD1 | BRCA1 (breast cancer 1) associated RING domain 1 | 3.34 | 2.62 | |
| AB013452 | ATP8A1 | ATPase, aminophospholipid transporter (APLT), class 1, type 8A, member 1 | 3.32 | 3.29 | |
| Y11525 | CEBPA | CCAAT/enhancer binding protein (C/EBP), alpha | 3.32 | 3.34 | |
| M34423 | GLB1 | Galactosidase, beta 1 | 3.31 | 4.04 | |
| AA203213 | NA | zx57e04.r1 | 3.31 | 3.49 | |
| X64624 | POU4F1 | POU (Pit, Oct, Unc) domain, class 4, transcription factor 1 | 3.31 | 3.92 | |
| AB018271 | BPAG1 | Bullous pemphigoid antigen 1 | 3.25 | 2.12 | |
| LI3435 | NA | Human chromosome 3p21.1 gene sequence | 3.24 | 2.92 | |
| AF055993 | SAP30 | Sin3-associated polypeptide | 3.18 | 3.47 | |
| U29332 | FHL2 | Four and a half LIM (Lin-1, Tsl-1, MEC-3) domains 2 | 3.16 | 3.60 | |
| AI540958 | NA | Homo sapiens cDNA, 5 end/clone_end=5 | 3.15 | 3.59 | |
| D38522 | SYT11 | Synaptotagmin XI | 3.09 | 3.58 | |
| U01038 | PLK1 | Polo-like kinase 1 | 3.07 | 3.40 | |
| X74262 | RBBP4 | Retinoblastoma binding protein 4 | 3.05 | 5.58 | |
| L23959 | TFDP1 | Transcription factor Dp-1 (E2F dimerization partner-1) | 3.03 | 3.76 | |
| M87434 | OAS2 | 2′-5′-oligoadenylate synthetase 2 | 3.02 | 2.96 | |
| M25280 | SELL | Selectin L (lymphocyte adhesion molecule 1) | 3.02 | 3.31 | |
| M63488 | RPA1 | Replication protein A1 | 3.01 | 3.39 | |
| U78190 | GCHFR | GTP cyclohydrolase I feedback regulatory protein | 3.01 | 2.09 | |
| AF067656 | ZWINT | ZW10 interactor | 2.97 | 2.50 | |
| M15024 | MYB | v-myb myeloblastosis viral oncogene homolog | 2.97 | 4.54 | |
| M81830 | SSTR2 | Somatostatin receptor 2 | 2.96 | 3.33 | |
| D28364 | ANXA11 | Annexin II, 5 | 2.96 | 6.05 | |
| M63838 | FI16 | Interferon, gamma-inducible protein 16 | 2.92 | 3.05 | |
| X72889 | NA | NA | 2.91 | 2.45 | |
| AC003108 | Anxa6 | Annexin VI | 2.84 | 3.79 | |
| L07541 | RFC3 | Replication factor C (activator 1) 3 | 2.83 | 2.67 | |
| D78261 | IRF4 | Interferon regulatory factor 4* | 2.83 | 3.14 | |
| AA005018 | NA | zh96a09.rl | 2.83 | 2.12 | |
| AA310786 | NA | EST181572 Homo sapiens cDNA | 2.78 | 2.29 | |
| M87284 | OAS2 | 2′-5′-oligoadenylate synthetase 2 | 2.77 | 3.11 | |
| U09813 | ATP5G3 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c (subunit 9) isoform 3 | 2.76 | 4.85 | |
| AC005253 | UBA52 | Ubiquitin A-52 residue ribosomal protein fusion product 1 | 2.75 | 2.85 | |
| AF070523 | JWA | Cytoskeleton related vitamin A responsive protein | 2.75 | 2.29 | |
| U38545 | PLD1 | Phospholipase D1, phophatidylcholine-specific | 2.74 | 2.17 | |
| U25182 | PRDX4 | Peroxiredoxin 4 | 2.73 | 3.10 | |
| AF022385 | PDCD10 | Programmed cell death 10 | 2.73 | 2.09 | |
| AF000416 | EXTL2 | Exostoses (multiple)-like 2 | 2.73 | 2.13 | |
| D26361 | KIF14 | Kinesin family member 14 | 2.72 | 2.47 | |
| AF064084 | ICMT | Isoprenylcysteine carboxyl methyltransferase | 2.70 | 4.31 | |
| X98248 | SORT1 | Sortilin 1 | 2.69 | 5.01 | |
| U65410 | MAD2L1 | MAD2 (mitotic arrest-deficient 2) mitotic arrest deficient-like 1 | 2.66 | 2.73 | |
| U51004 | HINT1 | Histidine triad nucleotide binding protein 1 | 2.64 | 2.09 | |
| M23379 | RASA1 | RAS p21 protein activator (GTPase activating protein) 1 | 2.64 | 2.43 | |
| X57398 | PM5 | pM5 protein | 2.63 | 2.65 | |
| X65550 | MK167 | Antigen identified by monoclonal antibody Ki-67 | 2.60 | 3.41 | |
| J05614 | PCNA | Proliferating cell nuclear antigen† | 2.59 | 2.50 | |
| X67951 | PRDX1 | Peroxiredoxin 1 | 2.58 | 2.16 | |
| M11507 | TFRC | Transferrin receptor (p90, CD71) | 2.57 | 2.59 | |
| D26599 | PSMB2 | Proteasome (prosome, macropain) subunit, beta type, 2 | 2.55 | 2.91 | |
| AF053551 | MTX2 | Metaxin 2 | 2.53 | 3.54 | |
| AF016898 | BATF | Basic leucine zipper transcription factor, AT-like | 2.50 | 2.04 | |
| AF055993 | SAP30 | sin3-associated polypeptide | 2.49 | 2.73 | |
| D14710 | ATP5A1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1 | 2.48 | 2.01 | |
| U09564 | NA | Human serine kinase | 2.48 | 2.51 | |
| AL109672 | P24B | Integral type I protein | 2.48 | 3.85 | |
| AI056696 | NA | oz26h05.x1 | 2.44 | 2.65 | |
| M13228 | MYCN | v-myc myelocytomatosis viral related oncogene, neuroblastoma derived | 2.44 | 2.78 | |
| AI566877 | NA | tn24f02.x1 | 2.44 | 2.25 | |
| L09235 | ATP6V1A | ATPase, H+ transporting, lysosomal | 2.42 | 5.18 | |
| X51688 | CCNA2 | Cyclin A2 | 2.36 | 2.45 | |
| U13695 | PMS1 | Postmeiotic segregation increased 1 | 2.34 | 2.17 | |
| AB007455 | TP53AP1 | TP53 activated protein 1 | 2.33 | 2.77 | |
| D44497 | CORO1A | Coronin, actin binding protein, 1A | 2.33 | 2.77 | |
| AA255502 | NA | zr85b06.r1 | 2.33 | 2.78 | |
| X73066 | NME1 | Non-metastatic cells 1, protein (NM23A) expressed in | 2.32 | 3.00 | |
| AI347088 | NA | qp60d09.x1 | 2.32 | 2.21 | |
| U37689 | POLR2H | Polymerase (RNA) II (DNA directed) polypeptide H | 2.32 | 2.13 | |
| U86529 | GSTZ1 | Glutathione transferase zeta 1 | 2.32 | 2.28 | |
| J03133 | SP1 | SP1 (SV40 promoter 1) transcription factor | 2.31 | 4.73 | |
| AB007447 | FLN29 | FLN29 gene product (TNF receptor factor-like protein) | 2.30 | 2.34 | |
| X74008 | PPP1CC | Protein phosphatase 1, catalytic subunit, gamma isoform | 2.30 | 2.07 | |
| U56833 | VBP1 | von Hippel-Lindau binding protein 1 | 2.29 | 2.16 | |
| AF020038 | IDH1 | Isocitrate dehydrogenase 1 (NADP+), soluble | 2.29 | 3.40 | |
| U63809 | PAWR | PRKC (protein kinase C), apoptosis, WT1, regulator | 2.29 | 2.35 | |
| AL050034 | ADPRTL3 | ADP-ribosyltransferase-like 3 | 2.25 | 2.09 | |
| M87339 | HUMACT1A | Human replication factor C, 37-kDa subunit | 2.25 | 2.98 | |
| AA056747 | NA | zk81f02.s1 | 2.24 | 2.95 | |
| U10886 | PTPRJ | Protein tyrosine phosphatase, receptor type, J | 2.23 | 2.52 | |
| X84194 | ACYP1 | A-cylphosphatase 1, erythrocyte (common) type | 2.23 | 4.40 | |
| AB024401 | ING1 | Inhibitor of growth family, member 1 | 2.20 | 14.85 | |
| M19481 | FST | Follistatin | 2.20 | 2.54 | |
| AF006084 | ARPC1B | Actin related protein 2/3 complex, subunit 1B, | 2.19 | 3.71 | |
| Y00097 | ANXA6 | Annexin A6 | 2.19 | 2.00 | |
| D00017 | ANXA2 | Annexin A2 | 2.18 | 2.81 | |
| U50079 | HDAC1 | Histone deacetylase 1 | 2.17 | 2.62 | |
| AA811338 | NA | ob81g05.sl | 2.17 | 5.19 | |
| M97676 | MSX1 | msh homeo box homolog 1 | 2.16 | 3.15 | |
| M63573 | PPIB | Peptidylprolyl isomerase B (cyclophilin B) | 2.16 | 2.85 | |
| J00314 | OK/SW-cl.56 | beta 5-tubulin | 2.16 | 2.19 | |
| U30872 | CENPF | Centromere protein F | 2.16 | 2.43 | |
| AA969267 | NA | on57d12.sl | 2.13 | 2.49 | |
| AF053306 | BUB1B | Budding uninhibited by benzimidazoles 1 homolog beta | 2.13 | 2.04 | |
| AF042384 | BC-2 | Putative breast adenocarcinoma marker | 2.12 | 2.52 | |
| U32849 | NMI | N-myc (and STAT) interactor | 2.12 | 2.90 | |
| AB028975 | KIAA1052 | KIAA1052 protein | 2.12 | 2.53 | |
| AJ000480 | C8FW | Phosphoprotein regulated by mitogenic pathways | 2.11 | 4.49 | |
| D50692 | MYCBP | c-myc binding protein | 2.10 | 3.00 | |
| M16279 | CD99 | CD99 antigen | 2.10 | 2.94 | |
| X69398 | CD47 | CD47 antigen (Rh-related antigen) | 2.09 | 2.17 | |
| AF054825 | VAMP5 | Vesicle-associated membrane protein 5 | 2.09 | 2.69 | |
| D37984 | RECQL | RecQ protein-like (DNA helicase Q1-like) | 2.09 | 2.27 | |
| U90549 | HMGN4 | High mobility group nucleosomal binding domain 4 | 2.09 | 2.16 | |
| AF087036 | MSC | Musculin (activated B-cell factor-1) | 2.08 | 2.67 | |
| J04058 | ETFA | Electron-transfer-flavoprotein, alpha polypeptide | 2.06 | 2.54 | |
| X62534 | HMGB2 | High-mobility group box 2 | 2.04 | 2.26 | |
| S57501 | NA | Protein phosphatase 1, alpha catalytic subunit | 2.04 | 2.71 | |
| AJ001612 | PSPHL | Phosphoserine phosphatase-like | 2.03 | 2.89 | |
| AJ245416 | LSM2 | LSM2 (Sm-like protein 2) homolog, U6 small nuclear RNA associated | 2.03 | 2.18 | |
| U91932 | AP3S1 | Adaptor-related protein complex 3, sigma 1 subunit | 2.03 | 2.89 | |
| U74612 | FOXM1 | Forkhead box M1 | 2.02 | 2.17 | |
| L42243 | IFNAR2 | Interferon (alpha, beta, and omega) receptor 2 | 2.02 | 2.88 | |
. | . | . | Fold change . | . | |
|---|---|---|---|---|---|
| Accession no. . | Symbol . | Gene . | CD4+ . | CD45RO+ . | |
| AF070648 | CAV1 | Caveolin 1* | 34.82 | 49.74 | |
| AF150241 | PGDS | Prostaglandin D2 synthase | 31.21 | 12.56 | |
| AL080181 | TSLC1 | Tumor suppressor in lung cancer 1 | 30.36 | 36.86 | |
| M36711 | TFAP2A | Transcription factor AP-2 alpha | 19.14 | 11.28 | |
| AF079529 | PDE8B | Phosphodiesterase 8B | 17.58 | 13.07 | |
| AB002305 | ARNT2 | Aryl-hydrocarbon receptor nuclear translocator 2 | 17.15 | 22.59 | |
| X58288 | PTPRM | Protein tyrosine phosphatase, receptor type, M | 15.89 | 5.12 | |
| Y00664 | NMYC | V-myc myelocytomatosis viral related oncogene, neuroblastoma | 15.30 | 12.11 | |
| J03802 | PTHLH | Parathyroid hormone-like hormone*† | 14.10 | 8.68 | |
| AF022797 | KCNN4 (hKCa4) | Potassium intermediate/small conductance calcium-activated | 12.78 | 6.05 | |
| X00948 | RLN2 | Relaxin 2 (H2) | 12.02 | 8.85 | |
| M25753 | CCNB1 | CyclinB1 | 11.53 | 5.33 | |
| W26466 | NA | 32f11 | 11.20 | 10.83 | |
| X65233 | ZNF80 | Zinc finger protein 80 (pT17) | 10.54 | 7.10 | |
| U49089 | DLG3 | Discs, large (Drosophila) homolog 3 | 9.60 | 6.79 | |
| AL080146 | CCNB2 | CyclinB2 | 9.56 | 7.18 | |
| AI760162 | NA | wg58e09.x1 | 9.19 | 4.34 | |
| D87119 | TRB2 | Tribbles homolog 2 | 9.18 | 4.24 | |
| AF030107 | RGS13 | Regulator of G-protein signaling 13 | 8.33 | 9.41 | |
| AF024714 | AIM2 | Interferon-inducible protein (AIM2) absent in melanoma | 8.26 | 5.74 | |
| D13633 | DLG7 | Discs large homolog 7 | 7.53 | 3.06 | |
| M37712 | CDC2L2 | Cell division cycle 2-like 2 | 7.16 | 7.26 | |
| X01057 | IL2RA | Interleukin 2 receptor, alpha*† | 6.78 | 3.73 | |
| AI375913 | NA | tc14c08.x1 | 6.62 | 4.23 | |
| AB020316 | UST | Uronyl-2-sulfotransferase | 6.58 | 3.72 | |
| L25876 | CDKN3 | CIP2 (cyclin-dependent interacting protein 2) cyclin-dependent kinase inhibitor 3 | 6.39 | 8.06 | |
| D38073 | MCM3 | Minichromosome maintenance deficient 3 | 6.34 | 2.71 | |
| AB014566 | DAAM1 | Disheveled associated activator of morphogenesis 1 | 6.27 | 5.55 | |
| AF007155 | LOC254531 | PISC domain containing hypothetical protein | 6.14 | 4.20 | |
| L47276 | HUMTOPATR | Homo sapiens alpha topoisomerase truncated-form mRNA | 6.12 | 4.03 | |
| U37426 | KIF11 | Kinesin family member 11 | 6.12 | 3.15 | |
| AB012911 | FZD6 | Frizzled homolog 6 | 6.12 | 5.48 | |
| AL080121 | DKFZp564O0823 | Highly similar to rat castration induced prostatic apoptosis related protein-1 | 6.10 | 6.96 | |
| X83490 | TNFRSF6 | Tumor necrosis factor receptor superfamily member 6* (Fas/APO-1) | 6.02 | 3.85 | |
| AL050107 | TAZ | Transcriptional co-activator with PDZ (PSD-95, DLG, and ZO-1)—binding motif | 5.81 | 5.67 | |
| J04088 | TOP2A | Topoisomerase (DNA) II alpha | 5.59 | 4.52 | |
| D42055 | NEDD4 | Neural precursor cell expressed, developmentally down-regulated 4 | 5.47 | 2.02 | |
| D38251 | POLR2E | Polymerase (RNA) II (DNA directed) polypeptide E | 5.40 | 8.51 | |
| M74089 | LOC91137 | Hypothetical protein BC017169 | 5.28 | 3.81 | |
| D13643 | DHCR24 | 24-dehydrocholesterol reductase | 5.17 | 5.52 | |
| D14657 | KIAA0101 | KIAA0101 gene product | 4.94 | 3.93 | |
| AA203476 | NA | zx55e01.rl | 4.92 | 2.31 | |
| M13755 | GIP2 | Interferon, alpha-inducible protein | 4.86 | 6.20 | |
| U63743 | KIF2C | Kinesin family member 2C | 4.69 | 3.78 | |
| J03473 | ADPRT | ADP-ribosyltransferase | 4.52 | 3.58 | |
| U43916 | EMP1 | Epithelial membrane protein 1 | 4.49 | 2.75 | |
| Z70519 | TNFRSF6 | FAS soluble protein* | 4.44 | 2.69 | |
| U73379 | UBE2C | Ubiquitin-conjugating enzyme E2C | 4.43 | 4.57 | |
| D86062 | C21orf33 | Chromosome 21 open reading frame 33 | 4.41 | 7.29 | |
| AI127424 | NA | qb75b02.x1 | 4.36 | 6.10 | |
| AC004528 | C19orf6 | Chromosome 19 open reading frame 6 | 4.31 | 4.16 | |
| AL050069 | DOK5 | Docking protein 5 | 4.27 | 5.45 | |
| U81561 | PTPRN2 | Protein tyrosine phosphatase, receptor type, N polypeptide 2 | 4.24 | 5.04 | |
| M15205 | TK1 | Thymidine kinase 1, soluble | 4.21 | 4.71 | |
| M84443 | GALK2 | Galactokinase 2 | 4.09 | 2.29 | |
| D63861 | PPID | Peptidylprolyl isomerase D (cyclophilin D) | 4.08 | 2.34 | |
| X02883 | HSTCRAC | Human gene for T-cell receptor alpha chain C region | 4.05 | 2.81 | |
| D00596 | TYMS | Thymidylate synthetase | 4.03 | 4.17 | |
| U03494 | TFCP2 | Transcription factor CP2 (CCAAT protein) | 3.99 | 3.15 | |
| U31601 | JAK3 | Janus kinase 3 (a protein tyrosine kinase, leukocyte) | 3.95 | 3.48 | |
| AF009426 | C18orf1 | Chromosome 18 open reading frame 1 | 3.93 | 2.87 | |
| AL022398 | ADORA2BP | Adenosine A2b receptor pseudogene | 3.90 | 2.80 | |
| M97935 | STAT1 | Signal transducer and activator of transcription 1 | 3.89 | 4.36 | |
| AA919102 | NA | o184h02.sl | 3.86 | 2.11 | |
| U88964 | ISG20 | Interferon-stimulated gene | 3.81 | 3.21 | |
| AJ010953 | ATP2CI | ATPase, Ca++ transporting, type 2C, member 1 | 3.73 | 2.87 | |
| U53003 | C21orf33 | Chromosome 21 open reading frame 33 | 3.69 | 2.76 | |
| M36711 | NA | DNA-binding protein Ap-2 | 3.67 | 3.09 | |
| M34641 | FGFR1 | Fibroblast growth factor receptor 1 | 3.62 | 2.37 | |
| AI652660 | wb30c10.x1 | 3.62 | 3.47 | ||
| AJ000534 | SGCE | Sarcoglycan, epsilon | 3.61 | 3.07 | |
| D50920 | THRAP4 | Thyroid hormone receptor associated protein 4 | 3.48 | 2.05 | |
| AB024704 | TPX2 | Microtubule-associated protein homolog | 3.45 | 3.20 | |
| X14798 | ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 | 3.44 | 3.36 | |
| Y11681 | Y11681 | Mitochondrial ribosomal protein S12 | 3.44 | 7.90 | |
| D26155 | SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | 3.42 | 3.10 | |
| AL031737 | NA | Human DNA sequence from clone 8B22 on chromosome 1p35.1-36.21 | 3.42 | 2.35 | |
| M86699 | TTK | TTK (threonine/tyrosine kinase) protein kinase | 3.40 | 2.38 | |
| U16799 | ATP1B1 | ATPase, Na+/K+ transporting, beta 1 polypeptide | 3.39 | 3.81 | |
| AB018270 | MYO1D | Myosin ID | 3.34 | 2.57 | |
| U76638 | BARD1 | BRCA1 (breast cancer 1) associated RING domain 1 | 3.34 | 2.62 | |
| AB013452 | ATP8A1 | ATPase, aminophospholipid transporter (APLT), class 1, type 8A, member 1 | 3.32 | 3.29 | |
| Y11525 | CEBPA | CCAAT/enhancer binding protein (C/EBP), alpha | 3.32 | 3.34 | |
| M34423 | GLB1 | Galactosidase, beta 1 | 3.31 | 4.04 | |
| AA203213 | NA | zx57e04.r1 | 3.31 | 3.49 | |
| X64624 | POU4F1 | POU (Pit, Oct, Unc) domain, class 4, transcription factor 1 | 3.31 | 3.92 | |
| AB018271 | BPAG1 | Bullous pemphigoid antigen 1 | 3.25 | 2.12 | |
| LI3435 | NA | Human chromosome 3p21.1 gene sequence | 3.24 | 2.92 | |
| AF055993 | SAP30 | Sin3-associated polypeptide | 3.18 | 3.47 | |
| U29332 | FHL2 | Four and a half LIM (Lin-1, Tsl-1, MEC-3) domains 2 | 3.16 | 3.60 | |
| AI540958 | NA | Homo sapiens cDNA, 5 end/clone_end=5 | 3.15 | 3.59 | |
| D38522 | SYT11 | Synaptotagmin XI | 3.09 | 3.58 | |
| U01038 | PLK1 | Polo-like kinase 1 | 3.07 | 3.40 | |
| X74262 | RBBP4 | Retinoblastoma binding protein 4 | 3.05 | 5.58 | |
| L23959 | TFDP1 | Transcription factor Dp-1 (E2F dimerization partner-1) | 3.03 | 3.76 | |
| M87434 | OAS2 | 2′-5′-oligoadenylate synthetase 2 | 3.02 | 2.96 | |
| M25280 | SELL | Selectin L (lymphocyte adhesion molecule 1) | 3.02 | 3.31 | |
| M63488 | RPA1 | Replication protein A1 | 3.01 | 3.39 | |
| U78190 | GCHFR | GTP cyclohydrolase I feedback regulatory protein | 3.01 | 2.09 | |
| AF067656 | ZWINT | ZW10 interactor | 2.97 | 2.50 | |
| M15024 | MYB | v-myb myeloblastosis viral oncogene homolog | 2.97 | 4.54 | |
| M81830 | SSTR2 | Somatostatin receptor 2 | 2.96 | 3.33 | |
| D28364 | ANXA11 | Annexin II, 5 | 2.96 | 6.05 | |
| M63838 | FI16 | Interferon, gamma-inducible protein 16 | 2.92 | 3.05 | |
| X72889 | NA | NA | 2.91 | 2.45 | |
| AC003108 | Anxa6 | Annexin VI | 2.84 | 3.79 | |
| L07541 | RFC3 | Replication factor C (activator 1) 3 | 2.83 | 2.67 | |
| D78261 | IRF4 | Interferon regulatory factor 4* | 2.83 | 3.14 | |
| AA005018 | NA | zh96a09.rl | 2.83 | 2.12 | |
| AA310786 | NA | EST181572 Homo sapiens cDNA | 2.78 | 2.29 | |
| M87284 | OAS2 | 2′-5′-oligoadenylate synthetase 2 | 2.77 | 3.11 | |
| U09813 | ATP5G3 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c (subunit 9) isoform 3 | 2.76 | 4.85 | |
| AC005253 | UBA52 | Ubiquitin A-52 residue ribosomal protein fusion product 1 | 2.75 | 2.85 | |
| AF070523 | JWA | Cytoskeleton related vitamin A responsive protein | 2.75 | 2.29 | |
| U38545 | PLD1 | Phospholipase D1, phophatidylcholine-specific | 2.74 | 2.17 | |
| U25182 | PRDX4 | Peroxiredoxin 4 | 2.73 | 3.10 | |
| AF022385 | PDCD10 | Programmed cell death 10 | 2.73 | 2.09 | |
| AF000416 | EXTL2 | Exostoses (multiple)-like 2 | 2.73 | 2.13 | |
| D26361 | KIF14 | Kinesin family member 14 | 2.72 | 2.47 | |
| AF064084 | ICMT | Isoprenylcysteine carboxyl methyltransferase | 2.70 | 4.31 | |
| X98248 | SORT1 | Sortilin 1 | 2.69 | 5.01 | |
| U65410 | MAD2L1 | MAD2 (mitotic arrest-deficient 2) mitotic arrest deficient-like 1 | 2.66 | 2.73 | |
| U51004 | HINT1 | Histidine triad nucleotide binding protein 1 | 2.64 | 2.09 | |
| M23379 | RASA1 | RAS p21 protein activator (GTPase activating protein) 1 | 2.64 | 2.43 | |
| X57398 | PM5 | pM5 protein | 2.63 | 2.65 | |
| X65550 | MK167 | Antigen identified by monoclonal antibody Ki-67 | 2.60 | 3.41 | |
| J05614 | PCNA | Proliferating cell nuclear antigen† | 2.59 | 2.50 | |
| X67951 | PRDX1 | Peroxiredoxin 1 | 2.58 | 2.16 | |
| M11507 | TFRC | Transferrin receptor (p90, CD71) | 2.57 | 2.59 | |
| D26599 | PSMB2 | Proteasome (prosome, macropain) subunit, beta type, 2 | 2.55 | 2.91 | |
| AF053551 | MTX2 | Metaxin 2 | 2.53 | 3.54 | |
| AF016898 | BATF | Basic leucine zipper transcription factor, AT-like | 2.50 | 2.04 | |
| AF055993 | SAP30 | sin3-associated polypeptide | 2.49 | 2.73 | |
| D14710 | ATP5A1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1 | 2.48 | 2.01 | |
| U09564 | NA | Human serine kinase | 2.48 | 2.51 | |
| AL109672 | P24B | Integral type I protein | 2.48 | 3.85 | |
| AI056696 | NA | oz26h05.x1 | 2.44 | 2.65 | |
| M13228 | MYCN | v-myc myelocytomatosis viral related oncogene, neuroblastoma derived | 2.44 | 2.78 | |
| AI566877 | NA | tn24f02.x1 | 2.44 | 2.25 | |
| L09235 | ATP6V1A | ATPase, H+ transporting, lysosomal | 2.42 | 5.18 | |
| X51688 | CCNA2 | Cyclin A2 | 2.36 | 2.45 | |
| U13695 | PMS1 | Postmeiotic segregation increased 1 | 2.34 | 2.17 | |
| AB007455 | TP53AP1 | TP53 activated protein 1 | 2.33 | 2.77 | |
| D44497 | CORO1A | Coronin, actin binding protein, 1A | 2.33 | 2.77 | |
| AA255502 | NA | zr85b06.r1 | 2.33 | 2.78 | |
| X73066 | NME1 | Non-metastatic cells 1, protein (NM23A) expressed in | 2.32 | 3.00 | |
| AI347088 | NA | qp60d09.x1 | 2.32 | 2.21 | |
| U37689 | POLR2H | Polymerase (RNA) II (DNA directed) polypeptide H | 2.32 | 2.13 | |
| U86529 | GSTZ1 | Glutathione transferase zeta 1 | 2.32 | 2.28 | |
| J03133 | SP1 | SP1 (SV40 promoter 1) transcription factor | 2.31 | 4.73 | |
| AB007447 | FLN29 | FLN29 gene product (TNF receptor factor-like protein) | 2.30 | 2.34 | |
| X74008 | PPP1CC | Protein phosphatase 1, catalytic subunit, gamma isoform | 2.30 | 2.07 | |
| U56833 | VBP1 | von Hippel-Lindau binding protein 1 | 2.29 | 2.16 | |
| AF020038 | IDH1 | Isocitrate dehydrogenase 1 (NADP+), soluble | 2.29 | 3.40 | |
| U63809 | PAWR | PRKC (protein kinase C), apoptosis, WT1, regulator | 2.29 | 2.35 | |
| AL050034 | ADPRTL3 | ADP-ribosyltransferase-like 3 | 2.25 | 2.09 | |
| M87339 | HUMACT1A | Human replication factor C, 37-kDa subunit | 2.25 | 2.98 | |
| AA056747 | NA | zk81f02.s1 | 2.24 | 2.95 | |
| U10886 | PTPRJ | Protein tyrosine phosphatase, receptor type, J | 2.23 | 2.52 | |
| X84194 | ACYP1 | A-cylphosphatase 1, erythrocyte (common) type | 2.23 | 4.40 | |
| AB024401 | ING1 | Inhibitor of growth family, member 1 | 2.20 | 14.85 | |
| M19481 | FST | Follistatin | 2.20 | 2.54 | |
| AF006084 | ARPC1B | Actin related protein 2/3 complex, subunit 1B, | 2.19 | 3.71 | |
| Y00097 | ANXA6 | Annexin A6 | 2.19 | 2.00 | |
| D00017 | ANXA2 | Annexin A2 | 2.18 | 2.81 | |
| U50079 | HDAC1 | Histone deacetylase 1 | 2.17 | 2.62 | |
| AA811338 | NA | ob81g05.sl | 2.17 | 5.19 | |
| M97676 | MSX1 | msh homeo box homolog 1 | 2.16 | 3.15 | |
| M63573 | PPIB | Peptidylprolyl isomerase B (cyclophilin B) | 2.16 | 2.85 | |
| J00314 | OK/SW-cl.56 | beta 5-tubulin | 2.16 | 2.19 | |
| U30872 | CENPF | Centromere protein F | 2.16 | 2.43 | |
| AA969267 | NA | on57d12.sl | 2.13 | 2.49 | |
| AF053306 | BUB1B | Budding uninhibited by benzimidazoles 1 homolog beta | 2.13 | 2.04 | |
| AF042384 | BC-2 | Putative breast adenocarcinoma marker | 2.12 | 2.52 | |
| U32849 | NMI | N-myc (and STAT) interactor | 2.12 | 2.90 | |
| AB028975 | KIAA1052 | KIAA1052 protein | 2.12 | 2.53 | |
| AJ000480 | C8FW | Phosphoprotein regulated by mitogenic pathways | 2.11 | 4.49 | |
| D50692 | MYCBP | c-myc binding protein | 2.10 | 3.00 | |
| M16279 | CD99 | CD99 antigen | 2.10 | 2.94 | |
| X69398 | CD47 | CD47 antigen (Rh-related antigen) | 2.09 | 2.17 | |
| AF054825 | VAMP5 | Vesicle-associated membrane protein 5 | 2.09 | 2.69 | |
| D37984 | RECQL | RecQ protein-like (DNA helicase Q1-like) | 2.09 | 2.27 | |
| U90549 | HMGN4 | High mobility group nucleosomal binding domain 4 | 2.09 | 2.16 | |
| AF087036 | MSC | Musculin (activated B-cell factor-1) | 2.08 | 2.67 | |
| J04058 | ETFA | Electron-transfer-flavoprotein, alpha polypeptide | 2.06 | 2.54 | |
| X62534 | HMGB2 | High-mobility group box 2 | 2.04 | 2.26 | |
| S57501 | NA | Protein phosphatase 1, alpha catalytic subunit | 2.04 | 2.71 | |
| AJ001612 | PSPHL | Phosphoserine phosphatase-like | 2.03 | 2.89 | |
| AJ245416 | LSM2 | LSM2 (Sm-like protein 2) homolog, U6 small nuclear RNA associated | 2.03 | 2.18 | |
| U91932 | AP3S1 | Adaptor-related protein complex 3, sigma 1 subunit | 2.03 | 2.89 | |
| U74612 | FOXM1 | Forkhead box M1 | 2.02 | 2.17 | |
| L42243 | IFNAR2 | Interferon (alpha, beta, and omega) receptor 2 | 2.02 | 2.88 | |
Listed genes were selected by expression rate greater than 100 and more than 2-fold expression in ATL cells more than in CD4+ and CD4+CD45RO+ T cells with significant differences (P < .01). For all genes scored, the fold change was calculated by the mean expression value of ATL cells by those of CD4+ or CD4+CD45RO+ T cells. NA indicates not applicable.
Genes were already reported as genes with high expression in HTLV-1—infected T cells.
Genes were already reported as genes with high expression in ATL cells.
Extremely high-level expression of CAV1, PGDS, and TSLC1 in ATL cells
Among the 192 genes up-regulated in acute-type ATL cells, 3 showed more than a 30-fold increase in expression in ATL cells compared with CD4+ and CD4+CD45RO+ T cells. These genes were caveolin 1 (CAV1), prostaglandin D2 synthase (PGDS), and tumor suppressor in lung cancer 1 (TSLC1). CAV1, an integral membrane protein, was shown to be expressed in portions of ATL cells or HTLV-1–immortalized T cells.24 PGDS was expressed in human T helper 2 (Th2) but not Th1 clones,25 and TSLC1 was originally identified as a tumor suppressor in non-small cell lung cancer.9
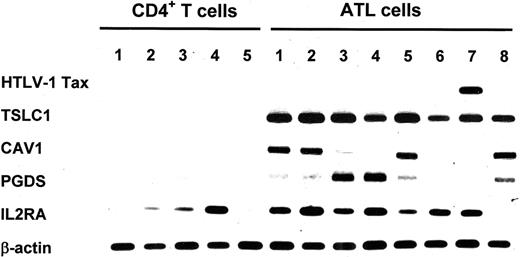
Thus, we focused on these 3 genes and confirmed them to be highly expressed in ATL cells by semiquantitative RT-PCR. The IL2RA and HTLV-1 Tax genes were also examined. As shown in Figure 1, TSLC1 was strongly expressed in all 8 primary ATL cells. CAV1 and PGDS were expressed in 6 and 7 of the 8, respectively. However, expression of the Tax gene was detected in only 1 case of acute ATL (case 7), suggesting that Tax does not directly enhance the expression of these genes in ATL cells. Since TSLC1 was expressed in extremely large amount in all ATL cases and is related to oncogenesis in lung cancer, we further characterized the expression of the TSLC1 gene in ATL cell lines and normal hematopoietic cells.
Semiquantitative PCR of 6 genes in primary ATL cells and control CD4+ T cells. Expression of up-regulated genes in the ATL cells was detected by DNA microarray analysis and confirmed by semiquantitative RT-PCR. cDNA prepared from total RNA of 8 primary ATL cells and 5 CD4+ T cells was amplified by PCR with the specific primer sets listed in “Patients, materials, and methods.” The expression of HTLV-1 Tax and IL2RA was also determined by RT-PCR. The expression level of β-actin is shown at the bottom of the figure as a control.
Semiquantitative PCR of 6 genes in primary ATL cells and control CD4+ T cells. Expression of up-regulated genes in the ATL cells was detected by DNA microarray analysis and confirmed by semiquantitative RT-PCR. cDNA prepared from total RNA of 8 primary ATL cells and 5 CD4+ T cells was amplified by PCR with the specific primer sets listed in “Patients, materials, and methods.” The expression of HTLV-1 Tax and IL2RA was also determined by RT-PCR. The expression level of β-actin is shown at the bottom of the figure as a control.
Ectopic expression of TSLC1 in ATL and HTLV-1–infected cell lines
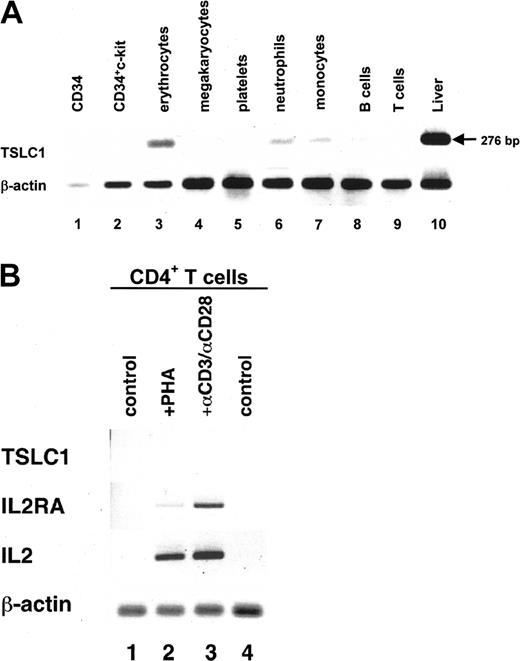
TSLC1 encodes a member of the immunoglobulin (Ig) superfamily of cell adhesion molecules (IgCAMs) and participates in cell-cell interactions.26 TSLC1 is expressed in almost all organs but not in lymphoid tissues.27 First, we examined the expression of TSLC1 in a series of hematopoietic cells by RT-PCR.12 As shown in Figure 2A, TSLC1 was weakly expressed in erythrocytes, followed by neutrophils, monocytes, and B cells and was not expressed in T cells. We next examined TSLC1 expression in resting or activated CD4+ T cells stimulated with PHA or an antibody mixture of anti-CD3 and anti-CD28. TSLC1 expression was not induced in response to both stimulants, although expression levels of IL2 and IL2RA were increased (Figure 2B). These results indicate that the expression of TSLC1 in ATL cells was not due to the cellular activation of normal T cells. Therefore, we investigated the biologic significance of ectopic expression of TSLC1 in ATL cells.
Expression of the TSLC1 gene in hematopoietic cells detected by RT-PCR. (A) Expression of the TSLC1 gene in cDNA from a series of hematopoietic cell fractions detected by RT-PCR. Each transcribed cDNA was amplified with the TSLC1-specific primer sets listed in “Patients, materials, and methods.” Each fraction of hematopoietic cells is indicated at the top of the lanes. Amplification of liver cDNA was used as a positive control (lane 10). (B) Expression of various genes in PHA-stimulated or unstimulated CD4+ T cells. Total RNA was prepared from CD4+ T cells stimulated with PHA or anti-CD3 and anti-CD28. cDNA was amplified using the primer sets for TSLC1, IL2RA, IL2, or β-actin as a quantitative control (described in “Patients, materials and, methods”). Control indicates unstimulated, +PHA, PHA-stimulated, and +αCD3/αCD28′, αCD3/αCD28′-stimulated CD4+ T cells.
Expression of the TSLC1 gene in hematopoietic cells detected by RT-PCR. (A) Expression of the TSLC1 gene in cDNA from a series of hematopoietic cell fractions detected by RT-PCR. Each transcribed cDNA was amplified with the TSLC1-specific primer sets listed in “Patients, materials, and methods.” Each fraction of hematopoietic cells is indicated at the top of the lanes. Amplification of liver cDNA was used as a positive control (lane 10). (B) Expression of various genes in PHA-stimulated or unstimulated CD4+ T cells. Total RNA was prepared from CD4+ T cells stimulated with PHA or anti-CD3 and anti-CD28. cDNA was amplified using the primer sets for TSLC1, IL2RA, IL2, or β-actin as a quantitative control (described in “Patients, materials and, methods”). Control indicates unstimulated, +PHA, PHA-stimulated, and +αCD3/αCD28′, αCD3/αCD28′-stimulated CD4+ T cells.
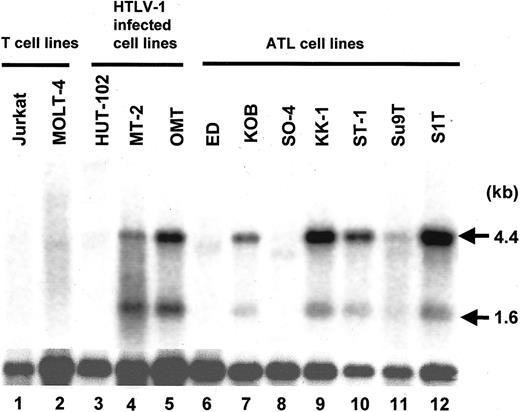
Next, we examined TSLC1 expression in cultured ATL and HTLV-1–infected T-cell lines by Northern blot analysis. As shown in Figure 3, signals corresponding to TSLC1 mRNA (4.4 and 1.6 kb) were detected in 5 of 7 ATL cell lines (KOB, KK-1, ST-1, Su9T, and S1T) as well as 2 of 3 HTLV-1–infected cell lines (MT-2 and OMT). In contrast, TSLC1 mRNA was not detected in the 2 T-cell lines unrelated to HTLV-1 (Jurkat and MOLT-4). Although in the remaining 2 ATL cell lines (ED and SO-4) and 1 HTLV-1–infected cell line (HUT-102) no signals were found, TSLC1 expression was detected by RT-PCR (data not shown). Moreover, TSLC1 expression was not detected by Northern blot analysis in 34 leukemia cell lines unrelated to HTLV-1 infection, consisting of 17 myelocytic/monocytic, 3 megakaryocytic, 2 erythrocytic, 4 B-lymphocytic, and 8 T-lymphocytic leukemia cell lines (“Patients, materials, and methods”) (data not shown). These findings suggest that TSLC1 is specifically expressed in ATL and HTLV-1–infected T cells.
Expression of the TSLC1 gene in T-cell lines detected by Northern hybridization. Poly(A)+ RNA was prepared from various T-cell lines with or without HTLV-1 infection and from ATL cell lines, and TSLC1 expression was analyzed by Northern hybridization. Two bands, 4.4 and 1.6 kb, for TSLC1 transcripts were detected in 2 of 3 T-cell lines with HTLV-1 infection (lanes 4 and 5) and 5 of 7 ATL cell lines (lanes 7, 9, 10, 11, and 12). β-actin expression was used as a control for mRNA quantity.
Expression of the TSLC1 gene in T-cell lines detected by Northern hybridization. Poly(A)+ RNA was prepared from various T-cell lines with or without HTLV-1 infection and from ATL cell lines, and TSLC1 expression was analyzed by Northern hybridization. Two bands, 4.4 and 1.6 kb, for TSLC1 transcripts were detected in 2 of 3 T-cell lines with HTLV-1 infection (lanes 4 and 5) and 5 of 7 ATL cell lines (lanes 7, 9, 10, 11, and 12). β-actin expression was used as a control for mRNA quantity.
Methylation of the TSLC1 promoter in CpG islands
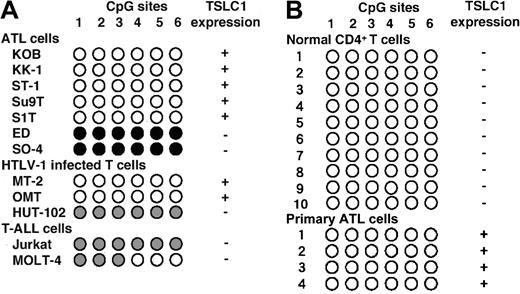
It is reported that TSLC1 is frequently inactivated by a combination of promoter hypermethylation and loss of heterozygosity (LOH) in non-small cell lung cancer (NSCLC) and many other primary human cancers.22,28-32 Therefore, we examined the state of the TSLC1 gene promoter in the cell lines using a bisulfite-SSCP analysis coupled with sequencing of the 6 cytosine-phosphorothioate-guanine (CpG) sites within the promoter. Among the HTLV-1–infected T-cell or ATL cell lines, 3 lines (ED, SO-4, and HUT-102) expressing no or very little TSLC1 showed hypermethylation or partial methylation of the promoter. In contrast, the other 7 cell lines expressing a large amount of TSLC1 showed unmethylated promoters (Figure 4A). Moreover, 2 T-cell lines without HTLV-1 infection and TSLC1 expression (Jurkat and MOLT-4) showed hypermethylation or partial methylation of the TSLC1 promoter. These results indicate that the absence or presence of TSLC1 expression in T-cell lines is well correlated with the state of the TSLC1 promoter. It is noteworthy, however, that promoter methylation was not observed in normal CD4+ T cells from 10 volunteers and 4 primary ATL cells (Figure 4B). Considering that TSLC1 is not physiologically expressed in any CD4+ T cells, the TSLC1 promoter does not need to be methylated in vivo like many other CpG islands of genes.
Methylation analysis of TSLC1. (A) Methylation state of the TSLC1 promoter in 6 CpG sites in 7 ATL, 3 HTLV-1–infected T-cell, and 2 T-ALL cell lines determined by bisulfite-SSCP and sequencing. (B) Methylation state of the TSLC1 promoter in 6 CpG sites in normal CD4+ T cells from 10 healthy volunteers and 4 patients with ATL determined by bisulfite-SSCP and sequencing. Black, gray, and white circles represent methylated, partially methylated, and unmethylated CpGs, respectively. Columns correspond to the 6 CpG sites just upstream of the predicted TATA box sequence.
Methylation analysis of TSLC1. (A) Methylation state of the TSLC1 promoter in 6 CpG sites in 7 ATL, 3 HTLV-1–infected T-cell, and 2 T-ALL cell lines determined by bisulfite-SSCP and sequencing. (B) Methylation state of the TSLC1 promoter in 6 CpG sites in normal CD4+ T cells from 10 healthy volunteers and 4 patients with ATL determined by bisulfite-SSCP and sequencing. Black, gray, and white circles represent methylated, partially methylated, and unmethylated CpGs, respectively. Columns correspond to the 6 CpG sites just upstream of the predicted TATA box sequence.
TSLC1 is mainly expressed on the cellular membrane of CTLL2 transformants
To investigate the subcellular localization of TSLC1 in T cells, a mouse IL2-dependent T-cell line, CTLL2,19 lacking endogenous TSLC1 expression, was transfected with TSLC1 fused with GFP and several stable transformants were obtained. Examination by laser scanning confocal microscopy revealed that TSLC1-GFP was present all along the cell membrane in the single-cell state (Figure 5B). However, when cells were attached to each other and formed aggregates, TSLC1 was preferentially expressed at cell-cell attachment sites (Figure 5C). A similar distribution of TSLC1 was reported in epithelial cells,26,32 suggesting that TSLC1 is possibly involved in cell-cell interaction.
Subcellular localization of TSLC-GFP in CTLL2 cells. Subcellular distribution of GFP-fused TSLC1 (TSLC-GFP) in CTLL2 cells. C/GFP (column A) and C/TSLC-GFP (column B and C) cells were observed under a laser scanning confocal microscope (model TSC4D; Leica Microsystems, Frankfurt, Germany), objective × 40/0.75 NA, with a filter set suitable for GFP detection and differential interference contrast (DIC). Bars represent 10 μm.
Subcellular localization of TSLC-GFP in CTLL2 cells. Subcellular distribution of GFP-fused TSLC1 (TSLC-GFP) in CTLL2 cells. C/GFP (column A) and C/TSLC-GFP (column B and C) cells were observed under a laser scanning confocal microscope (model TSC4D; Leica Microsystems, Frankfurt, Germany), objective × 40/0.75 NA, with a filter set suitable for GFP detection and differential interference contrast (DIC). Bars represent 10 μm.
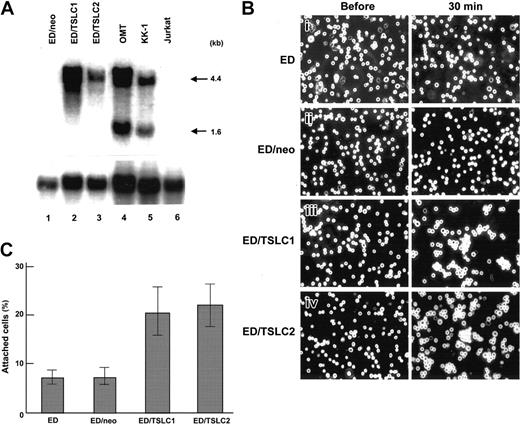
Since one of the characteristic features of ATL is the invasion by malignant cells of lymph nodes, skin, or various other organs, we examined the possible role of TSLC1 in these processes. For this purpose, a human ATL cell line, ED, lacking TSLC1 expression, was transduced with a retroviral vector carrying the TSLC1 cDNA. Two cell lines (ED/TSLC1 and ED/TSLC2) expressing a significant amount of TSLC1 that is comparable with that in the human ATL cell lines OMT and KK-1 were subsequently obtained (Figure 6A). The exogenous TSLC1 mRNA of about 4.4 kb observed in ED/TSLC1 and ED/TSLC2 cells contained a full-length TSLC1 cDNA, the neomycin-resistance gene and poly(A) signal sequences.33 The growth of ED/TSLC1 and ED/TSLC2 cells in vitro was not affected compared with that of ED cells transduced with the neoR gene alone (ED/neo) (data not shown). However, when we used a cell aggregation assay by dissociating and resuspending the cells, both ED/TSLC1 and ED/TSLC2 cells formed aggregates within 30 minutes (Figure 6B). In contrast, parental ED cells or ED/neo cells showed little aggregation in this time period, suggesting that TSLC1 mediates the intercellular adhesion of ATL cells via homophilic interaction, although the pathologic significance of the self-aggregation of ATL cells in vivo was not clear. The expression of TSLC1 in ED/TSLC2 cells was lower than that of ED/TSLC1 cells. However, no significant difference in cell adhesion was observed between ED/TSLC1 and ED/TSLC2 cells, implying that the cell adhesion capacity of ED cells may be affected by low-level TSLC1 as previously described.34
TSLC1 in ATL cell line plays an important role in adhesion to the vascular endothelial cells and self-aggregation. (A) Northern hybridization of TSLC1 in ED transformants. ED cells (ED/TSLC1 and ED/TSLC2) were transduced with a retrovirus vector containing the TSLC1 gene. A 4.4-kb band of TSLC1 transcript was detected in ED/TSLC1 and ED/TSLC2 cells (lanes 3 and 4). ED/neo (mock transformants) and Jurkat cells (lanes 1 and 6) were used as negative controls. OMT and KK-1 (lanes 4 and 5) were ATL cell lines as positive controls. (B) Cell aggregation experiments with TSLC1 transformants. To examine the aggregation activity of TSLC1 in parental ED cells, ED/TSLC1 and ED/TSLC2 transformants were well suspended just before the experiment and kept for 30 minutes in a 3.5-cm2 dish. Photographs were taken with an inverted microscope (model CKX41; Olympus, Tokyo, Japan) at a magnification of 100 × (objective × 10/0.25 NA). (C) Cell adhesion experiments with TSLC1 transformants. To examine the adhesion activity of TSLC1, parental ED, ED/neo, ED/TSLC1, and ED/TSLC2 were incubated with monolayer HUVECs for 30 minutes. Each value represents the average obtained from 3 independent experiments. Bars indicate standard deviation errors.
TSLC1 in ATL cell line plays an important role in adhesion to the vascular endothelial cells and self-aggregation. (A) Northern hybridization of TSLC1 in ED transformants. ED cells (ED/TSLC1 and ED/TSLC2) were transduced with a retrovirus vector containing the TSLC1 gene. A 4.4-kb band of TSLC1 transcript was detected in ED/TSLC1 and ED/TSLC2 cells (lanes 3 and 4). ED/neo (mock transformants) and Jurkat cells (lanes 1 and 6) were used as negative controls. OMT and KK-1 (lanes 4 and 5) were ATL cell lines as positive controls. (B) Cell aggregation experiments with TSLC1 transformants. To examine the aggregation activity of TSLC1 in parental ED cells, ED/TSLC1 and ED/TSLC2 transformants were well suspended just before the experiment and kept for 30 minutes in a 3.5-cm2 dish. Photographs were taken with an inverted microscope (model CKX41; Olympus, Tokyo, Japan) at a magnification of 100 × (objective × 10/0.25 NA). (C) Cell adhesion experiments with TSLC1 transformants. To examine the adhesion activity of TSLC1, parental ED, ED/neo, ED/TSLC1, and ED/TSLC2 were incubated with monolayer HUVECs for 30 minutes. Each value represents the average obtained from 3 independent experiments. Bars indicate standard deviation errors.
The initial step in the invasion by ATL cells of various human organs would be their interaction with vascular endothelial cells.35 Therefore, we examined the possible involvement of TSLC1 in the adhesion of ATL cells to human umbilical vein endothelial cells (HUVECs) in vitro. When fluorescence-labeled ED cells and their derivatives were seeded onto the HUVECs, incubated for 30 minutes, and then washed with medium, the numbers of attached ATL cells increased 2.9- and 3.1-fold in ED/TSLC1 and ED/TSLC2 cells, respectively, compared with parental ED or ED/neo cells (Figure 6C). These results suggest that the ectopic expression of TSLC1 in ATL cells may promote their invasion of various organs via interaction with surface molecules in vascular endothelial cells.
Discussion
Numerous attempts have been made to identify the key molecules for ATL development by using various methods, including differential display and microarray analysis. These studies identified a number of genes whose expression was up-regulated in ATL cells or induced in HTLV-1–infected T cells. Most, however, used cultured ATL cell lines or HTLV-1–infected T-cell lines as resources for a comparative analysis with activated normal CD4+ T cells. Although such studies have elucidated important molecular mechanisms of ATL development as well as several key molecules of leukemogenesis, knowledge regarding clinically useful markers is still very limited. In the present study, we investigated the expression profiles of 8 acute-type ATL cells using a GeneChip microarray to identify useful molecular markers for the preclinical or early diagnosis of the disease. We identified 192 genes that were up-regulated more than 2-fold in acute ATL cells compared with CD4+ or CD4+CD45RO+ T cells. Surprisingly, only 5 of 36 genes previously reported to be up-regulated in ATL and 3 of 72 genes in HTLV-1–infected T cells were included in the list of 192 (Table 2). However, these genes appear to be very important to the pathogenesis of ATL. IL2RA and parathyroid hormone–like hormone (PTHLH) were present in both groups. IL2RA as a surface marker for ATL, which is known to be overexpressed in HTLV-1–infected cells, plays a pivotal role in the proliferation of HTLV-1–transformed cells.21 PTHLH was implicated in ATL-associated hypercalcemia.36 As shown previously, significantly lower levels of HTLV-I tax mRNA were present in the fresh ATL cells than in the cultured ATL cells.37 Moreover, the gene expression profile easily changed under the culture conditions in vitro. Therefore, our approach to examining primary ATL should provide valuable information on molecular markers.
The expression of 3 genes, TSLC1, CAV1, and PGDS, was up-regulated more than 30-fold in acute-type ATL cells. Among them, TSLC1 was highly expressed in all acute ATL cases. TSLC1 is a tumor suppressor in non-small cell lung cancer, which is frequently deleted from the 11q23 region and compliments the tumor progression of lung cancer cells with this deletion in nude mice.9 Through RT-PCR, Northern hybridization, and promoter methylation assay, we found that the tumor suppressor gene TSLC1 is specifically expressed in acute-type ATL and HTLV-1–infected cells via an unknown mechanism. In this study, TSLC1 expression was detected by leukemia cells from 8 cases of acute-type ATL. Furthermore, we analyzed an additional 8 cases of acute-type ATL, along with 3 each of chronic-type and lymphoma-type ATL. The expression of TSLC1 was detected by RT-PCR for all cases (data not shown). If the overexpression of TSLC1 is a feature of ATL, additional cases involving other types of ATL should be investigated with regard to TSLC1 expression. To investigate whether TSLC1 mRNA overexpressed in primary ATL cells has any coding mutations, complete TSLC1 cDNA was isolated from 2 leukemia cells of ATL patients by RT-PCR (data not shown). The cDNA had 2 nucleotide exchanges as single-nucleotide polymorphisms (461G>A and 732A>T), which were reported in the National Center for Biotechnology Information (NCBI) database. The nucleotide change 461G>A lead to an amino acid exchange for Lys151Arg, but the functional differences between the 2 TSLC1 proteins are not known.
TSLC1 is a member of the immunoglobulin superfamily (IGSF4) and nectinlike protein family (necl-2).38 TSLC1/necl-2 shows Ca2+-independent heterophilic or homophilic cell-cell adhesion activity and has a band 4.1-binding motif in the juxtamembrane region that binds to the tumor suppressor DAL-1, which in turn connects TSLC1 to the actin cytoskeleton.26,39 The expression of TSLC1 inhibits the growth of lung cancer cells in vitro and in vivo, suggesting that cell-cell or cell-to-substrate interactions mediated by TSLC1 are critical for the suppression of tumorigenicity.34 However, overexpression of TSLC1 in ATL cells imparts the ability to adhere to vascular endothelial cells. Since ATL cells easily invade various organs, TSLC1 expression in ATL cells may play a role in organ involvement. To investigate the relationship, the expression of TSLC1 in control cases without organ involvement should be examined. However, the 3 cases of chronic-type ATL had hepatosplenomegaly and/or skin involvement. Therefore, the TSLC1 expression in many other ATL cases should be investigated. Instead, we are also now investigating TSLC1 expression in leukemia cells to determine their ability to infiltrate tissue using an in vivo cell transplantation system in immune deficient mice.
Along with the functional characterization of TSLC1 expression in ATL cells, elucidation of the mechanism responsible for ectopic TSLC1 expression is required. Since several transcription factors are involved in the pathogenesis of leukemia through retroviral insertions or chromosomal translocations, analysis of TSLC1 expression may provide insight into one of the major molecular events that occurs in ATL cells. Furthermore, since HTLV-1–infected cells expressed TSLC1, it is possible that the tax protein mediates TSLC1 expression during the early stages of infection. To elucidate the mechanism responsible for TSLC1 expression, the expression status of TSLC1 in HTLV-1–infected cells from HTLV-1 carriers should be investigated. However, TSLC1 expression in HTLV-1–infected cells is difficult to detect, because the percentage of HTLV-1–infected cells in peripheral blood is low. Therefore, we are developing a specific TSLC1 antibody for immunostaining or fluorescence activated cell sorting (FACS) of HTLV-1–infected cells. In summary, the ectopic expression of TSLC1 could provide a novel marker for acute-type ATL and may participate in tissue invasion, one of characteristic features of the malignant ATL cells.
Prepublished online as Blood First Edition Paper, October 7, 2004; DOI 10.1182/blood-2004-03-1222.
Supported by Grants-in-Aid for Scientific Research of Priority Area and for 21st Century COE program (life science) from the Ministry of Education, Culture, Sports, Science and Technology in Japan and for the Second Comprehensive 10-Year Strategy for Cancer in Japan from the Ministry of Health, Labor, and Welfare of Japan and by the Miyazaki Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, JST.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Y. Yamada, M. Maeda, and N. Arima for cell lines and T. Maruyama for technical assistance.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal