Abstract
FMS-like tyrosine kinase 3 (FLT3) is almost universally expressed in B-precursor childhood acute lymphoblastic leukemia (ALL). Cases of ALL with MLL gene rearrangements and those with high hyperdiploidy (> 50 chromosomes) express the highest levels of FLT3, and activating mutations of FLT3 occur in 18% of MLL-rearranged and 28% of hyperdiploid ALL cases. We determined the antileukemic activity of CEP-701, a potent and selective FLT3 inhibitor, in 8 ALL cell lines and 39 bone marrow samples obtained at diagnosis from infants and children with various subtypes of ALL. CEP-701 induced pronounced apoptotic responses in a higher percentage of samples that expressed high levels of FLT3 (74%, n = 23) compared with samples with low levels of expression (8%, n = 13; P = .0003). Sensitivity to FLT3 inhibition was particularly high in samples with MLL gene rearrangements (82%, n = 11; P = .0005), high hyperdiploidy (100%, n = 5; P = .0007), and/or FLT3 mutations (100%, n = 4; P = .0021). Seven of 7 sensitive samples examined by immunoblotting demonstrated constitutively phosphorylated FLT3 that was potently inhibited by CEP-701, whereas 0 of 6 resistant samples expressed constitutively phosphorylated FLT3. We conclude that the FLT3 inhibitor CEP-701 effectively suppresses FLT3-driven leukemic cell survival. Clinical testing of CEP-701 as a novel molecularly targeted agent for the treatment of childhood ALL is warranted.
Introduction
FMS-like tyrosine kinase 3 (FLT3) is a receptor tyrosine kinase that plays an important role in the pathogenesis of leukemia.1-4 FLT3 is expressed in most acute leukemias, including 94% of B-lineage acute lymphoblastic leukemia (ALL), 32% of T-lineage ALL, and 89% of acute myelogenous leukemia (AML).1,5-7 Moreover, constitutive activation of FLT3 can occur in leukemia cells through 2 mechanisms: coexpression of FLT3 ligand (FL), which leads to constitutive activation through autocrine, paracrine, or intracrine signaling,8-11 and mutations of FLT3, which result in ligand-independent autophosphorylation and activation of the FLT3 receptor.12-14 FLT3 mutations include juxtamembrane domain mutations (FLT3/JMD, comprising internal tandem duplication [FLT3/ITD] and deletion/insertion mutations) and kinase domain mutations (FLT3/KD, most commonly single–amino acid substitutions at positions 835 or 836).12-14 Activation of the FLT3 receptor, whether through ligand binding or mutation, results in activation of downstream targets, including proteins in the signal transducers and activators of transcription (STAT), mitogen-activated protein (MAP) kinase, and AKT pathways that regulate proliferation, differentiation, and survival.15-25 In many model systems, constitutively activated FLT3 has been shown to contribute to the leukemic phenotype.25-30
FLT3-activating mutations were initially discovered in AML and represent the most common somatic genetic alteration in that disease, with FLT3/ITD mutations occurring in approximately 25% of adult AML cases and 15% of pediatric AML cases and kinase domain mutations occurring in another 8% of both.1,12,13 The presence of an FLT3/ITD mutation is associated with a poor prognosis in both adult and pediatric AML cases.1,2,31 Small-molecule FLT3 inhibitors induce cytotoxic responses in AML blasts with FLT3/ITD mutations, and several of these agents are being tested in clinical trials in adults.28,32-42 One of these is CEP-701 (West Chester, PA), an orally bioavailable indolocarbazole derivative. CEP-701 inhibits mutant and wild-type (WT) FLT3 phosphorylation with an inhibitory concentration of 50% (IC50) of 3 nM, whereas 500 to 1000 nM or greater is required to inhibit the most closely related kinases (platelet-derived growth factor receptor β [PDGFR-β], FMS, and KIT).32
FLT3 has recently been implicated in the pathogenesis of infant and childhood ALL. In addition to the near-universal FLT3 expression in B-precursor ALL, gene expression analyses have shown that some of the highest levels of FLT3 expression occur in infant and childhood ALL with rearrangements of the MLL gene at chromosome 11q23 and in childhood ALL with hyperdiploidy with more than 50 chromosomes (high hyperdiploidy).43,44 FLT3/KD mutations occur in 18% of MLL-rearranged infant and childhood leukemia cases and in 16% of childhood ALL cases with hyperdiploidy.45,46 Small insertion/deletion mutations in the juxtamembrane domain have also been reported in an additional 12% of high-hyperdiploid ALL cases,45 for a total FLT3 mutation rate of 28% in hyperdiploid ALL cases. By contrast, only 1% of all childhood ALL cases have FLT3/ITD mutations, and these mutations do not appear to be more common in MLL-rearranged or hyperdiploid ALL cases.1,47,48 Preliminary studies have shown antileukemic activity of the small-molecule inhibitor PKC-412 (Basel, Switzerland) against 3 primary ALL samples and one ALL cell line with MLL rearrangements,49 suggesting a potential therapeutic role for FLT3 inhibitors in the treatment of infant and childhood ALL.
To define the potential role of FLT3 inhibition as a therapeutic strategy for infant and childhood ALL, we tested the antileukemic activity of the FLT3 inhibitor CEP-701 in 8 ALL cell lines and 39 primary ALL samples representative of the spectrum of phenotypes and genotypes characteristic of childhood ALL.
Patients, materials, and methods
Cells
Primary leukemic cells were enriched from the diagnostic bone marrow samples of 39 patients with childhood ALL by Ficoll-Hypaque centrifugation. All samples were collected under Johns Hopkins and St Jude children's research hospitals institutional review board (IRB)–approved cell procurement protocols from newly diagnosed infants and children with ALL. Informed consent was obtained in accordance with the Helsinki protocol. Samples from patient numbers J-1 to J-30 (Table 2) were cryopreserved within 5 hours of collection and stored in the Saint Jude Children's Research Hospital Tissue Resource Laboratory and are a subset of samples from a cohort of patients that has been previously described.44 Samples from patient numbers H-1 to H-9 (Table 2) were collected from patients at the Johns Hopkins Children's Center and assayed fresh. The diagnosis of ALL was based on morphology and flow cytometric analysis of immunophenotype. Cytogenetics were determined by standard procedures.
Information on primary patient samples examined
Patient no. . | FLT3 expression . | Cytogenetics/ALL subtype . | FLT3 genotype . | Child/infant . |
|---|---|---|---|---|
| J-1 | High | MLL | WT | Infant |
| J-2 | High | MLL | WT | Infant |
| J-3 | High | MLL | D835E | Infant |
| J-4 | High | MLL | WT | Infant |
| J-5 | High | MLL | WT | Infant |
| J-6 | High | MLL | WT | Child |
| J-7 | High | MLL | WT | Child |
| J-8 | High | MLL | WT | Child |
| J-9 | High | MLL | WT | Child |
| J-10 | High | MLL | WT | Child |
| J-11 | Borderline | Hyper > 50 | WT | Infant |
| J-12 | High | Hyper > 50 | WT | Child |
| J-13 | High | Hyper > 50 | D835E | Child |
| J-14 | High | Hyper > 50 | ITD | Child |
| J-15 | High | Hyper > 50 | WT | Child |
| J-16 | High | Hypo | WT | Child |
| J-17 | High | Pseudo | WT | Child |
| J-18 | High | Pseudo | JM del/ins | Child |
| J-19 | High | Hyper 47-50 | WT | Infant |
| J-20 | High | Normal | WT | Child |
| J-21 | High | Normal | WT | Child |
| J-22 | Low | E2A-PBX1 | WT | Child |
| J-23 | Low | E2A-PBX1 | WT | Child |
| J-24 | Low | TEL-AML1 | WT | Child |
| J-25 | Low | TEL-AML1 | WT | Child |
| J-26* | Low | T-ALL | WT | Child |
| J-27 | Low | T-ALL | WT | Child |
| J-28 | Low | BCR-ABL | WT | Child |
| J-29 | Low | BCR-ABL | WT | Child |
| J-30 | Low | Pseudo | WT | Child |
| H-1 | High | MLL | WT | Infant |
| H-2 | High | Inv(19) | WT | Child |
| H-3 | High | Hyper > 50 | WT | Child |
| H-4 | ND | Normal | WT | Infant |
| H-5 | Low | T-ALL | WT | Child |
| H-6 | Low | T-ALL | WT | Child |
| H-7 | Low | Normal | WT | Child |
| H-8 | Low | MYC-IGH | WT | Child |
| H-9 | Low | Normal | WT | Child |
| H-10 | Low | Normal | WT | Child |
Patient no. . | FLT3 expression . | Cytogenetics/ALL subtype . | FLT3 genotype . | Child/infant . |
|---|---|---|---|---|
| J-1 | High | MLL | WT | Infant |
| J-2 | High | MLL | WT | Infant |
| J-3 | High | MLL | D835E | Infant |
| J-4 | High | MLL | WT | Infant |
| J-5 | High | MLL | WT | Infant |
| J-6 | High | MLL | WT | Child |
| J-7 | High | MLL | WT | Child |
| J-8 | High | MLL | WT | Child |
| J-9 | High | MLL | WT | Child |
| J-10 | High | MLL | WT | Child |
| J-11 | Borderline | Hyper > 50 | WT | Infant |
| J-12 | High | Hyper > 50 | WT | Child |
| J-13 | High | Hyper > 50 | D835E | Child |
| J-14 | High | Hyper > 50 | ITD | Child |
| J-15 | High | Hyper > 50 | WT | Child |
| J-16 | High | Hypo | WT | Child |
| J-17 | High | Pseudo | WT | Child |
| J-18 | High | Pseudo | JM del/ins | Child |
| J-19 | High | Hyper 47-50 | WT | Infant |
| J-20 | High | Normal | WT | Child |
| J-21 | High | Normal | WT | Child |
| J-22 | Low | E2A-PBX1 | WT | Child |
| J-23 | Low | E2A-PBX1 | WT | Child |
| J-24 | Low | TEL-AML1 | WT | Child |
| J-25 | Low | TEL-AML1 | WT | Child |
| J-26* | Low | T-ALL | WT | Child |
| J-27 | Low | T-ALL | WT | Child |
| J-28 | Low | BCR-ABL | WT | Child |
| J-29 | Low | BCR-ABL | WT | Child |
| J-30 | Low | Pseudo | WT | Child |
| H-1 | High | MLL | WT | Infant |
| H-2 | High | Inv(19) | WT | Child |
| H-3 | High | Hyper > 50 | WT | Child |
| H-4 | ND | Normal | WT | Infant |
| H-5 | Low | T-ALL | WT | Child |
| H-6 | Low | T-ALL | WT | Child |
| H-7 | Low | Normal | WT | Child |
| H-8 | Low | MYC-IGH | WT | Child |
| H-9 | Low | Normal | WT | Child |
| H-10 | Low | Normal | WT | Child |
J indicates St Jude patient; MLL, rearranged MLL gene; WT, wild type FLT3; D835E, substitution of glutamate for aspartate at position 835 of FLT3; Hyper > 50, high hyperdiploid (> 50 chromosomes); ITD, FLT3 internal tandem duplication; Hypo, hypodiploid; Pseudo, pseudodiploid; JM del/ins, FLT3 juxtamembrane region deletion/insertion; Hyper 47-50, hyperdiploid (47-50 chromosomes); E2A-PBX1, t(1;19); TEL-AML1, t(12;21); T-ALL, T-cell lineage ALL; BCR-ABL, t(9;22); H, Johns Hopkins patient; ND, not determined; and MYC-IGH, t(8;14).
Sample not evaluable
Information on cell lines examined
Cell line . | Phenotype . | Cytogenetics . |
|---|---|---|
| HB-1119 | Pre-B ALL | t(11;19)(q23;p13)→ MLL/ENL fusion |
| SEM-K2 | Pro-B ALL | t(4;11)(q21;q23)→ MLL/AF4 fusion |
| B1 | Pro-B ALL | t(4;11)(q21;q23)→ MLL/AF4 fusion |
| 697 | Pre-B ALL | t(1;19)(q23;p13)→ E2A/PBX fusion |
| LAZ-221 | Pre-B ALL | Hypodiploid |
| Karpas-45 | T-ALL | t(X;11)(q13;q23)→ MLL/AFX fusion |
| Nalm-6 | Pre-B ALL | t(5;12)(q33;p13) |
| Reh | Pre-B ALL | t(12;21)(p13;q22)→ TEL/AML1 fusion |
Cell line . | Phenotype . | Cytogenetics . |
|---|---|---|
| HB-1119 | Pre-B ALL | t(11;19)(q23;p13)→ MLL/ENL fusion |
| SEM-K2 | Pro-B ALL | t(4;11)(q21;q23)→ MLL/AF4 fusion |
| B1 | Pro-B ALL | t(4;11)(q21;q23)→ MLL/AF4 fusion |
| 697 | Pre-B ALL | t(1;19)(q23;p13)→ E2A/PBX fusion |
| LAZ-221 | Pre-B ALL | Hypodiploid |
| Karpas-45 | T-ALL | t(X;11)(q13;q23)→ MLL/AFX fusion |
| Nalm-6 | Pre-B ALL | t(5;12)(q33;p13) |
| Reh | Pre-B ALL | t(12;21)(p13;q22)→ TEL/AML1 fusion |
Reagents
CEP-701 (Cephalon, West Chester, PA) was stored as a 4-mM stock solution in dimethyl sulfoxide (DMSO) at -20°C and was diluted into medium just before use. For every experiment described in this report, each sample, including the controls, contained an identical amount of DMSO. All cell culture reagents were purchased from Invitrogen (Carlsbad, CA) except for FBS, which was from Gemini (Woodland, CA), and hydrocortisone, which was from Sigma (St Louis, MO). Phycoerythrin (PE)–conjugated annexin V and fluorescein isothiocyanate (FITC)–conjugated CD19 and CD3 were obtained from BD Biosciences (San Diego, CA), anti-FLT3 S-18 antibody was from Santa Cruz Biotechnology (Santa Cruz, CA), and 4G10 antiphosphotyrosine antibody and protein A agarose were from Upstate Biotechnology (Lake Placid, NY).
Normal human bone marrow feeder layer coculture system
Normal human bone marrow samples were obtained from healthy volunteer donors enrolled on a Johns Hopkins IRB-approved cell procurement protocol. The procedure for establishing confluent stromal feeder layers into 96-well plates has been described elsewhere.51 Briefly, mononuclear cells were isolated by centrifugation over a Ficoll-Hypaque gradient and cultured in RPMI-1640 medium supplemented with 10% FBS, penicillin/streptomycin, L-glutamine, and 1 μM hydrocortisone at 33°C, 5% CO2, for 4 to 6 weeks with weekly medium changes. Cells were then trypsinized and cultured in 96-well plates for an additional 1 to 2 weeks. Just prior to the experiments, the adherent cells were washed 7 times in AIM V serum-free lymphocyte medium (Invitrogen, Carlsbad, CA).
Cytotoxicity and apoptosis assays
Cytotoxicity assays were performed using MTT (3-4,5-dimethylthiazol-2,5-diphenyltetrazolium). For the cryopreserved samples (patient numbers J-1 to J-30 in Table 2), 50 μL of AIM V medium with 2 × concentration of CEP-701 was aliquoted into triplicate wells of bone marrow stromal feeder layer 96-well plates for each CEP-701 concentration point. Fifty microliters of cells suspended in AIM V medium was then added to each well (200 000-400 000 viable cells/well). Plates were incubated for 48 hours at 37°C, 5% CO2, then the MTT assay was performed according to the manufacturer's instructions (Roche, Indianapolis, IN). To control for the MTT metabolized by the bone marrow stroma, and for any potential cytotoxic effect of CEP-701 on the stroma itself, the optical density (OD) of control wells containing only bone marrow stroma was subtracted from the OD of the wells with both stroma and leukemic blasts after treatment with the same concentration of CEP-701 for the 48-hour assay period. No cytotoxicity was observed in the stroma-only control wells at concentrations up to 100 nM CEP-701 (maximum dose used in the assay). For the fresh primary samples (patient numbers H-1 to H-9 in Table 2) and the cells lines (Table 1), 96-well plates without stromal feeder layers were used.
Apoptosis assays were performed using annexin-V binding (AVB). Cells were incubated for 48 hours in increasing CEP-701 concentrations as described for the MTT assay, then harvested and costained with PE-conjugated annexin V and either FITC-conjugated CD19 (for B-lineage samples) or FITC-conjugated CD3 (for T-lineage samples). The cells were then subjected to flow cytometry (FACSCalibur; BD Biosciences) and analyzed with CellQuest software (BD Biosciences). The leukemic cells were isolated by gating on the CD19+ or CD3+ cells. The percent apoptosis for each sample was defined as the percent of the gated cells that were annexin-V positive.
Immunoprecipitation and immunoblotting
Cells were incubated for one hour with 50 nM CEP-701 or DMSO control (untreated). The cells were then lysed with lysis buffer (1% nonidet P–40 [NP-40], 0.2 M Tris [tris(hydroxymethyl)aminomethane], 0.15 M NaCl, 0.1 M NaF, 0.01 M EDTA [ethylenediaminetetraacetic acid] with mini-Complete protease inhibitor cocktail tablet [Roche, Mannheim, Germany], and 5 mM sodium orthovanadate) and the lysate was clarified by centrifugation. Five hundred micrograms of protein was immunoprecipitated with anti-FLT3 antibody overnight at 4°C, and then protein A agarose was added for 2 hours. After electrophoresis through 8% polyacrylamide gels and transfer to nitrocellulose, membranes were blotted sequentially with antiphosphotyrosine antibody (4G10), followed by stripping and reblotting with anti-FLT3 antibody. Protein bands were visualized using chemiluminescence (ECL; Amersham, Piscataway, NJ) and scanned with an AGFA Arcus 1200 laser scanner (AGFA, Ridgefield Park, NJ).
Gene expression profiling
The preparation of mononuclear cell suspensions from diagnostic bone marrow aspirates, extraction of total RNA, and preparation of hybridization solutions for microarray analysis have been previously described.44,52 Hybridization solutions were hybridized to Affymetrix U95Av2 oligonucleotide microarrays (Affymetrix, Santa Clara, CA) according to Affymetrix protocols. The primary gene expression data has been previously published and is available at www.stjuderesearch.org/data/ALL1.53 Arrays were scanned using a laser confocal scanner (Agilent, Palo Alto, CA) and then analyzed with Affymetrix Microarray Suite 5.0 (MAS 5.0). Detection values (present, marginal, or absent) were determined by default parameters, and signal values were scaled by global methods to a target value of 500. Quantitation of the level of expression of the FLT3 gene was assessed using probes 34 583_at and 1065_at. High expression was defined as an expression value of greater than 2000 for each probe and low expression as an expression value less than 1000 for 34 583_at and less than 700 for 1065_at. Expression between these levels was considered borderline. The thresholds of high, low, and borderline FLT3 expression were generated by the MAS 5.0 software based on statistical analysis of the expression data and were therefore established independent of the results of the cytotoxicity assays.
Screening of the ITD and D835 mutation of the FLT3 gene
Genomic DNA was extracted from leukemic samples with Trizol (Invitrogen). The presence of a FLT3/ITD was determined by amplifying a region spanning exons 14 and 15 using primers 14F (5′-CAA TTT AGG TAT GAA AGC C-3′) and 15R (5′-GTA CCT TTC AGC ATT TTG AC-3′), followed by electrophoresis on 2.5% agarose gel. To detect mutations at D835/I836, genomic DNA was used to amplify FLT3 exon 20 using primers 20F (5′-CCG CCA GGAACG TGC TTG-3′) and 17R (5′-GCA GCC TCA CAT TGC CCC-3′). The amplified products were then digested with EcoRV and subjected to electrophoresis on agarose gels. An EcoRV restriction site exists within the WT FLT3 sequence in codon 835/836. Any mutation that alters the coding region of this region results in the elimination of this restriction site. All undigested bands seen following digestion were cut from the gel, purified with QIAquick gel extraction kit (Qiagen, Chatsworth, CA), and directly sequenced.
Statistical methods
Statistical significance of the difference in mean MTT cytotoxicity responses by group was determined by Student t test. The degree of correlation of MTT cytotoxicity and AVB apoptosis assays was determined using Pearson correlation coefficient.
Results
Sensitivity of ALL cell lines to FLT3 inhibition
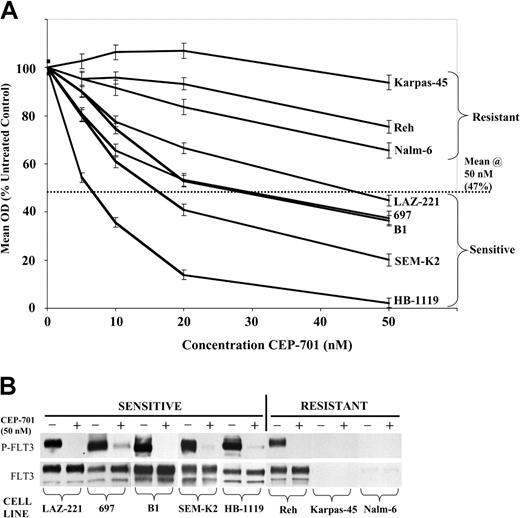
Eight ALL cell lines (Table 1) were examined with 48-hour MTT cytotoxicity assays using increasing doses of CEP-701 and with immunoprecipitation and immunoblotting after treatment for 1 hour with 50 nM CEP-701 (+) or diluent control (-). In the MTT assay, the mean cytotoxic response for all 8 cell lines to CEP-701 at 50 nM was a mean OD of 47% of untreated controls. Hence, a cell line was considered to be sensitive to FLT3 inhibition if the mean OD for the cell line was less than 47% of untreated controls at 50 nM CEP-701. By this criterion, 5 of the 8 cell lines were sensitive (Figure 1A). Immunoprecipitation and immunoblotting revealed that all 5 sensitive cell lines express high levels of FLT3 that is constitutively phosphorylated and potently inhibited by 50 nM CEP-701 (Figure 1B). It is notable that the 3 most sensitive cell lines (HB-1119, SEM-K2, and B1) have rearrangements of the MLL gene at 11q23. In the 3 resistant cell lines, expression and activation of FLT3 was variable. One cell line (Karpas-45) is a T-lineage ALL line that does not express appreciable amounts of FLT3, consistent with previous reports of low-frequency FLT3 expression in T-ALL.5,9 The other 2 resistant cell lines (Nalm-6 and Reh) are B-precursor ALL cell lines. Nalm-6 cells express very low levels of FLT3 that is not constitutively phosphorylated. Reh cells express high levels of constitutively active FLT3 that is potently inhibited by CEP-701. However, FLT3 inhibition alone is insufficient to induce significant cytotoxicity in these cells.
ALL cell lines with high-level expression and constitutive phosphorylation of FLT3 are differentially sensitive to FLT3 inhibition. (A) MTT dose-response curves showing mean cytotoxic response to CEP-701 for indicated cell lines, normalized to untreated controls. Error bars represent standard error of the mean (SEM) of triplicate wells. The dotted line indicates mean cytotoxic response of all 8 cell lines at 50 nM CEP-701. (B) FLT3 immunoprecipitation/immunoblotting showing FLT3 phosphorylation status (P-FLT3) and total protein expression (FLT3) for each cell line treated for 1 hour with 50 nM CEP-701 (+) or diluent control (-), grouped by CEP-701 response.
ALL cell lines with high-level expression and constitutive phosphorylation of FLT3 are differentially sensitive to FLT3 inhibition. (A) MTT dose-response curves showing mean cytotoxic response to CEP-701 for indicated cell lines, normalized to untreated controls. Error bars represent standard error of the mean (SEM) of triplicate wells. The dotted line indicates mean cytotoxic response of all 8 cell lines at 50 nM CEP-701. (B) FLT3 immunoprecipitation/immunoblotting showing FLT3 phosphorylation status (P-FLT3) and total protein expression (FLT3) for each cell line treated for 1 hour with 50 nM CEP-701 (+) or diluent control (-), grouped by CEP-701 response.
Primary ALL cells with high levels of FLT3 expression are differentially sensitive to FLT3 inhibition
The cryopreserved primary ALL samples (Table 2; patients J-1 to J-30) were selected based on quantitative FLT3 expression, cytogenetics, and FLT3 mutation status, which had previously been determined. The fresh primary ALL samples (Table 2; patients H-1 to H-9) were consecutive cases of childhood ALL enrolled on a Johns Hopkins IRB-approved cell procurement protocol. Fortyeight–hour MTT cytotoxicity assays were performed in triplicate and AVB apoptosis assays were performed in duplicate on all samples using increasing doses of CEP-701. Samples were considered evaluable if there was adequate leukemic cell viability in the untreated control wells at the end of the 48-hour assay period (ie, a net mean optical density of at least 0.100 in the MTT assay). By this criterion, 38 (97%) of 39 samples were evaluable (sample J-26, from a child with T-lineage ALL and WT FLT3, was not evaluable). Two other samples were excluded from the following analysis. Sample H-4 was excluded because there were inadequate numbers of cells to assess level of FLT3 expression. Sample J-11 was excluded because the FLT3 expression level was borderline. Thus, a total of 36 samples were included in this analysis.
We hypothesized that high-level expression of FLT3 in a leukemic sample may signify a high degree of dependence on FLT3 signaling for the survival of that sample's leukemic blasts. If so, inhibition of FLT3 signaling would be expected to result in significant and selective cytotoxicity in these samples. Figure 2A and Table 3 show the results of the MTT assays grouped by level of FLT3 expression. The dose-response curves represent the mean MTT cytotoxic response of all samples within each group to CEP-701, normalized to untreated controls. There was more cytotoxicity induced in samples with high-level FLT3 expression than in samples with low-level FLT3 expression. This difference was statistically significant at all CEP-701 dose levels (P value range, < .0001-< .0024) (Figure 2A; Table 3). There was no significant difference in CEP-701–induced cytotoxicity between the fresh samples and the cryopreserved samples in each group (data not shown).
Primary ALL blasts with high-level FLT3 expression, particularly those with MLL gene rearrangements, high hyperdiploidy, and FLT3 juxtamembrane domain mutations, are differentially sensitive to FLT3 inhibition. (A) MTT dose-response curves showing mean cytotoxic response to CEP-701 for infant and childhood ALL blasts, grouped by level of FLT3 expression, normalized to untreated controls. Error bars represent standard error of the mean (SEM). (B) Same data as in panel A but with the “FLT3 HIGH” group further stratified by cytogenetics. (C) Dot plot of individual samples' cytotoxic response (mean of triplicate wells) to CEP-701 at the 50-nM dose level, grouped by level of FLT3 expression and cytogenetics. All samples are FLT3 wild-type except where indicated. KD indicates FLT3 kinase domain mutation (ie, D835E substitution); ITD, FLT3 internal tandem duplication; JM del/ins, FLT3 juxtamembrane domain deletion/insertion. Error bars show mean ± SEM for each group. The dotted line indicates mean cytotoxic response of all 37 samples at 50 nM CEP-701 (59% untreated control). (D) Same data as in panel C, except with “FLT3 HIGH” samples only, grouped by FLT3 mutation status. WT indicates wild type; KD, kinase domain point mutation; JMD, juxtamembrane domain mutation; MUT, any mutation.
Primary ALL blasts with high-level FLT3 expression, particularly those with MLL gene rearrangements, high hyperdiploidy, and FLT3 juxtamembrane domain mutations, are differentially sensitive to FLT3 inhibition. (A) MTT dose-response curves showing mean cytotoxic response to CEP-701 for infant and childhood ALL blasts, grouped by level of FLT3 expression, normalized to untreated controls. Error bars represent standard error of the mean (SEM). (B) Same data as in panel A but with the “FLT3 HIGH” group further stratified by cytogenetics. (C) Dot plot of individual samples' cytotoxic response (mean of triplicate wells) to CEP-701 at the 50-nM dose level, grouped by level of FLT3 expression and cytogenetics. All samples are FLT3 wild-type except where indicated. KD indicates FLT3 kinase domain mutation (ie, D835E substitution); ITD, FLT3 internal tandem duplication; JM del/ins, FLT3 juxtamembrane domain deletion/insertion. Error bars show mean ± SEM for each group. The dotted line indicates mean cytotoxic response of all 37 samples at 50 nM CEP-701 (59% untreated control). (D) Same data as in panel C, except with “FLT3 HIGH” samples only, grouped by FLT3 mutation status. WT indicates wild type; KD, kinase domain point mutation; JMD, juxtamembrane domain mutation; MUT, any mutation.
Mean cytotoxic response of the FLT3-low and FLT3-high groups at each dose level
Dose level, nM . | P . |
|---|---|
| 5 | .0024 |
| 10 | .0007 |
| 20 | < .0001 |
| 50 | < .0001 |
| 100 | < .0001 |
Dose level, nM . | P . |
|---|---|
| 5 | .0024 |
| 10 | .0007 |
| 20 | < .0001 |
| 50 | < .0001 |
| 100 | < .0001 |
P values generated by Student t test comparing the mean cytotoxic response of the 2 groups at each dose level. All P values are significant.
Primary ALL cells with MLL gene rearrangements, high hyperdiploidy, and/or FLT3 mutations are particularly sensitive to FLT3 inhibition
Given that infant and childhood ALL specimens with either MLL gene rearrangements or high hyperdiploidy have been shown to express particularly high levels of FLT3 and have a relatively high rate of FLT3-activating mutations, we determined whether these cytogenetic subgroups were particularly sensitive to FLT3 inhibition. The mean cytotoxic response for all 36 samples to CEP-701 at 50 nM was a mean OD of 59% of untreated controls. Hence, a sample was considered to be sensitive to FLT3 inhibition if the mean OD for the sample was less than 59% of untreated controls at 50 nM CEP-701. As shown in Tables 4 and 5 and in Figure 2B-C, among samples with high-level expression of FLT3, the most pronounced cytotoxicity was observed with high-hyperdiploid and MLL-rearranged samples compared with those with other cytogenetic abnormalities or normal cytogenetics. Notably, all 3 of the high-FLT3–expressing cytogenetic subgroups, including the samples lacking MLL rearrangements or high hyperdiploidy, demonstrated significantly greater cytotoxicity than the low-FLT3–expressing group (P value range, < .0001-< .009) (Table 5). Thus, high-FLT3 expression, even in the absence of MLL rearrangement or high hyperdiploidy, is associated with increased sensitivity to FLT3 inhibition.
CEP-701 sensitivity by FLT3 expression level and cytogenetics
. | Sensitive . | Resistant . | % Sensitive . | P, versus low . |
|---|---|---|---|---|
| FLT3-low | 1 | 12 | 8 | — |
| FLT3-high | 17 | 6 | 74 | .0003* |
| MLL | 9 | 2 | 82 | .0005* |
| High hyper | 5 | 0 | 100 | .0007* |
| Other | 3 | 4 | 43 | .1011 |
. | Sensitive . | Resistant . | % Sensitive . | P, versus low . |
|---|---|---|---|---|
| FLT3-low | 1 | 12 | 8 | — |
| FLT3-high | 17 | 6 | 74 | .0003* |
| MLL | 9 | 2 | 82 | .0005* |
| High hyper | 5 | 0 | 100 | .0007* |
| Other | 3 | 4 | 43 | .1011 |
P value determined by Fisher exact test.
—Indicates comparison not possible; MMLL, samples with rearranged MLL gene; High hyper, samples with hyperdiploidy (> 50 chromosomes); Other, samples with normal cytogenetics or other abnormalities (see Table 2).
Significant
Comparison of cytotoxic responses of FLT3 low, MLL, high hyper, and other groups
Group . | P . |
|---|---|
| Low versus high | < .0001* |
| Low versus MLL | < .0001* |
| Low versus hyper | < .0001* |
| Low versus other | .009* |
| MLL versus high | — |
| MLL versus MLL | — |
| MLL versus hyper | .0446* |
| MLL versus other | .1313 |
| Hyper versus high | — |
| Hyper versus MLL | — |
| Hyper versus hyper | — |
| Hyper versus other | .0113* |
Group . | P . |
|---|---|
| Low versus high | < .0001* |
| Low versus MLL | < .0001* |
| Low versus hyper | < .0001* |
| Low versus other | .009* |
| MLL versus high | — |
| MLL versus MLL | — |
| MLL versus hyper | .0446* |
| MLL versus other | .1313 |
| Hyper versus high | — |
| Hyper versus MLL | — |
| Hyper versus hyper | — |
| Hyper versus other | .0113* |
P values generated by Student t test comparing cytotoxic responses of each group.
Significant
Our sample set contained 4 samples with activating mutations of FLT3, including 2 with kinase domain mutations (FLT3/KD, both D835E substitutions) and 2 with mutations in the juxtamembrane domain (FLT3/JMD, one internal tandem duplication [ITD] and one insertion/deletion mutation), as shown in Table 2. We hypothesized that among samples with high-level FLT3 expression, samples with activating mutations may be especially dependent on FLT3 signaling for their survival and may therefore demonstrate differential sensitivity to FLT3 inhibition. While the mutant FLT3 group as a whole demonstrated increased sensitivity to FLT3 inhibition compared with those with wild-type FLT3 (P = .0402), this difference was due primarily to the marked sensitivity of the 2 FLT3/JMD samples (P = .0219; Figure 2D; Table 6). The 2 FLT3/KD samples did not demonstrate increased sensitivity compared with FLT3 wild-type samples (P = .2994).
The mechanism of cytotoxicity in CEP-701–sensitive primary ALL cells is the induction of apoptosis
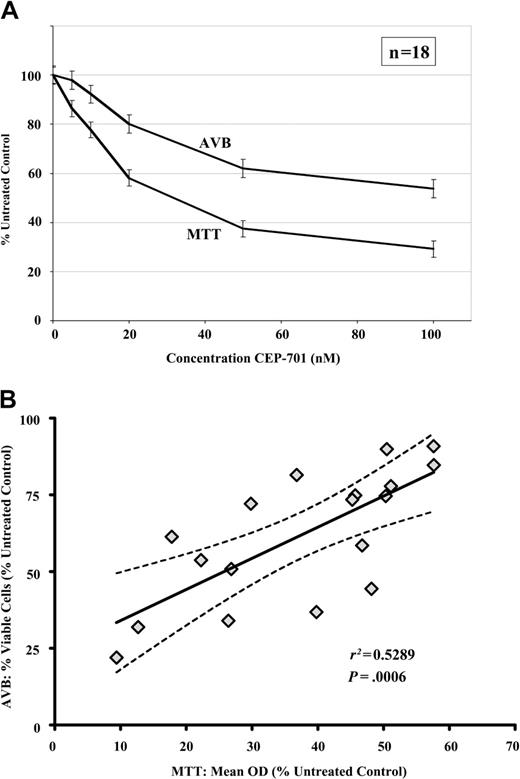
The MTT assay does not provide information on the mechanisms underlying cytotoxicity.54 To determine whether CEP-701 cytotoxicity was due to induction of apoptosis, we performed 48-hour annexin-V binding (AVB) flow cytometric assays in parallel with MTT assays on all samples. The percentage of viable cells was normalized to the untreated controls and plotted. As shown in Figure 3A-B, there was a significant linear correlation between the MTT and AVB assays for the CEP-701–sensitive samples (n = 18), confirming that the predominant mechanism of cytotoxicity of CEP-701 in these samples is the induction of apoptosis.
The primary mechanism of the cytotoxic effects of CEP-701 on primary ALL cells is the induction of apoptosis. (A) Dose-response curves showing the mean MTT and annexin-V binding (AVB) apoptosis assays for all CEP-701–sensitive samples (n = 18). Error bars indicate standard error of the mean (SEM). (B) Correlation of MTT and AVB results at the CEP-701 50-nM dose level. r2 indicates Pearson correlation coefficient; the solid line, best-fit line by linear regression; and the dotted lines, 95% confidence interval of best-fit line.
The primary mechanism of the cytotoxic effects of CEP-701 on primary ALL cells is the induction of apoptosis. (A) Dose-response curves showing the mean MTT and annexin-V binding (AVB) apoptosis assays for all CEP-701–sensitive samples (n = 18). Error bars indicate standard error of the mean (SEM). (B) Correlation of MTT and AVB results at the CEP-701 50-nM dose level. r2 indicates Pearson correlation coefficient; the solid line, best-fit line by linear regression; and the dotted lines, 95% confidence interval of best-fit line.
MLL gene rearrangement, and not patient age, is the primary determinant of sensitivity to FLT3 inhibition in infant ALL
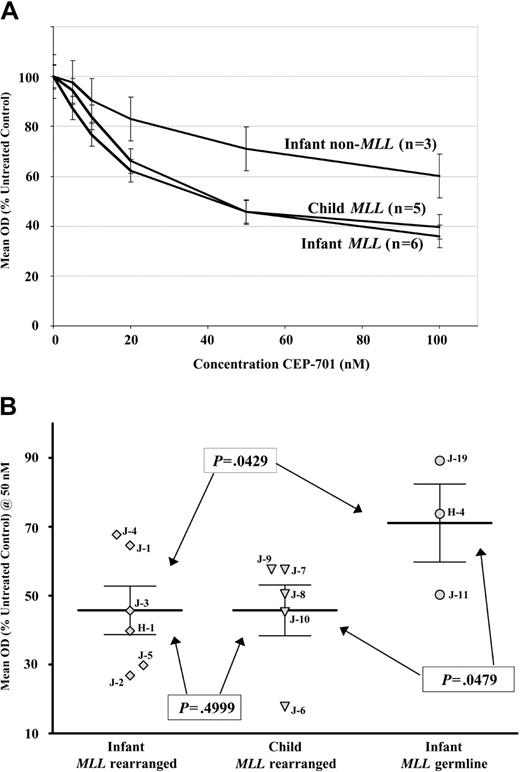
Rearrangements of the MLL gene at 11q23 occur in both infants and older children with ALL. While such rearrangements are a poor prognostic factor in ALL at any age, this is particularly true of infants, where 5-year event-free survival (EFS) is approximately 20%.55,56 Older children with MLL rearrangements fare somewhat better, with a 5-year EFS of approximately 50%.55,56 Since infants and children with MLL-rearranged ALL have significantly different outcomes, we sought to determine whether this difference might be partly attributable to differences in dependence on FLT3 signaling in the blasts of infants versus children with MLL rearrangements. Additionally, infants with ALL who have germ line (ie, nonrearranged) MLL also have a poor prognosis, although not as dismal as their MLL-rearranged counterparts (5-year EFS of approximately 45% for germ line MLL versus 20% for rearranged MLL).57 We therefore also sought to determine the relative FLT3 dependence of the blasts from infants with and without MLL rearrangements. As shown in Figure 4A-B, both infants and children with MLL rearrangements demonstrate marked cytotoxicity with FLT3 inhibition, with no significant difference between the 2 groups. Further, infants with MLL rearrangements demonstrate significantly more cytotoxicity with FLT3 inhibition than infants with germ line MLL.
MLL gene rearrangement, and not patient age, is the primary determinant of sensitivity to FLT3 inhibition in infant ALL. (A) MTT dose-response curves showing mean cytotoxic response to CEP-701 for infant and childhood ALL blasts, grouped by MLL gene status and age, normalized to untreated controls. Error bars represent standard error of the mean (SEM). (B) Dot plot of individual samples' MTT cytotoxic response at 50 nM CEP-701. Error bars show the group mean ± SEM. P values are from Student t test. Patient numbers (from Table 2) are noted next to each curve. H indicates Johns Hopkins patient; and J, St Jude patient.
MLL gene rearrangement, and not patient age, is the primary determinant of sensitivity to FLT3 inhibition in infant ALL. (A) MTT dose-response curves showing mean cytotoxic response to CEP-701 for infant and childhood ALL blasts, grouped by MLL gene status and age, normalized to untreated controls. Error bars represent standard error of the mean (SEM). (B) Dot plot of individual samples' MTT cytotoxic response at 50 nM CEP-701. Error bars show the group mean ± SEM. P values are from Student t test. Patient numbers (from Table 2) are noted next to each curve. H indicates Johns Hopkins patient; and J, St Jude patient.
Sensitivity of primary ALL cells to FLT3 inhibition strongly correlates with high-level expression of constitutively activated FLT3
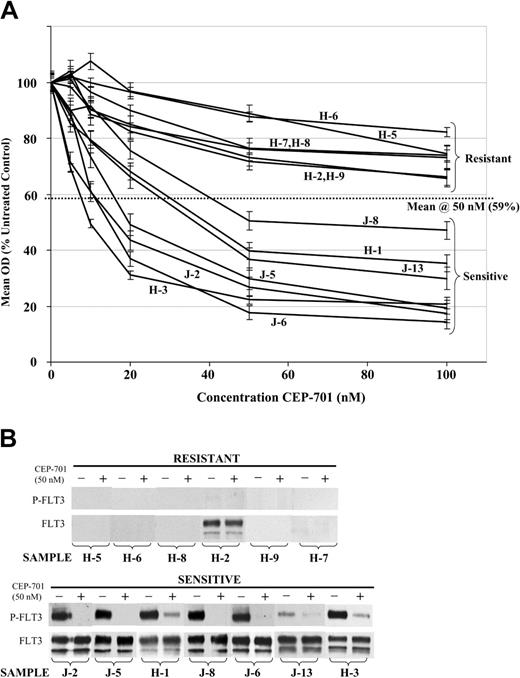
To further investigate the molecular basis for the variability in cytotoxic response to CEP-701, we performed FLT3 immunoprecipitation and immunoblotting on CEP-701–treated and untreated infant and childhood ALL primary samples for which sufficient cells were available (Figure 5A-B). Of the 6 samples not sensitive to CEP-701, 5 did not express appreciable levels of FLT3. The one sample that did express a high level of FLT3 had no constitutive FLT3 phosphorylation. By contrast, all 7 sensitive samples expressed high levels of FLT3 that was constitutively activated. As shown in Table 2, all but one of these samples (J-13, which contained a FLT3/KD mutation) expressed wild-type FLT3. This suggests the possibility of autocrine activation of FLT3 by FL coexpression, as has been demonstrated in some cases of wild-type FLT3–expressing AML samples.8 In all 7 cases, phosphorylation of FLT3 was potently inhibited by 50 nM CEP-701. Thus, there appears to be a strong correlation between cytotoxic response to CEP-701 and high-level expression of constitutively activated FLT3.
In primary infant and childhood ALL blasts, cytotoxic response to FLT3 inhibition is strongly correlated with high-level expression of constitutively activated FLT3. (A) MTT dose-response curves showing mean cytotoxic response to CEP-701 for primary infant and childhood ALL samples, normalized to untreated controls. Error bars represent standard error of the mean (SEM) of triplicate wells. Patient numbers (from Table 2) are noted next to each curve. H indicates Johns Hopkins patient; and J, St Jude patient. (B) FLT3 immunoprecipitation/Western blotting showing FLT3 phosphorylation status (P-FLT3) and total protein expression (FLT3) for each sample from panel A treated for 1 hour with 50 nM CEP-701 (+) or diluent control (-), grouped by CEP-701 responsiveness.
In primary infant and childhood ALL blasts, cytotoxic response to FLT3 inhibition is strongly correlated with high-level expression of constitutively activated FLT3. (A) MTT dose-response curves showing mean cytotoxic response to CEP-701 for primary infant and childhood ALL samples, normalized to untreated controls. Error bars represent standard error of the mean (SEM) of triplicate wells. Patient numbers (from Table 2) are noted next to each curve. H indicates Johns Hopkins patient; and J, St Jude patient. (B) FLT3 immunoprecipitation/Western blotting showing FLT3 phosphorylation status (P-FLT3) and total protein expression (FLT3) for each sample from panel A treated for 1 hour with 50 nM CEP-701 (+) or diluent control (-), grouped by CEP-701 responsiveness.
Discussion
We have demonstrated that FLT3 inhibition induces cytotoxicity in ALL cells, including both cell lines and primary infant and childhood ALL lymphoblasts. The sensitivity to CEP-701–induced cytotoxicity in these cells is directly related to levels of FLT3 expression. Among samples with high-FLT3 expression, those with hyperdiploidy with more than 50 chromosomes, rearrangements in the MLL gene at 11q23, and mutations in the juxtamembrane domain of FLT3 are particularly sensitive. In samples obtained from infants, those with MLL rearrangements are significantly more sensitive than those with germ line MLL. Finally, high-level expression of constitutively activated FLT3 strongly correlates with sensitivity to CEP-701–induced cytotoxicity.
Since CEP-701 and other small-molecule inhibitors of FLT3 are in various stages of clinical development in adult patients,33,35,38,58 these findings have clear therapeutic implications for infant and childhood leukemia. This is particularly true in the case of infants with MLL-rearranged ALL, whose prognosis with standard therapy is dismal55-57,59-64 and does not appear to be significantly improved by hematopoietic stem cell transplantation.55,56 Novel therapies are clearly needed for these patients. Our results show that leukemic blasts from these patients express high levels of FLT3 that is in a constitutively phosphorylated state, presumably through coexpression of FLT3 ligand in most cases where FLT3 is wild type. FLT3 phosphorylation in these cases is potently inhibited by CEP-701, and this inhibition results in pronounced cytotoxic and apoptotic responses, suggesting that the blasts from these patients are highly dependent on FLT3 signaling for their survival. Given that blasts from infants with MLL-rearranged ALL have been shown to be intrinsically resistant to multiple chemotherapeutic agents,65,66 the therapeutic strategy of FLT3 inhibition is an exciting one that should be investigated without delay. Furthermore, while older children with MLL-rearranged ALL fare better than their infant counterparts, they have a significantly worse prognosis than children with germ line MLL.67 Our data indicate that the leukemic blasts from older children with MLL rearrangements are just as FLT3 dependent as those from infants. Therefore, these patients are also likely to benefit from the incorporation of FLT3 inhibitors into their therapy.
Unlike rearrangements of the MLL gene, hyperdiploidy with more than 50 chromosomes is a favorable prognostic feature in childhood ALL.68,69 Blasts from these patients have been shown to represent a distinct biologic subset with propensity to apoptosis and pronounced chemosensitivity in vitro.70,71 Nonetheless, approximately 20% of these patients will suffer relapse of their disease.68,69 Our data indicate that FLT3 delivers crucial survival signals in high-hyperdiploid cells, as indicated by their exquisite sensitivity to FLT3 inhibition in cytotoxicity and apoptosis assays. FLT3 inhibitors, therefore, may have a role in the treatment of high-hyperdiploid ALL, particularly in the setting of relapsed disease.
The potential benefits of novel molecularly targeted therapies are not limited to poor-prognosis leukemias. As outcome for children with ALL has improved, the focus of clinical research in this disease has shifted to identifying low-risk populations where less toxic therapy can be given without compromising the chances of cure.72 Targeted therapies, such as FLT3 inhibitors, have the potential of producing far fewer short- and long-term toxicities than standard chemotherapeutics due to their relative specificity for malignant cells.73,74 As such, the successful incorporation of these agents into the treatment of childhood ALL may enable the decreased use of chemotherapeutics (with their significant short- and long-term toxicities) while maintaining high cure rates. Children with high-hyperdiploid ALL present an opportunity to test this hypothesis.
Our data show that cases of ALL with high levels of FLT3 expression are likely to respond clinically to inhibition of FLT3 signaling. This is particularly true in the 2 subsets of ALL with the highest levels of FLT3 expression, those with MLL rearrangements and those with high hyperdiploidy. Strategies to test the clinical efficacy of FLT3 inhibitors should be based upon these data. Standard cytogenetics can identify MLL rearrangements and hyperdiploidy, and a reasonable approach might be to initially test FLT3 inhibitors in these patients. Subsequent studies could use molecular techniques to prospectively quantify the level of FLT3 expression, FLT3 mutation status, and FLT3 phosphorylation status. This might more rationally identify patients likely to benefit from FLT3 inhibitors.
Regardless of how FLT3 inhibitors are used clinically, they are unlikely to be curative as monotherapy. While we have seen no evidence of intrinsic resistance of activated FLT3 to inhibition by CEP-701 in cell lines or primary samples, resistance to FLT3 inhibitors given as monotherapy is likely to develop by a variety of mechanisms. For example, the CEP-701–resistant cell line Reh is shown in Figure 1 to express high levels of constitutively activated FLT3 that is potently inhibited by CEP-701. Inhibition of activated FLT3 in this cell line is, by itself, insufficient to induce apoptosis, implying the presence of FLT3-independent survival signals in these cells. Treatment of acute leukemia with a FLT3 inhibitor alone may select for subclones with relatively strong FLT3-independent survival signals, leading to the development of resistance. In addition, prolonged exposure to FLT3 inhibitors is likely to select for mutations in FLT3 that reduce the binding affinity of these drugs for the FLT3 receptor, as has been seen in some cases of imatinib-resistant chronic myeloid leukemia (CML).75 Therefore, it will be important to rationally incorporate FLT3 inhibitors into existing multiagent combination chemotherapy regimens. Synergy between FLT3 inhibitors and various chemotherapy agents has been reported in studies of AML blasts.76,77 Similar studies should be used to guide the initial design of clinical trials of FLT3 inhibitors for infant and childhood ALL.
Prepublished online as Blood First Edition Paper, September 16, 2004; DOI 10.1182/blood-2004-06-2498.
Supported by grants from the National Cancer Institute (NCI; CA60441, P.B.; CA095600, M.L.; CA91177, CA90668, and CA70970, D.S.), American Society of Clinical Oncology (M.L.), Sidney Kimmel Foundation (M.L.), Leukemia and Lymphoma Society (D.S.), and Burroughs Wellcome Fund (D.S.). D.S. is the Douglas Kroll Research Foundation Translational Researcher of the Leukemia and Lymphoma Society and the recipient of the Kyle Haydock Professorship in Oncology. P.B. is a Richard S. Ross Clinician Scientist Award recipient.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Michael Cleary for providing the HB-1119 cell line, Carolyn Felix for providing the SEM-K2 cell line, and Bruce Ruggeri and Susan Jones-Bolin of Cephalon, Inc for providing CEP-701.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal