Abstract
The chromosomal translocation t(7; 11)(p15;p15), observed in human myeloid leukemia, results in a NUP98 and HOXA9 gene fusion. We generated a transgenic mouse line that specifically expressed the chimeric NUP98-HOXA9 gene in the myeloid lineage. While only 20% of the transgenic mice progressed to leukemia after a latency period, myeloid progenitor cells from nonleukemic transgenic mice still exhibited increased proliferative potential. This suggested that the NUP98-HOXA9 fusion induced a preleukemic phase, and other factors were required for complete leukemogenesis. NUP98-HOXA9 expression promoted the onset of retrovirus-induced BXH2 myeloid leukemia. This phenomenon was used to identify cooperative disease genes as common integration sites (CISs). Meis1, a known HOX cofactor, was identified as a CIS with a higher integration frequency in transgenic than in wild-type BXH2 mice. By the same means we identified further 4 candidate cooperative genes, Dnalc4, Fcgr2b, Fcrl, and Con1. These genes cooperated with NUP98-HOXA9 in transforming NIH 3T3 cells. The system described here is a powerful tool to identify cooperative oncogenes and will assist in the clarification of the multistep process of carcinogenesis.
Introduction
Carcinogenesis requires mutations in multiple genetic loci, and many genes involved in regulation of the cell cycle, apoptosis, transcription, and signaling have been identified as critical disease genes.1 Previous studies have found that combinations of genetic abnormalities are required to make normal cells become cancer cells in vitro.2 However, it remains unclear how individual genetic mutations cooperate with each other in human cancer. Retroviral insertional mutagenesis is a powerful technique to isolate causative disease genes, and a variety of important genes have already been identified using this method.3-5 Studies have applied this technique to the identification of oncogenes that collaborate with cell cycle regulators, such as Cdkn2a and p27Kip1.6,7 Therefore, the combination of retroviral insertional mutagenesis and an appropriate animal model with a phenotype similar to that of a human cancer should provide an ideal experimental system to identify cooperative cancer genes. These genes are extremely important, as they could be used to prevent the cancer progression of premalignant lesions or neoplasms with low malignant potential that contain known primary genetic events.
Chromosomal translocations and genetic chimerism in human hematopoietic and soft tissue neoplasms are early genetic events. However, several reports have indicated that chromosomal translocation and chimeric protein expression alone are insufficient for complete carcinogenesis.8 Therefore, it is of great interest to discover which genes cooperate with chimeric oncogenes in carcinogenesis and to what genetic pathways these cooperative oncogenes belong.
The nucleoporin gene NUP98 on human chromosome 11p15 is involved in a series of relatively rare but recurrent translocations observed in de novo acute and chronic myeloid leukemias, as well as in therapy-related myelodysplastic syndrome (t-MDS) and acute myeloid leukemia (t-AML). The major targets of NUP98 as fusion partners are clustered and nonclustered homeobox genes, including HOXA9,9,10 HOXA11, HOXA13,11 HOXC11,12 HOXC13,13 HOXD11,14 HOXD13,15 and PMX1.16 HOXA9 is the most frequent fusion partner of NUP98 in chromosomal translocations.17 The NUP98-HOXA9 oncoprotein retains the homeodomain of HOXA9 and the phenylalanine-glycine (FG) repeat region of NUP98.9,10 Enforced expression of NUP98-HOXA9 in NIH 3T3 cells results in transformation. The oncogenic potential of NUP98-HOXA9 is thought to involve the FG repeats, which are capable of interacting with the transcriptional co-activator, CREB (cyclic adenosine monophosphate response element binding protein) binding protein (CBP)/p300.18
The leukemogenic potential of murine Hoxa9 was directly assessed in hemopoietic chimeras where its overexpression in bone marrow cells induced AML after a latency period.19 In addition, Meis1, together with Hoxa7 or Hoxa9, was a frequent target of endogenous retroviral insertional activation in BXH2 mouse leukemic cells, which suggested leukemogenic cooperation between Meis1 and the Hox genes.20 In support of this hypothesis, Meis1 coexpression dramatically reduced the latency period of Hoxa9-induced AML.19 Also, the trimeric DNA binding complex consisting of Hoxa9 and MEINOX family homeoproteins such as Meis and PBX was observed in leukemic cells, Hoxa9 transcriptional activity requires Meis and PBX interaction domains. Meis1 also cooperated with HOXB3, a homeobox gene functionally divergent from HOXA9, which suggested that Meis1 might be a common leukemogenic collaborator.21 However, as in vitro immortalization of myeloid progenitor cells by Hoxa9 was reported to occur in the absence of Meis expression and Hoxa9-PBX interaction,22 a subset of HOX-induced transformations may occur independently of Meis or involve other factors able to replace Meis function.
To investigate the leukemogenic activity of NUP98-HOXA9, we generated transgenic mice in which the NUP98-HOXA9 fusion gene was specifically expressed in the myeloid lineage under the control of the cathepsin G promoter. Approximately 20% of the transgenic (Tg) mice developed AML after long latency periods, which indicated that additional cofactors were required for complete leukemogenesis.
After crossing the Tg mice onto the BXH2 strain, we used retroviral insertional mutagenesis to identify novel cofactors for NUP98-HOXA9 in addition to Meis1. Cooperation between NUP98-HOXA9 and novel cofactors were then confirmed in a transformation assay using NIH 3T3 cells. Our study provides a model for the application of retroviral insertional mutagenesis to the discovery of cooperative oncogenes.
Materials and methods
Generation of NUP98-HOXA9 transgenic mice
To generate the NUP98-HOXA9 transgene, the entire coding sequence of the chimeric NUP98-HOXA910 was amplified by reverse transcriptase-polymerase chain reaction (RT-PCR) and inserted into the human cathepsin G gene cassette.23 The purified construct was microinjected into (C57BL/6J × DBA/2J)F2 fertilized eggs, and transgenic founders were identified by Southern blot hybridization as previously described.24
NUP98-HOXA9 Tg mice (line 1589) were backcrossed to BXH2 mice over 3 generations to introduce the transgene into the BXH2 genetic background, and the leukemia-free survival rates were compared between Tg-positive and Tg-negative BXH2 mice at the third generation of the cross. Mice were monitored daily for evidence of disease, and smear samples of peripheral blood were examined every month. The survival rate of each group was evaluated using the Kaplan-Meier test.
RNA extraction and RT-PCR
Total RNA was extracted from mouse and Sca-1-enriched bone marrow (BM) cells tissues using RNAzol (TelTest, Friendswood, TX). RT-PCR was performed as previously described.10 The PCR primers used for RT-PCR were as follows: Fusgt2, 5′-AAAGTTGAGGGGGGGCATTAG-3′; Cggt2, 5′-TCATTCTGGATGGTCCGCTG-3′; Dnalc4 forward, 5′-GGAGAGACTGAAGGGAAGAAAGA-3′; Dnalc4 reverse, 5′-TTGCAGCACTCTCATTGTTG-3′; Fcgr2b forward, 5′-CAGGTGCTCAAGGAAGACAC-3′; Fcgr2b reverse, 5′-TCCCATTTCCCTGTGATCAG-3′; Fcrl forward, 5′-TCAGCGTGTCCTATGACTGG-3′; Fcrl reverse, 5′-GTTTCTGAAGAGCCTGTACCCA-3′; Con1 forward, 5′-CTCAAGGCACCTTGCAAGTC-3′; Con1 reverse, 5′-GTTCCCATCCTGAAAGTCCA-3′; Con2 forward, 5′-ACGTCACTCCCCGAGAGTCT-3′; and Con2 reverse, 5′-TAGTCGGAGAGGTGCCTCAG-3′.
Histology and fluorescence-activated cell sorting (FACS)
Tissues were fixed in 10% buffered formalin embedded in paraffin, and sections were stained with hematoxylin and eosin using standard methodologies. For flow cytometry, single-cell suspensions of 1 × 105 cells were incubated with fluorescein isothiocyanate (FITC)-conjugated antibodies, Gr-1 (RB6-8C5), Mac-1 (M1/70), B220 (RA3-6B2), or CD3 (145-2C11) (PharMingen, San Diego, CA) for 20 minutes on ice. Cells were then washed 3 times in 1 × phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin and applied to a FACS-calibur flow cytometer (Becton Dickinson, Mountain View, CA).
Replating assay of bone marrow cells and G-CSF stimulation
Bone marrow was flushed from the femurs of Tg and wild-type mice. Bone marrow cells were washed in PBS, and 1 × 104 cells were plated onto 35-mm Petri dishes in Methocult M3434 methylcellulose medium (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with cytokines (10 ng/mL recombinant mouse [rm] interleukin-3 [IL-3], 10 ng/mL recombinant human [rh] IL-6, 50 ng/mL rm stem cell factor, and 3 U/mL rh erythropoietin). Cultures were incubated at 37°C in a 5% CO2 humidified atmosphere, and colonies were enumerated after 7 to 11 days. Colonies were then harvested, and 1 × 104 cells were replated in the same way.
For granulocyte colony-stimulating factor (G-CSF) stimulation, Tg and wild-type mice were injected intraperitoneally with 200 μg/kg/d rh G-CSF (Kirin, Tokyo, Japan) for 5 days. White blood cells (WBCs) were counted with Turk solution (MERCK, Darmstadt, Germany), and peripheral blood progenitor cells (PBPCs) were enumerated as the number of colonies formed after 7 days in cultures of 20 μL blood in Methocult methylcellulose medium as described in the preceding paragraph.
Cloning of retroviral integration sites
The cloning of viral integration sites from tumor DNA is described elsewhere.3 Briefly, NUP98-HOXA9/BXH2 tumor DNA samples were digested completely with SacII or BamHI, self-ligated, and subjected to nested inverse PCR (IPCR). PCR conditions were 10 cycles of 94°C for 15 seconds, 63°C for 30 seconds, and 68°C for 15 minutes, followed by 23 cycles of 94°C for 15 seconds, 63°C for 30 seconds, and 72°C for 15 minutes with a 20-second extension per cycle. PCR products were analyzed by agarose gel electrophoresis, and tumor-specific products were subcloned and sequenced.
DNA extraction and Southern blot analysis
High-molecular-weight genomic DNA was extracted from frozen leukemic cell suspensions. Samples consisting of 5 μg genomic DNA were subjected to restriction endonuclease digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization as described previously.25 Appropriate genomic DNA fragments derived from flanking sequences of each common integration sites were used as probes.
DNA sequencing and sequence comparison
DNA sequencing was performed using T7 and SP6 primers, integrated DNA, and the Dye Terminator Cyclesequencing Kit (Beckman Coulter, Fullerton, CA). Sequencing reactions were analyzed using a CEQ8000 DNA sequencer (Beckman Coulter).
We compared retroviral integration site sequences with GenBank and UCSC (University of California at Santa Cruz) databases by BLAST (basic local alignment search tool) analysis and located genes in the vicinity of viral integrations for further analysis.
Preparation of a Sca-1-enriched BM cell population by positive selection
Sca-1-enriched BM cells from nonleukemic transgenic and wild-type mice were purified on magnetic cell sorting (MACS) columns using microbead-conjugated Sca-1 antibody. Separation was performed according to the manufacturer's recommendations (MidiMACS; Miltenyi Biotec, Bergisch-Gladbach, Germany).
Retroviral constructs
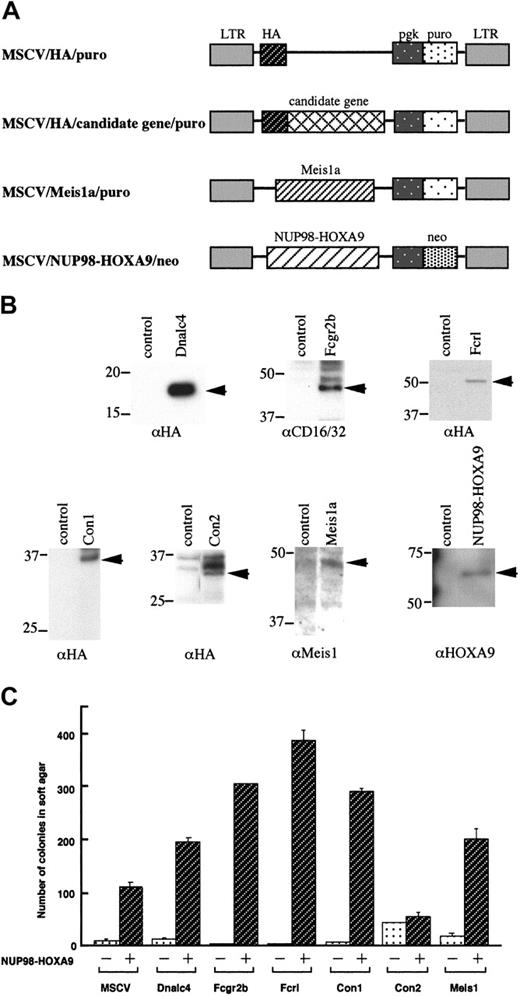
Retroviral constructs encoding NUP98-HOXA9 and Meis1 were described previously.19,26 Hemagglutinin (HA) epitope-tagged constructs of the candidate cooperative genes were engineered by fusing the PCR products of each gene in-frame to the 3′ end of the HA epitope in the pcDNA3.1 vector (Invitrogen, Carlsbad, CA). The HA-tagged constructs were subcloned into the polylinker of the murine stem cell proviral vector, murine stem cell virus-phosphoglycerate kinase-puromycin resistance gene (MSCV-pgk-puro), as shown in Figure 7A. MSCV-pgk-puro without insert was used as a vector control.
Expression of candidate cooperative genes in NUP98-HOXA9/BXH2 leukemias. RT-PCR analyses of the cooperative genes as indicated. The leukemia samples with retroviral integrations at each loci are indicated as +. Gene expression in Sca-1-enriched bone marrow cells derived from nonleukemic transgenic mice and wild-type littermates were also shown. dw indicates negative control.
Expression of candidate cooperative genes in NUP98-HOXA9/BXH2 leukemias. RT-PCR analyses of the cooperative genes as indicated. The leukemia samples with retroviral integrations at each loci are indicated as +. Gene expression in Sca-1-enriched bone marrow cells derived from nonleukemic transgenic mice and wild-type littermates were also shown. dw indicates negative control.
Retroviral stocks and infection of NIH 3T3 cells
Retroviral supernatants were produced as previously described using the plat E packaging cell line (a kind gift from Dr Toshio Kitamura, University of Tokyo).27 Briefly, plasmid DNA was transfected by lipofectamine 2000 (Invitrogen) into plat E cells, and virus-containing supernatants were collected 48 hours after transfection. For infection, 1 × 105 NIH 3T3 cells were plated in 10-cm dishes and were cultured for 24 hours with virus-containing supernatants (∼ 5 × 105 colony-forming unit [CFU]/mL as assessed by G-418 or puromycin resistance in NIH 3T3 cells) in the presence of 8 μg/mL Polybrene (Sigma, St Louis, MO). Cells were then washed with PBS, incubated for 24 hours with Dulbecco modified Eagle medium (DMEM) supplemented with 10% calf serum, and exposed to selection for 10 days in medium containing 1 mg/mL G-418 (GIBCO, Burlington, ON, Canada), and/or 10 μg/mL puromycin (Sigma) according to the selectable marker in each construct.
Transformation assay
To determine the ability of candidate genes to confer anchorage independent growth, colony formation in soft agar was assessed. Stably transduced cells were seeded at 104 cells per dish in 0.3% Agar Noble (Difco, Detroit, MI) over a 0.6% agar bottom layer. After 3 weeks colonies consisting of greater than 50 cells were counted microscopically. The experiment was repeated 3 times, and mean values with standard deviations were calculated.
Western blot analysis
Expression of the retroviral constructs was verified by Western blot analysis. Transiently expressed plat-E cells were lysed in sodium dodecyl sulfate (SDS) sample buffer containing 50 mM tris(hydroxymethyl)aminomethane (Tris) HCl (pH 6.8), 2% SDS, 10% glycerol, 100 mM dithiothreitol (DTT), and 0.1% bromophenol blue, and proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Size-fractionated proteins were blotted onto polyvinylidenefluoride membranes (Immobilon-P, Bedford, MA). Membranes were blocked in Tris-buffered saline (pH 7.5) with 0.1% Tween-20 (TBS-T) containing 5% skim milk for 1 hour at room temperature and then incubated with monoclonal anti-HA (Roche, Mannheim, Germany), anti-CD16/32 (2.4G2) (for Fcgr2b; PharMingen), polyclonal anti-Meis1 (kindly provided by Michael Cleary, Stanford University) or anti-HOXA9 antibodies.26 After 3 washes with TBS-T, bound antibodies were detected with horseradish peroxidase-conjugated anti-rat antibody and enhanced chemiluminescence (ECL) Western blotting detection reagents (Amersham, Buckinghamshire, United Kingdom).
Results
Generation of NUP98-HOXA9 transgenic mice
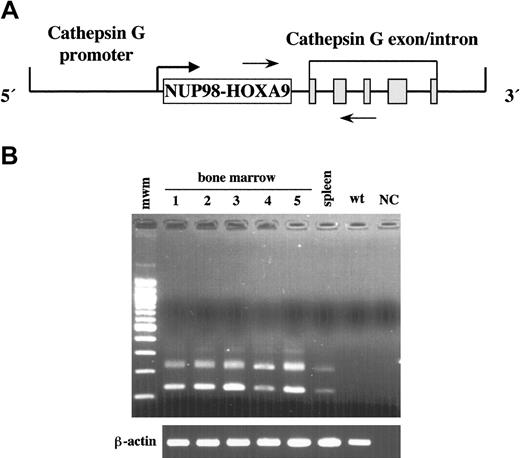
To determine whether overexpression of NUP98-HOXA9 induced leukemia, we constructed a transgene containing NUP98-HOXA9 cDNA under the control of sequences that regulate the promyelocyte-specific expression of the human Cathepsin G (hCG) gene (Figure 1A).23 Five independent founder lines were generated, and bone marrow-specific expression of the transgene was confirmed in all the lines by RT-PCR (Figure 1B). Transgene expression was limited to the bone marrow and the spleen at the lower level, with no expression detected in any of the other tissues examined. The NUP98-HOXA9 protein was not detected in the bone marrow by Western blotting, probably due to low expression level (data not shown). Two of 5 lines (1589 and 131 in Figure 1B) were maintained, and leukemia developed in the subset of mice of both lines. Line 1589 was selected for further analysis, since the leukemia incidence in this line was a little higher than in line 131 (10% versus 7% at 1 year of age).
Generation of NUP98-HOXA9 transgenic mice. (A) The hCG-NUP98-HOXA9 transgenic construct. The NUP98-HOXA9 cDNA was inserted at the transcriptional start site of the hCG gene. The solid gray boxes represent the 5 exons of the hCG gene. The polyadenylation signal is provided by the hCG gene. The horizontal arrows indicate the location of RT-PCR primers. (B) RT-PCR analysis of bone marrow RNAs obtained from the transgenic mice of 5 lines (1, line 60; 2, line 131; 3, line 1514; 4, line 1583; 5, line 1589) and spleen of line 1589. Lines 131 and 1589 were used for further analysis. Two different kinds of transcripts were created due to alternative splicing of cathepsin G exons. β-actin was amplified from the same samples to check for RNA quality. mwm indicates 100 base pair (bp) ladder; wt, bone marrow of the wild-type mouse; and NC, negative control.
Generation of NUP98-HOXA9 transgenic mice. (A) The hCG-NUP98-HOXA9 transgenic construct. The NUP98-HOXA9 cDNA was inserted at the transcriptional start site of the hCG gene. The solid gray boxes represent the 5 exons of the hCG gene. The polyadenylation signal is provided by the hCG gene. The horizontal arrows indicate the location of RT-PCR primers. (B) RT-PCR analysis of bone marrow RNAs obtained from the transgenic mice of 5 lines (1, line 60; 2, line 131; 3, line 1514; 4, line 1583; 5, line 1589) and spleen of line 1589. Lines 131 and 1589 were used for further analysis. Two different kinds of transcripts were created due to alternative splicing of cathepsin G exons. β-actin was amplified from the same samples to check for RNA quality. mwm indicates 100 base pair (bp) ladder; wt, bone marrow of the wild-type mouse; and NC, negative control.
Development of myeloid leukemia in NUP98-HOXA9 transgenic mice
Ten percent of the transgenic mice developed chronic myeloproliferation and subsequent AML by 1 year of age, and 22% by 15 months (Figure 2A; Table 1). Diseased mice showed pale bone marrow as well as lymphadenopathy and hepatosplenomegaly. Blood smears from transgenic mice in the myeloproliferative stage showed increased numbers of abnormal myelomonocytic cells (Figure 2B), and at their leukemic phase more than 20% blasts were shown in bone marrow (Figure 2C). Histologic examination of diseased animals revealed that the normal hematopoietic system in the bone marrow was completely replaced by blastic cells, and that these blastic cells had infiltrated to the lymph nodes, spleen, and liver (Figure 2D). Leukemia of each mice could be classified into myeloid leukemia with or without maturation, myelomonocytic leukemia, or megakaryocytic leukemia, according to the classification of the Bethesda proposals (Figure 2E-G).28
Development and characteristics of leukemias in the Tg mice. (A) Leukemia-free survival curve of the Tg mice. (B) Wright-Giemsa-stained smear of peripheral blood showing leukemia cells displaying abnormal myelomonocytic morphology with immature nuclear segmentation at the myeloproliferative stage (magnification, × 1000). Images were visualized at indicated original magnification using an Olympus BX40 microscope equipped with a 40 ×/0.75 or a 100 ×/1.30 objective lens (Olympus, Tokyo, Japan). Images were photographed with an Olympus C4040 digital camera and acquired with Adobe Photoshop 4.0 J software (Adobe Systems, San Jose, CA). (C) Bone marrow smear of myeloid leukemia with maturation showing many myeloblasts (Wright-Geimsa; magnification, × 1000). (D) Histologic analysis showing leukemic blasts infiltrating in the liver (hematoxylin and eosin [H&E]; magnification, × 200). (E) Myeloid leukemia with maturation (H&E; magnification, × 400). (F) Myeloid leukemia without maturation (H&E; magnification, × 400). (G) Megakaryocytic leukemia (H&E; magnification, × 400). (H) Flow cytometric analysis of leukemic Tg spleen cells compared with wild-type littermates. Cells in suspension were stained with Gr-1, Mac-1, B220, and 2C11 antibodies conjugated with fluorescein isothiocyanate and depicted by the solid lines.
Development and characteristics of leukemias in the Tg mice. (A) Leukemia-free survival curve of the Tg mice. (B) Wright-Giemsa-stained smear of peripheral blood showing leukemia cells displaying abnormal myelomonocytic morphology with immature nuclear segmentation at the myeloproliferative stage (magnification, × 1000). Images were visualized at indicated original magnification using an Olympus BX40 microscope equipped with a 40 ×/0.75 or a 100 ×/1.30 objective lens (Olympus, Tokyo, Japan). Images were photographed with an Olympus C4040 digital camera and acquired with Adobe Photoshop 4.0 J software (Adobe Systems, San Jose, CA). (C) Bone marrow smear of myeloid leukemia with maturation showing many myeloblasts (Wright-Geimsa; magnification, × 1000). (D) Histologic analysis showing leukemic blasts infiltrating in the liver (hematoxylin and eosin [H&E]; magnification, × 200). (E) Myeloid leukemia with maturation (H&E; magnification, × 400). (F) Myeloid leukemia without maturation (H&E; magnification, × 400). (G) Megakaryocytic leukemia (H&E; magnification, × 400). (H) Flow cytometric analysis of leukemic Tg spleen cells compared with wild-type littermates. Cells in suspension were stained with Gr-1, Mac-1, B220, and 2C11 antibodies conjugated with fluorescein isothiocyanate and depicted by the solid lines.
Characteristics of diseased NUP98-HOXA9 transgenic mice
Mouse ID . | Sex . | Time of leukemia onset, d . | Organs with leukemic cell invasion . | Diagnosis . | % blast in BM . |
|---|---|---|---|---|---|
| 1688 | F | 471 | BM, LN, Sp, Th, Lv, Lu, P, U | Myeloid leukemia with maturation | 37.0 |
| 2484 | F | 303 | BM, Sp, Lu | Myelomonocytic leukemia | 36.0 |
| 2539 | F | 309 | BM, LN, Sp, Th, Lv | Myeloid leukemia without maturation | 85.2 |
| 10001 | F | 394 | BM, LN, Sp, Th, Lv, Lu, Sa, A, Tr, K, H, GI | Myeloid leukemia with maturation | 29.2 |
| 10003 | M | 413 | BM, Sp, Lv, Lu, A, Te | Megakaryocytic leukemia | 49.8 |
| 10005 | M | 415 | BM, LN, Sp, Lv, K | Myeloid leukemia with maturation | 40.6 |
| 10009 | F | 196 | BM, LN, Sp, Lv, A, K, GI | Myeloid leukemia with maturation | 27.0 |
| 11010 | M | 221 | BM, LN, Sp, Lv, Lu, K | Myeloid leukemia without maturation | 88.1 |
| 11011 | M | 417 | BM, LN, Sp, Th, Lv, Lu, Sa, A, K, H, GI | Myeloid leukemia without maturation | 84.6 |
| 11043 | M | 401 | BM, LN, Sp, Lv, K | Myeloid leukemia with maturation | 69.0 |
| 11050 | F | 389 | BM, Sp, Lv, A | Myeloid leukemia with maturation | 25.8 |
| 11061 | F | 356 | BM, LN, Sp, Lv, Lu, Sa | Myeloid leukemia without maturation | 92.2 |
| 11062 | F | 344 | BM, LN, Sp, Th, Lv, Lu, Sa, U | Myeloid leukemia without maturation | 90.6 |
| 11075 | M | 383 | BM, LN, Sp, Lv, Sa, A, K, H, GI | Myeloid leukemia with maturation | 78.6 |
Mouse ID . | Sex . | Time of leukemia onset, d . | Organs with leukemic cell invasion . | Diagnosis . | % blast in BM . |
|---|---|---|---|---|---|
| 1688 | F | 471 | BM, LN, Sp, Th, Lv, Lu, P, U | Myeloid leukemia with maturation | 37.0 |
| 2484 | F | 303 | BM, Sp, Lu | Myelomonocytic leukemia | 36.0 |
| 2539 | F | 309 | BM, LN, Sp, Th, Lv | Myeloid leukemia without maturation | 85.2 |
| 10001 | F | 394 | BM, LN, Sp, Th, Lv, Lu, Sa, A, Tr, K, H, GI | Myeloid leukemia with maturation | 29.2 |
| 10003 | M | 413 | BM, Sp, Lv, Lu, A, Te | Megakaryocytic leukemia | 49.8 |
| 10005 | M | 415 | BM, LN, Sp, Lv, K | Myeloid leukemia with maturation | 40.6 |
| 10009 | F | 196 | BM, LN, Sp, Lv, A, K, GI | Myeloid leukemia with maturation | 27.0 |
| 11010 | M | 221 | BM, LN, Sp, Lv, Lu, K | Myeloid leukemia without maturation | 88.1 |
| 11011 | M | 417 | BM, LN, Sp, Th, Lv, Lu, Sa, A, K, H, GI | Myeloid leukemia without maturation | 84.6 |
| 11043 | M | 401 | BM, LN, Sp, Lv, K | Myeloid leukemia with maturation | 69.0 |
| 11050 | F | 389 | BM, Sp, Lv, A | Myeloid leukemia with maturation | 25.8 |
| 11061 | F | 356 | BM, LN, Sp, Lv, Lu, Sa | Myeloid leukemia without maturation | 92.2 |
| 11062 | F | 344 | BM, LN, Sp, Th, Lv, Lu, Sa, U | Myeloid leukemia without maturation | 90.6 |
| 11075 | M | 383 | BM, LN, Sp, Lv, Sa, A, K, H, GI | Myeloid leukemia with maturation | 78.6 |
BM indicates bone marrow; LN, lymph node; Sp, spleen; Th, thymus; Lv, liver; Lu, lung; P, pancreas; U, uterus; Sa, salivary gland; A, adrenal; Tr, thyroid; K, kidney; H, heart; GI, gastrointestinal tract; Te, testis.
FACS analysis of bone marrow and spleen cells was performed to examine cell-surface antigen expression. Samples from transgenic leukemic mice showed a markedly increased population of cells positive for Gr-1 and Mac-1 antigens (Figure 2H), which clearly indicated a leukemia of granulocytic origin. This myeloid lineage expansion was not observed in wild-type littermates and nonleukemic transgenic mice.
Alteration of the proliferative ability of progenitor cells expressing NUP98-HOXA9
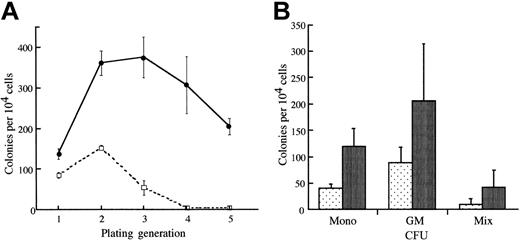
Given the relatively low incidence of leukemia in the transgenic mice, we examined whether the transgenic bone marrow cells exhibited any abnormal growth properties. To evaluate the self-renewal capacity of progenitor cells expressing NUP98-HOXA9, we performed replating assays using bone marrow cells derived from nonleukemic transgenic mice in methylcellulose cultures. Seven to 11 days after plating, entire methylcellulose cultures from NUP98-HOXA9 transgenic mice and littermate mice were harvested, and 1 × 104 cells were replated in methylcellulose under optimal conditions for the differentiation of multipotential progenitors (see “Materials and methods”). Primary cultures of transgenic bone marrow cells exhibited no difference in terms of number or types of colonies generated as compared with wild-type mice. However, significant differences between transgenic and wild-type littermates were observed on serial replating of the colonies. After 2 to 3 generations in methylcellulose, cultures from wild-type mice no longer formed colonies. In contrast, colonies derived from mice expressing NUP98-HOXA9 continued to grow and to generate colonies (Figure 3). These results indicated that BM cells from transgenic mice had a significantly higher self-renewal capacity than those from nontransgenic littermates.
Replating analysis of BM cells derived from Tg mice. (A) Bone marrow was harvested from femurs of Tg mice or wild-type littermate controls, and 1 × 104 cells were plated in culture dishes containing methylcellulose and appropriate medium. Bulk cultures were harvested after 7 to 11 days in culture, and 1 × 104 cells were replated for each sample. Each point represents the number of colonies generated per 1 × 104 cells seeded. Cells (1 × 104) were plated in duplicate dishes, and the mean numbers of colonies for a representative experiment are shown. These data are representative of 5 similar experiments. (B) Classification of colonies in methylcellulose. Colonies at the second replating were analyzed morphologically. Granulocyte-macrophage colony-forming unit (CFU-GM), CFU of monocyte (CFU-mono), and CFU of mixed lineages (CFU-Mix) are increased in the transgenic bone marrow.
Replating analysis of BM cells derived from Tg mice. (A) Bone marrow was harvested from femurs of Tg mice or wild-type littermate controls, and 1 × 104 cells were plated in culture dishes containing methylcellulose and appropriate medium. Bulk cultures were harvested after 7 to 11 days in culture, and 1 × 104 cells were replated for each sample. Each point represents the number of colonies generated per 1 × 104 cells seeded. Cells (1 × 104) were plated in duplicate dishes, and the mean numbers of colonies for a representative experiment are shown. These data are representative of 5 similar experiments. (B) Classification of colonies in methylcellulose. Colonies at the second replating were analyzed morphologically. Granulocyte-macrophage colony-forming unit (CFU-GM), CFU of monocyte (CFU-mono), and CFU of mixed lineages (CFU-Mix) are increased in the transgenic bone marrow.
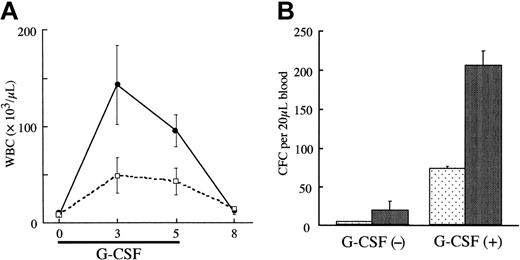
We also compared the responses of NUP98-HOXA9 transgenic mice and wild-type mice with G-CSF. As shown in Figure 4A-B, after 3 days of G-CSF injections wild-type mice demonstrated marked neutrophilic leukocytosis, with a 30-fold increase in peripheral blood WBC and PBPC numbers. WBCs and PBPCs derived from the transgenic mice showed even higher increases, 2.9-fold and 2.8-fold compared with wild-type, respectively, on day 3. These results clearly indicated that NUP98-HOXA9 expression induced G-CSF hypersensitivity.
G-CSF hypersensitivity of Tg bone marrow cells. (A) The numbers of white blood cells induced by G-CSF (200 μg/kg/d for 5 days) in Tg mice and their wild-type littermate controls. (B) The peripheral blood progenitor cell numbers in Tg mice and their wild-type littermate controls (n = 2 for each) after G-CSF injection (200 μg/kg/d for 3 days). These data are representative of 3 similar experiments.
G-CSF hypersensitivity of Tg bone marrow cells. (A) The numbers of white blood cells induced by G-CSF (200 μg/kg/d for 5 days) in Tg mice and their wild-type littermate controls. (B) The peripheral blood progenitor cell numbers in Tg mice and their wild-type littermate controls (n = 2 for each) after G-CSF injection (200 μg/kg/d for 3 days). These data are representative of 3 similar experiments.
Taken together, these results demonstrate that expression of NUP98-HOXA9 significantly enhances the proliferative potential of myeloid progenitors. As a result, NUP98-HOXA9-expressing bone marrow cells might obtain a sufficient window of time to obtain sequential genetic alterations, so that they could acquire even more growth advantages.
NUP98-HOXA9 accelerates BXH2 myeloid disease
To identify cooperative genes for NUP98-HOXA9 in leukemogenesis, we transferred the NUP98-HOXA9 transgene into the BXH2 genetic background for 3 generations by backcross mating. BXH2 mice offer a valuable model system for identifying cooperative genes, as virtually 100% of BXH2 mice develop acute myeloid leukemia by 1 year of age.25,29 The high incidence of leukemia in BXH2 mice is causally associated with the expression of a horizontally transmitted B-ecotropic murine leukemia virus. Ecotropic viruses induce disease by insertional activation of proto-oncogenes or insertional inactivation of tumor suppressor genes.30 Genes that cause leukemia in BXH2 mice have already been systematically identified using a proviral tagging technique.3,4
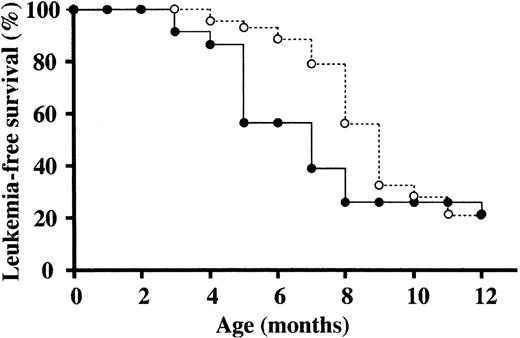
The incidence and latency of myeloid leukemias in NUP98-HOXA9/BXH2, wild-type BXH2 and C57BL/6J background NUP98-HOXA9 transgenic mice were compared. As shown in Figure 5, NUP98-HOXA9/BXH2 mice developed acute myeloid leukemia faster than BXH2 and transgenic mice at the time point between 4 and 8 months. This result suggested that NUP98-HOXA9 expression accelerated the progression of BXH2 myeloid leukemia, and that cooperative genes for NUP98-HOXA9 may be located near common retroviral integration sites.
NUP98-HOXA9 accelerates BXH2 myeloid leukemia. Leukemia-free survival rates of BXH2 mice (○) and BXH2/Tg mice (•) were plotted against the age (months after birth). The NUP98-HOXA9 tg mice were backcrossed to the BXH2 strain to introduce the transgene into BXH2 genetic background, and survival rates of Tg-positive BXH2 mice were compared with those of Tg-negative littermates at the third generation.
NUP98-HOXA9 accelerates BXH2 myeloid leukemia. Leukemia-free survival rates of BXH2 mice (○) and BXH2/Tg mice (•) were plotted against the age (months after birth). The NUP98-HOXA9 tg mice were backcrossed to the BXH2 strain to introduce the transgene into BXH2 genetic background, and survival rates of Tg-positive BXH2 mice were compared with those of Tg-negative littermates at the third generation.
Identification of NUP98-HOXA9 cooperative genes
To characterize the retroviral integration sites, to identify nearby cooperative genes, host-virus junction sequences were amplified by inverse PCR using NUP98-HOXA9/BXH2 leukemia DNAs and retrovirus-specific primers.3 From a panel of 19 tumors, we isolated and sequenced a total of 67 retroviral integration sites. The clonality of retroviral integrations at these loci was confirmed by the Southern blot analysis (Figure 6) to exclude the possibility that these sites are mere minor clones incidentally identified as PCR artifacts. Homology searches of integration site flanking DNA sequences using the BLAST alignment tool established by GenBank and the UCSC genome browser (October 2003; http://genome.ucsc.edu/cgi-bin/hgGateway) allowed us to determine the location of the individual integration sites. The data will be deposited on the Mouse Retroviral Tagged Cancer Gene Database (MRTCGD; http://genome2.ncifcrf.gov/RTCGD/). Of the 67 integration sites, 49 were unique, and a summary of the 49 candidate genes is given in Table 2. Integration sites in each tumor are shown in Supplemental Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article.
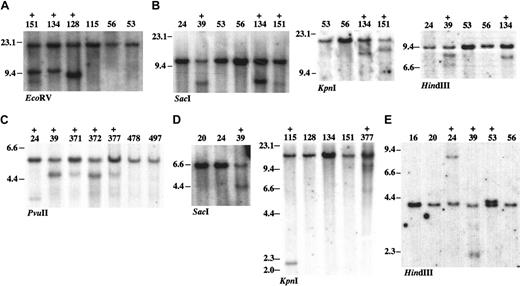
DNA rearrangement at common retroviral integration sites. Southern blot analysis was carried out using 5 μg genomic DNAs extracted from Tg/BXH2 leukemias. The leukemia samples with rearrangements are indicated as + above the sample ID. Restriction enzymes used in each blots are indicated. The size of DNA was indicated in kilobase. (A) Meis1 using the Meis1 3′ probe. Tumors with Meis1 integration at the 3′ region show DNA rearrangement. (B) Dnalc4. (C) Fcgr2b/Fcrl. (D) Con1. (E) Con2.
DNA rearrangement at common retroviral integration sites. Southern blot analysis was carried out using 5 μg genomic DNAs extracted from Tg/BXH2 leukemias. The leukemia samples with rearrangements are indicated as + above the sample ID. Restriction enzymes used in each blots are indicated. The size of DNA was indicated in kilobase. (A) Meis1 using the Meis1 3′ probe. Tumors with Meis1 integration at the 3′ region show DNA rearrangement. (B) Dnalc4. (C) Fcgr2b/Fcrl. (D) Con1. (E) Con2.
Retroviral integration sites in NUP98-HOXA9/BXH2 leukemia
Gene . | Protein family . | Chromosome location, chromosome No., cM . | Tumor ID . |
|---|---|---|---|
| Transcription | |||
| Meis1 | Homeodomain protein | 11, 18.9 | 14, 15, 16, 53, 128, 134, 151, 478, 497 |
| Sox4 | HMG box protein | 13, 28.4 | 151 |
| C/EBPα | bZIP protein | 7, 25.6 | 53 |
| C/EBPϵ | bZIP protein | 14, 45.8 | 15 |
| Ap-4 | bHLH-ZIP protein | 16, 4.0 | 24 |
| Myb | Myb transcription factor | 10, 21.0 | 134 |
| Tsga | Testis-specific zinc finger | 6, 72.5 | 371 |
| MII3 | Trithorax homologue | 5, 23.8 | 372 |
| Ncoa6ip | Co-repressor complex protein | 4, 3.0 | 115 |
| Signaling | |||
| Fcgr2b | IgG Fc receptor | 1, 172.2 | 24, 39, 151, 371, 372, 377 |
| Fcrl | IgG Fc receptor | 1, 172.2 | 24, 39, 151, 371, 372, 377 |
| Fcrn | IgG Fc receptor | 7, 34.6 | 478 |
| Snx6 | Ser-Thr kinase | 12, 49.1 | 377 |
| Pdgfrb | Receptor tyrosine kinase | 18, 61.3 | 134 |
| Gnas | G protein | 2, 175.5 | 39 |
| Gng3 | G protein | 19, 8.2 | 128 |
| Itpr1 | Inositol-triphosphate receptor | 6, 109.2 | 371 |
| Aps | Substrate for insulin receptor | 5, 134.8 | 115 |
| Agtrp | Angiotensin II receptor, type 1 | 4, 143.4 | 478 |
| Cav2 | Caveolin 2 | 6, 17.1 | 497 |
| Fdft1 | Farnesyl transferase | 14, 54.2 | 56 |
| Rin3 | Ras and Rab interacting protein | 12, 96.5 | 16 |
| Metabolism | |||
| XM138092 | Mitochondrial ATP synthase | 12, 78.9 | 128 |
| Moat-b | ABC transporter 4 | 14, 109.8 | 371 |
| Srd5a2I | Steroid reductase | 5, 75.6 | 39 |
| Kcnd3 | Potassium gate channel | 3, 106.1 | 372 |
| Glu decarboxylase | Glu decarboxylase | 10, 99.6 | 56 |
| Apoptosis | |||
| Bad | Pro-apoptotic molecule | 19, 3.1 | 377 |
| Cytoskeleton/membrane traffic | |||
| Dnalc4 | Dynein motor complex | 15, 80.5 | 39, 128, 134, 151 |
| Dctn3 | Activator of dynein | 4, 41.4 | 377 |
| Transitin | Cytoskeletal protein | 7, 53.5 | 377 |
| Transgelin2 | Muscle contractile protein | 1, 173.8 | 134 |
| Pex16 | Peroxysomal protein | 2, 93.3 | 371 |
| Tuba2 | α-tubulin 2 | 15, 99.8 | 16 |
| Prefoldin subunit 6 | Microtubule-associated chaperonin | 13, 51.3 | 39 |
| Tspan2 | Tetraspanin 2 | 3, 102.8 | 375 |
| Zinc transporter 5 | 15 transmembrane protein | 13, 98.2 | 128 |
| Miscellaneous | |||
| Tgfbi | TGF-β–induced gene | 13, 56.1 | 377 |
| MN1 | Fused to TEL in AML | 5, 109.1 | 372 |
| Osa1 | Chromatin remodeling complex | 4, 131.2 | 151 |
| Rrm1 | Ribonucleotide reductase | 7, 92.2 | 128 |
| Mms2 | Ubiquitin conjugating enzyme | 15, 31.6 | 56 |
| Cyclophilin 18 | Peptidyl-prolyl cis-trans isomerase | 3, 134.8 | 128 |
| Rpl44 | Ribosomal protein L44 | 12, 63.8 | 478 |
| Agrp | Agouti-related | 8, 105.4 | 478 |
| Ptma | Prothymoshin α | 1, 86.9 | 478 |
| Mrvil | Jaw1 homologue | 7, 100.5 | 371 |
| Unknown | |||
| BI526426 (Con1) | Not determined | 13, 48.9 | 39, 115, 377 |
| AI854024 (Con2) | Not determined | 3, 121.7 | 24, 39, 53 |
Gene . | Protein family . | Chromosome location, chromosome No., cM . | Tumor ID . |
|---|---|---|---|
| Transcription | |||
| Meis1 | Homeodomain protein | 11, 18.9 | 14, 15, 16, 53, 128, 134, 151, 478, 497 |
| Sox4 | HMG box protein | 13, 28.4 | 151 |
| C/EBPα | bZIP protein | 7, 25.6 | 53 |
| C/EBPϵ | bZIP protein | 14, 45.8 | 15 |
| Ap-4 | bHLH-ZIP protein | 16, 4.0 | 24 |
| Myb | Myb transcription factor | 10, 21.0 | 134 |
| Tsga | Testis-specific zinc finger | 6, 72.5 | 371 |
| MII3 | Trithorax homologue | 5, 23.8 | 372 |
| Ncoa6ip | Co-repressor complex protein | 4, 3.0 | 115 |
| Signaling | |||
| Fcgr2b | IgG Fc receptor | 1, 172.2 | 24, 39, 151, 371, 372, 377 |
| Fcrl | IgG Fc receptor | 1, 172.2 | 24, 39, 151, 371, 372, 377 |
| Fcrn | IgG Fc receptor | 7, 34.6 | 478 |
| Snx6 | Ser-Thr kinase | 12, 49.1 | 377 |
| Pdgfrb | Receptor tyrosine kinase | 18, 61.3 | 134 |
| Gnas | G protein | 2, 175.5 | 39 |
| Gng3 | G protein | 19, 8.2 | 128 |
| Itpr1 | Inositol-triphosphate receptor | 6, 109.2 | 371 |
| Aps | Substrate for insulin receptor | 5, 134.8 | 115 |
| Agtrp | Angiotensin II receptor, type 1 | 4, 143.4 | 478 |
| Cav2 | Caveolin 2 | 6, 17.1 | 497 |
| Fdft1 | Farnesyl transferase | 14, 54.2 | 56 |
| Rin3 | Ras and Rab interacting protein | 12, 96.5 | 16 |
| Metabolism | |||
| XM138092 | Mitochondrial ATP synthase | 12, 78.9 | 128 |
| Moat-b | ABC transporter 4 | 14, 109.8 | 371 |
| Srd5a2I | Steroid reductase | 5, 75.6 | 39 |
| Kcnd3 | Potassium gate channel | 3, 106.1 | 372 |
| Glu decarboxylase | Glu decarboxylase | 10, 99.6 | 56 |
| Apoptosis | |||
| Bad | Pro-apoptotic molecule | 19, 3.1 | 377 |
| Cytoskeleton/membrane traffic | |||
| Dnalc4 | Dynein motor complex | 15, 80.5 | 39, 128, 134, 151 |
| Dctn3 | Activator of dynein | 4, 41.4 | 377 |
| Transitin | Cytoskeletal protein | 7, 53.5 | 377 |
| Transgelin2 | Muscle contractile protein | 1, 173.8 | 134 |
| Pex16 | Peroxysomal protein | 2, 93.3 | 371 |
| Tuba2 | α-tubulin 2 | 15, 99.8 | 16 |
| Prefoldin subunit 6 | Microtubule-associated chaperonin | 13, 51.3 | 39 |
| Tspan2 | Tetraspanin 2 | 3, 102.8 | 375 |
| Zinc transporter 5 | 15 transmembrane protein | 13, 98.2 | 128 |
| Miscellaneous | |||
| Tgfbi | TGF-β–induced gene | 13, 56.1 | 377 |
| MN1 | Fused to TEL in AML | 5, 109.1 | 372 |
| Osa1 | Chromatin remodeling complex | 4, 131.2 | 151 |
| Rrm1 | Ribonucleotide reductase | 7, 92.2 | 128 |
| Mms2 | Ubiquitin conjugating enzyme | 15, 31.6 | 56 |
| Cyclophilin 18 | Peptidyl-prolyl cis-trans isomerase | 3, 134.8 | 128 |
| Rpl44 | Ribosomal protein L44 | 12, 63.8 | 478 |
| Agrp | Agouti-related | 8, 105.4 | 478 |
| Ptma | Prothymoshin α | 1, 86.9 | 478 |
| Mrvil | Jaw1 homologue | 7, 100.5 | 371 |
| Unknown | |||
| BI526426 (Con1) | Not determined | 13, 48.9 | 39, 115, 377 |
| AI854024 (Con2) | Not determined | 3, 121.7 | 24, 39, 53 |
Of these 49 integration site loci, 5 were common and resulted in the identification of 6 candidate genes (Table 3). The MRTCGD search revealed only the Meis1 gene as a common integration site in wild-type BXH2 leukemia. Meis1 is a known cofactor for the wild-type HOXA9 as well as chimeric NUP98-HOXA9,19,26 and the incidence of integration at the Meis1 gene in NUP98-HOXA9/BXH2 leukemia was much higher than that in wild-type BXH2 leukemia. This result confirmed that this experimental system was able to efficiently identify cooperative genes for NUP98-HOXA9, such as Meis1. Moreover, the result also indicated that acceleration of BXH2 leukemia is caused by NUP98-HOXA9 expression, not by other modifier genes in the transgenic line.
Common integration sites and cooperative genes in NUP98-HOXA9/BXH2 leukemia
Chromosome locus, No. cM . | Candidate gene . | Distance from integrations . | Incidence in Tg/BXH2, % (n = 19) . | Incidence in BXH2, %* . |
|---|---|---|---|---|
| 11, 18.9 | Meis1 | Intragenic 3′ or 1 kb upstream | 47.4 | 15 |
| 15, 80.5 | Dnalc4 | Downstream 6-10 kb | 21.1 | None |
| 13, 48.9 | Con1 (BI526426) | Intron 1 | 15.8 | None |
| 1, 172.2 | Fcgr2b, Fcrl | Downstream 10-15 kb (Fcgr2b), upstream 15-20 kb (Fcrl) | 31.6 | None |
| 3, 121.7 | Con2 (AI854024) | Intron 1 | 15.8 | None |
Chromosome locus, No. cM . | Candidate gene . | Distance from integrations . | Incidence in Tg/BXH2, % (n = 19) . | Incidence in BXH2, %* . |
|---|---|---|---|---|
| 11, 18.9 | Meis1 | Intragenic 3′ or 1 kb upstream | 47.4 | 15 |
| 15, 80.5 | Dnalc4 | Downstream 6-10 kb | 21.1 | None |
| 13, 48.9 | Con1 (BI526426) | Intron 1 | 15.8 | None |
| 1, 172.2 | Fcgr2b, Fcrl | Downstream 10-15 kb (Fcgr2b), upstream 15-20 kb (Fcrl) | 31.6 | None |
| 3, 121.7 | Con2 (AI854024) | Intron 1 | 15.8 | None |
The incidence of integration in wild-type BXH2 was described previously (Moskow et al31 and Mouse Retroviral Tagged Cancer Gene Database (MRTCGD; http://genome2.ncifcrf.gov/RTCGD/)
Of the 4 other common integration sites, Fcgr2b and Fcrl are located within the immunoglobulin G Fc receptor gene cluster on chromosome 1. Since all viral integrations involving this region were located between Fcgr2b and Fcrl, either or both of these genes may be involved.
The Dnalc4 gene encodes the dynein light chain 4 that is a component of the dynein motor complex. Dynein complexes are involved in the cytoplasmic transportation machinery and have been reported to interact with important tumor-related gene products, including p53 and β-catenin.32,33 More important, studies have shown that other dynein light chain proteins, such as Dlc2 and Dlc8, interact with BH3-only proteins, including Bim and Bmf, and may block apoptotic signals by suppressing Bim/Bmf functions.34,35
The Con1 gene (cooperative gene for NUP98-HOXA91) encodes a novel protein that contains an N-terminal short consensus repeat (SCR) domain consisting of 60 amino acid residues. The SCR domain (or Sushi domain) contains 4 cysteines that form 2 disulfide bonds and is thought to be involved in protein-protein interactions. The domain is important for the functional activities of receptor proteins such as soluble IL15R and complement receptors.36,37 However, it remains to be clarified what proteins interact with the Con1 SCR domain and how abnormal Con1 expression promotes NUP98-HOXA9-positive AML.
The Con2 gene has a C-terminal domain homologous to MurG, a key enzyme in peptidoglycan biosynthesis in bacteria.38 However, its role in mammalian cell systems is unclear.
Expression of candidate cooperative genes in NUP98-HOXA9/BXH2 leukemias
Expression of candidate cooperative genes in NUP98-HOXA9/BXH2 leukemic cells was analyzed by RT-PCR (Figure 7). All leukemic cells with retroviral integrations at each candidate gene expressed these genes, and leukemias with Dnalc4, Con1, and Con2 integrations showed enhanced expression compared with leukemias without integration. Fcgr2b was expressed in leukemic cells both with and without integrations, although the former showed slightly enhanced expression. Expression of these 4 genes was very weak in Sca-1-positive normal bone marrow cells. However, the expression level of Fcrl was not very different among the leukemia samples. Fcrl was also expressed in Sca-1-enriched bone marrow cells, suggesting that Fcrl may be expressed in early hematopoietic progenitors. Overall, these observations indicated that most of the candidate cooperative genes are up-regulated by retroviral integration.
Cooperative transforming activity for NIH 3T3 cells
We tested whether the coexpression of NUP98-HOXA9 and each candidate cooperative gene could transform NIH 3T3 cells in soft agar. NIH 3T3 cells were transduced with retroviral vectors bearing individual cooperative candidate genes and colony numbers compared with or without the addition of the NUP98-HOXA9 vector (Figure 8A). Consistent with a previous report,18 NIH 3T3 cells overexpressing NUP98-HOXA9 alone formed colonies in soft agar more efficiently than control cells (Figure 8C). When Meis1 was coexpressed with NUP98-HOXA9, transforming activity was significantly enhanced, consistent with the previous finding of cooperative activity in bone marrow cells.26 While NIH 3T3 cells overexpressing Dnalc4, Fcgr2b, Fcrl, and Con1 alone did not lead to enhanced colony formation, the coexpression of NUP98-HOXA9 led to an additional 2- to 3-fold increase in colony formation compared with NIH 3T3 cells overexpressing NUP98-HOXA9 only. However, coexpression of Con2 and NUP98-HOXA9 did not enhance the colony-forming ability of NIH 3T3 cells. Thus, Dnalc4, Fcgr2b, Fcrl, and Con1 cooperatively transformed NIH 3T3 cells with NUP98-HOXA9, such that, while these genes were not transforming genes in the NIH 3T3 assay system in their own right, the genes cooperated with NUP98-HOXA9 to promote oncogenesis.
Anchorage-independent growth of NIH 3T3 cells coexpressing NUP98-HOXA9 and cooperative genes. (A) Structure of retroviral constructs for NUP98-HOXA9 and cooperative genes. (B) Western blot analysis. Arrows indicate exogenously expressed proteins detected with the antibodies indicated at the bottom of each panel. (C) The numbers of colonies per 1 × 104 cells seeded were determined 3 weeks after plating in soft agar. Values are the means with standard deviations of 3 experiments. Infected retroviral vectors are indicated at the bottom. Open bars and hatched bars indicate presence of the empty MSCV vector (-) or NUP98-HOXA9 retrovirus (+), respectively.
Anchorage-independent growth of NIH 3T3 cells coexpressing NUP98-HOXA9 and cooperative genes. (A) Structure of retroviral constructs for NUP98-HOXA9 and cooperative genes. (B) Western blot analysis. Arrows indicate exogenously expressed proteins detected with the antibodies indicated at the bottom of each panel. (C) The numbers of colonies per 1 × 104 cells seeded were determined 3 weeks after plating in soft agar. Values are the means with standard deviations of 3 experiments. Infected retroviral vectors are indicated at the bottom. Open bars and hatched bars indicate presence of the empty MSCV vector (-) or NUP98-HOXA9 retrovirus (+), respectively.
Discussion
Chromosomal translocation with the formation of a chimeric oncogene is a critical and early genetic event in myeloid leukemogenesis.39,40 In this report, to verify a biologic role for NUP98-HOXA9 in leukemogenesis, we generated transgenic mice in which chimeric NUP98-HOXA9 fusion cDNA is specifically expressed in promyelocytes. Approximately 20% of the transgenic mice progresses into AML after a long latent period, although nonleukemic transgenic mice exhibited an increased G-CSF response and a high self-renewal capacity of myeloid progenitors compared with wild-type mice. In many studies, it has been demonstrated that many other fusion genes are not sufficient to induce leukemia.8 Our transgenic model fits well into this natural history of leukemia following translocation, and some cofactors seem to be needed for complete leukemogenesis. We identified novel genes that cooperate with NUP98-HOXA9 for leukemogenesis using retroviral insertional mutagenesis.
It is interesting to speculate as to how these newly cooperative genes promote leukemogenesis. In BXH2 mouse leukemias, 90% of cases with viral integration at Hoxa7 or Hoxa9 showed cooperative activation of Meis1.20 In other studies, both wild-type Hoxa9 and chimeric NUP98-HOXA9 were able to cooperate with Meis1 in myeloid leukemogenesis.19,26 However, in our study, only half of the leukemias showed cooperation between NUP98-HOXA9 and Meis1. This suggested that some of the cooperative genes identified in this study may have replaced the leukemogenic function of Meis1. An example of this might be Con1, as Con1 integration sites never overlapped with Meis1 integration sites in a single tumor. In contrast, leukemias exhibiting Meis1 integration also showed integration at the Dnalc4 locus (Table 2), which suggested that the 2 genes functioned in different leukemogenic pathways. Like other dynein light chain proteins, it is very likely that Dnalc4 binds to proapoptotic proteins such as Bim and Bmf. Our preliminary experiments showed interaction in vivo between Dnalc4 and the extra-long isoform of Bim protein (Daisuke Takahashi and T.N., unpublished data, September 2004). In this case, overexpression of Dnalc4 may block Bim function to give an antiapoptotic potential to preleukemic cells.41 This hypothesis is supported at least in part by the findings in a previous study that Meis1 did not enhance the antiapoptotic activity of myeloid cells. Moreover, we also identified Dctn3, encoding dynactin 3 as a single integration site in this study. Since dynactin is an upstream regulator of dynein,42,43 our findings strongly suggested the importance of the Dynein/Dynactin pathway in NUP98-HOXA9 leukemogenesis.
Interestingly, Fcgr2b is a target for the 1q21 translocation in human follicular lymphoma.44,45 While the functional significance of Fcgr2b up-regulation remains to be clarified, overexpression of Fcgr2b has been reported to enhance nonlymphoid cancer cell growth both in vitro and in vivo.46,47 As a possibility, it has been demonstrated that Fcgr2b protein seems to be a potent regulator of antibody-dependent cell-mediated cytotoxicity in vivo.48 Thus, dysregulation of Fcgr2b may play an important role in tumor progression. Fcrl is preferentially expressed in germinal center centroblasts and in a subset of diffuse large B-cell lymphoma cells, and the gene has also been reported to serve as a unique marker for the characterization for B-cell malignancy.49,50 We also identified another Fc receptor gene, Fcrn, on chromosome 7 as an integration site.51 Although Fcrn was not a common integration site, our results suggested that signaling pathways involving Fc receptors may be critical for NUP98-HOXA9-induced myeloid leukemogenesis.
Studies have demonstrated that BCR-ABL cooperates with NUP98-HOXA9 to cause chronic myeloid leukemia (CML) blast crisis in a mouse model.52,53 However, cooperation between 2 different chimeras is practically very rare in human leukemia. In addition, since there is a circumstance where NUP98-HOXA9 has already been expressed in chronic-phase patients, it is not applied to all cases. Therefore, we have attempted to identify some major cooperative genes, such as FLT3 in human AML,54 for NUP98-HOXA9 in leukemogenesis.
We tested cooperative transforming activity between NUP98-HOXA9 and the genes identified in retroviral insertional mutagenesis using NIH 3T3 cells. It has been exhibited that NUP98-HOXA9 alone could transform NIH 3T3 cells,18 suggesting that NUP98-HOXA9 might have common activity to transform both hematopoietic cells and nonhematopoietic fibroblasts, and that this is a reliable if not perfect assay system to evaluate oncogene cooperation. Therefore, Dnalc4, Fcgr2b, Fcrl, and Con1 are the real cooperative genes cooperating with NUP98-HOXA9 to promote oncogenesis. It might be useful to introduce those genes into bone marrow cells and then repopulate the cells into irradiated hosts to check cooperative transforming activity in vivo. In this case Con2 that did not cooperatively transform NIH 3T3 with NUP98-HOXA9 may be found as a cooperative gene in this system.
In conclusion, our strategy of combining retroviral insertional mutagenesis with a mouse tumor model allowed the efficient identification of cooperative genes with the NUP98-HOXA9 chimera in leukemogenesis. Our approach can be used to isolate second, third, and subsequent genetic events for known oncogenes and will assist in the clarification of the molecular pathways of carcinogenesis.
Prepublished online as Blood First Edition Paper, September 28, 2004; DOI 10.1182/blood-2004-04-1508.
Supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology, Japan; and by the National Cancer Institute (NCI), Department of Health and Human Services.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Miki Takuwa, Deborah Swing, and Bryn Eagleson for technical assistance; Jay Grisolano and Timothy Ley for the cathepsin G promoter; Toshio Kitamura for plat E cells; and Michael Cleary for anti-Meis1.


![Figure 2. Development and characteristics of leukemias in the Tg mice. (A) Leukemia-free survival curve of the Tg mice. (B) Wright-Giemsa-stained smear of peripheral blood showing leukemia cells displaying abnormal myelomonocytic morphology with immature nuclear segmentation at the myeloproliferative stage (magnification, × 1000). Images were visualized at indicated original magnification using an Olympus BX40 microscope equipped with a 40 ×/0.75 or a 100 ×/1.30 objective lens (Olympus, Tokyo, Japan). Images were photographed with an Olympus C4040 digital camera and acquired with Adobe Photoshop 4.0 J software (Adobe Systems, San Jose, CA). (C) Bone marrow smear of myeloid leukemia with maturation showing many myeloblasts (Wright-Geimsa; magnification, × 1000). (D) Histologic analysis showing leukemic blasts infiltrating in the liver (hematoxylin and eosin [H&E]; magnification, × 200). (E) Myeloid leukemia with maturation (H&E; magnification, × 400). (F) Myeloid leukemia without maturation (H&E; magnification, × 400). (G) Megakaryocytic leukemia (H&E; magnification, × 400). (H) Flow cytometric analysis of leukemic Tg spleen cells compared with wild-type littermates. Cells in suspension were stained with Gr-1, Mac-1, B220, and 2C11 antibodies conjugated with fluorescein isothiocyanate and depicted by the solid lines.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/2/10.1182_blood-2004-04-1508/6/m_zh80020572400002.jpeg?Expires=1769083051&Signature=Zy9AN9BFmHcaGEReorCdKuMGZq10ZmmSt6hdt6~QP2Hf83aAX9zgzrTdheNNhMHqjfAMj-Oymso7s4N1Rctaj4mK6zfUKXCE0S1rpay4JxufZkJ9bqAAwoKqKesb4DQvliiHn7dHSf7j3zRzJEN3hi0QKYtelM2etiRcTY--nXvygrLrzdrBe5B3Ci~7IawMLEJ3iQvWIgo5LaZWDNd~4noiB3uHzDT5qs9pnNO396Q9C-BM6xVuHCpDnZIoHz2orExqcTnT57DPOnEGrxftENNvRsHL9k1qXs1M8oWnDQfYxze-pto8VBY11Kd3DvBISlb7Fc0V66DAlucO~1g70w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal