Abstract
Patients with inherited bleeding disorders who received clotting factor concentrates before 1987 have high rates of hepatitis C virus (HCV) or HCV/HIV infection. We evaluated HCV quasispecies evolution in longitudinally collected specimens comparing those from patients with progression to end-stage liver disease (ESLD; cases) to those with compensated chronic hepatitis (controls). Plasma samples were obtained from the National Cancer Institute Multicenter Hemophilia Cohort Study. Controls were matched for age, sex, infection duration, and presence/absence of HIV. Samples from early infection were compared to those obtained after onset of ESLD in the cases. The first hypervariable (HVR1) and core proteincoding regions were amplified, subcloned, and sequenced. Complexity and diversity were determined. More than 700 sub-clones from 10 pairs of patients (8 with HIV) followed over approximately 9.3 years were evaluated. HVR1 complexity narrowed over time in the cases, whereas it increased in controls (P = .01). Similar trends were observed for diversity within HVR1 and the core region (P = .04). HCV-infected patients with inherited bleeding disorders undergo quasispecies evolution over time. Evolution patterns differ for progressors and nonprogressors. Further understanding of these mechanisms may help identify factors related to progression rate and treatment response.
Introduction
Hepatitis C virus (HCV) infection is associated with progression to end-stage liver disease (ESLD) in a significant subset of chronically infected patients. Hemophiliacs and other patients with inherited bleeding disorders who received clotting factor concentrates prior to 1987 are at particular risk for acquiring HCV infection from recurrent exposure to concentrates made from plasma pools prepared from 20 000 or more blood donors at a time when single-unit risk of infection approached 5%.1 Through the 1980s the HCV infection risk was compounded by transmission of HIV in blood products. Increased rates of progression to ESLD in those with chronic HCV infection have been well documented in those with hemophilia and in other populations with HIV coinfection.2-4
HCV replicates at very high levels with production of virions estimated at up to 1012 copies/d.5 This high rate of replication, combined with lack of an error correction mechanism and ongoing but variable immune selection, results in development of both synonymous and nonsynonymous mutations. Immune selection puts pressure on key antigen-recognition sites and drives the emergence of a closely related virus family that can be isolated from the serum of infected patients. Previous evaluation of viral population dynamics has provided critical insights into short-term outcomes including early spontaneous viral clearance,6 interferon-associated viral clearance,7,8 and HCV emergence following liver transplantation.9,10 However, lack of suitable long-term longitudinal cohorts has limited study of the relationship between HCV quasispecies evolution and development of ESLD. Furthermore, most studies assessing quasispecies heterogeneity have focused on the first hypervariable region (HVR1) within the E2/NS1 portion of the HCV genome. However, the core region has been associated with multiple functions, including regulation of tumor suppressor genes,11-13 modification of cell susceptibility to apoptosis,14,15 and inhibition of HIV-1 and hepatitis B virus (HBV) replication.16,17 Therefore, studies of the core region may provide important insights into the relationship between quasispecies and disease progression.
In this report, we evaluated the emergence of HCV quasispecies in a longitudinal cohort of patients with hemophilia with either HCV or HCV/HIV coinfection among subjects with clear clinical progression to ESLD compared to emergence in matched controls without evidence of clinical progression.
Patients, materials, and methods
Study population
Plasma samples from patients with known HCV and HIV status were collected from the National Cancer Institute (NCI) Multicenter Hemophilia Cohort Study (MHCS) and stored at –80°C. This study cohort was initiated for the study of HIV infections in 1982 and included periodic clinical evaluation and testing for HIV antibodies beginning in 1984 and HCV antibodies beginning in 1990.4 All individuals provided informed consent at the time of enrollment, and the protocol was approved by the Institutional Review Board at each of the participating clinical sites. Laboratory evaluation included complete blood count, serum transaminases (aspartate aminotransferase [AST] and alanine aminotransferase [ALT]), hepatitis B antigen and antibody status, and CD4+ counts. These were collected approximately annually over a minimum of 5 years for 2 comparison groups of patients comprising cases and controls. Cases consisted of patients with either HCV/HIV coinfection or HCV monoinfection with liver failure, defined as persistent ascites, variceal bleeding, hepatic encephalopathy, or death attributed to liver disease.3,4 The control group included coinfected or monoinfected patients without liver failure and with stable liver function tests. Cases and controls were matched by age, sex, HIV status, and estimated duration of HCV infection at first and last samples tested herein. Dates of initial HCV infection were estimated using history of use of clotting factor concentrate (first use equals infection) and plasma or cryoprecipitate (midpoint between first plasma/cryoprecipitate and first concentrate use or 1990). There were 10 matched pairs overall: 8 pairs were HCV/HIV coinfected and 2 pairs were HCV monoinfected. The preponderance of coinfected pairs is representative of the proportional difference in coinfected versus monoinfected patients with liver failure within the cohort.
HCV RNA isolation, HVR1, and core region amplification
HCV RNA was extracted from 140 μL patient plasma using a QIAamp Viral RNA Mini kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. Amplification of the E2/NS1 was carried out using a nested reverse transcription-polymerase chain reaction (RT-PCR) method described by Sherman et al.18 The first set of primers used were: external forward (EF), position 1048 to 1067, 5′-GGTGCTCACTGGGGAGTCCT-3′; external reverse (ER), position 1291c to 1269c, 5′-CATTGCAGTTCAGGGCCGTGCTA-3′; internal forward (IF), position 1087 to 1106, 5′-TCCATGGTGGGGAACTGGGC-3′; and internal reverse (IR), position 1262c to 1243c, 5′-TGCCAACTGCCGTTGGTGTT-3′, with the primer positions corresponding to the HCV sequence described by Choo et al.19,20 These primers flank a 136-bp region of the E1/E2 envelope–coding domain that includes the HVR1. The second set of primers, used to amplify a 1270-bp product that includes the HCV core protein, were: EF, position –62 to –42, 5′-CTTGTGGTACTGCCTGATAGG-3′; ER position 1291c to 1269c, 5′-CATTGCAGTTCAGGGCCGTGCTA-3′; IF, position –51 to –29, 5′-GCCTGATAGGGTGCTTGCGAGTG-3′, and IR, position 1262c to 1243c, 5′-TGCCAACTGCCGTTGGTGTT-3′.
Cloning and sequencing of cDNA
The HCV cDNA products were ligated into pGEM-T Easy Vector and used to transform JM109 high-efficiency competent cells according to the manufacturer's protocol (Promega, Madison, WI). Each transformation culture was plated out on Luria-Bertani (LB)/ampicillin/isopropylthiogalactoside (IPTG)/X-gal plates and grown overnight at 37°C. White colonies containing the inserts were selected and grown overnight in 5 mL LB broth containing ampicillin in a shaking incubator at 37°C. The plasmid DNA was then purified using the QIAprep Spin Miniprep kit (QIAGEN) and the presence of the inserts confirmed by digestion with EcoR1. Sequencing was performed on approximately 10 clones per patient at the early (first available) and late (first available after progression to ESLD) time points and their matched control samples. Due to limited sample availability, it was not possible to clone and sequence samples of matched pairs at interim time points.
DNA sequence manipulation and phylogenetic analysis
Sequences were aligned manually and gaps created where any additions or deletions occurred. Phylogenetic and molecular evolutionary analyses were conducted using Molecular Evolutionary Genetic Analysis software (MEGA version 2.1).21 The MEGA algorithms were used to calculate the mean Kimura 2-parameter pairwise distance for all clones as well as a matrix of Kimura 2-parameter pairwise distances for each patient. The numbers of synonymous mutations (dS) and the numbers of nonsynonymous mutations (dN) were determined using the Nei-Gojobori method with the Jukes-Cantor correction for multiple substitutions.22 The mean dN/dS ratio, a measure of selective pressure, was also calculated for each patient. Phylogenetic trees of each patient, illustrating genetic distance between clonal sets, were generated using the Neighbor Joining method of Saitou and Nei.23 Samples from early infection in both cases and controls were compared to those obtained at follow-up from cases and controls, after onset of ESLD in the cases.
Statistical analysis
Demographic comparisons were performed using the χ2 test and the paired t test. Nonlinear and linear regression models, the paired t test, and a 2-way mixed model ANOVA were used to assess differences in complexity and distance matrices between case/control pairs over time. These analyses were conducted for the 136-bp product amplified from the IF and IR primers and the truncated 81-bp sequence corresponding to the HVR1 region, and for the core region from the core IF through 573 bp. The difference in amino acid diversity between pairs was evaluated using a paired t test by amino acid position. Specific amino acid differences and hydropathic character differences were evaluated using the χ2 test. Two-tailed P < .05 was considered statistically significant. Statistical analyses were conducted using either Stata version 7.0 (Stata, College Station, TX) or SAS version 8 (SAS Institute, Cary, NC).
Nucleotide sequence accession numbers
Results
Patient demographics
Table 1 shows the demographic characteristics of the cases and controls. All patients were men, and all were HCV genotype 1. Nineteen of 20 patients (95%) had prior, resolved coinfection with HBV. One patient was negative for HBV marker. Baseline CD4+ and CD8+ T-cell counts, AST and ALT levels, and HCV/HIV RNA titers were not available for all patients. Baseline values were used when available; otherwise, the next measurement after baseline was used.
Demographic characteristics of progressors to ESLD (cases) and nonprogressors (controls)
. | Progressors (n = 10) . | Nonprogressors (n = 10) . | P . |
|---|---|---|---|
| Race, no. (%) | |||
| Caucasian | 10 (100) | 6 (60) | NS |
| Other | — | 4 (40) | |
| HIV+, no. (%) | 8 (80) | 8 (80) | NS |
| HCV viral load, bDNA, mean copies/mL (range) | 2.27e + 07 (5.19e + 05 to 6.88e + 07) | 2.38e + 07 (5.55e + 05 to 8.96e + 07) | NS |
| HCV subtypes* | |||
| 1 | 1 | 1 | NS |
| 1a | 4 | 7 | |
| 1b | 5 | 2 | |
| Baseline CD4+, mean cells/mL (range) | 525 (249-1388) | 417 (212-680) | NS |
| Baseline CD8+, mean cells/mL (range) | 698 (277-1379) | 922 (133-2592) | NS |
| Time from baseline to follow-up, mean y, (range) | 7.82 (4.7-13.9) | 10.71 (6.1-14.3) | .01 |
| Baseline AST, mean U/L (range) | 70 (38-128) | 47.6 (29-64) | NS |
| Baseline ALT, mean U/L (range) | 90 (48-130) | 87 (34-197) | NS |
. | Progressors (n = 10) . | Nonprogressors (n = 10) . | P . |
|---|---|---|---|
| Race, no. (%) | |||
| Caucasian | 10 (100) | 6 (60) | NS |
| Other | — | 4 (40) | |
| HIV+, no. (%) | 8 (80) | 8 (80) | NS |
| HCV viral load, bDNA, mean copies/mL (range) | 2.27e + 07 (5.19e + 05 to 6.88e + 07) | 2.38e + 07 (5.55e + 05 to 8.96e + 07) | NS |
| HCV subtypes* | |||
| 1 | 1 | 1 | NS |
| 1a | 4 | 7 | |
| 1b | 5 | 2 | |
| Baseline CD4+, mean cells/mL (range) | 525 (249-1388) | 417 (212-680) | NS |
| Baseline CD8+, mean cells/mL (range) | 698 (277-1379) | 922 (133-2592) | NS |
| Time from baseline to follow-up, mean y, (range) | 7.82 (4.7-13.9) | 10.71 (6.1-14.3) | .01 |
| Baseline AST, mean U/L (range) | 70 (38-128) | 47.6 (29-64) | NS |
| Baseline ALT, mean U/L (range) | 90 (48-130) | 87 (34-197) | NS |
NS indicates not significant.
Three patients exhibited genotype shifts during the study period: one from 1a to 1b, one from 1b to 1a, and one from 1b to 1.24
Analysis of the HVR1
HCV quasispecies complexity. A total of 342 clones from 20 patients were sequenced and analyzed. The mean time between early and late samples was 9.3 years (SE, 0.69 years). Mean numbers of clones per patient were 9.9 (SE, 0.76) for the early time point and 7.4 (SE, 0.34) for the follow-up time point. Complexity was assessed by the proportion of unique nucleotide sequences to the number of clones assessed at each time point for each patient (Table 2). Regardless of HIV status, the mean early time point frequency of unique nucleotide sequences within HVR1 for progressors was 0.73 (SE, 0.04) sequence/clone for progressors compared to 0.59 (SE, 0.08) for nonprogressors (P = .23). At the late time point, the mean frequencies were 0.61 (SE, 0.04) and 0.77 (SE, 0.07) for progressors versus nonprogressors, respectively (P = .22). The differences between early and late frequencies indicate a narrowing of the quasispecies population complexity for progressors (–0.12 sequence/clone) and an increase in complexity (0.18 sequence/clone) for nonprogressors. The changes in complexity over time were statistically significantly different for progressors compared with nonprogressors (P = .01).
Mean frequencies of unique HVR1 nucleotide sequences for each patient subpopulation
. | Mean complexity . | . | . | ||
|---|---|---|---|---|---|
| Time point . | Progressors (n = 10) . | Nonprogressors (n = 10) . | Difference: progressor-nonprogressor . | ||
| Coinfected (n = 16) | |||||
| Early | 0.78 (SE, 0.02) | 0.54 (SE, 0.09) | 0.24 | ||
| Late | 0.65 (SE, 0.07) | 0.72 (SE, 0.07) | -0.07 | ||
| Monoinfected (n = 4) | |||||
| Early | 0.53 (SE, 0.03) | 0.81 (SE, 0.18) | -0.28 | ||
| Late | 0.48 (SE, 0.23) | 1.0 (SE, 0.0) | -0.52 | ||
| Total | |||||
| Early | 0.73 (SE, 0.04) | 0.59 (SE, 0.08) | 0.14 | ||
| Late | 0.61 (SE, 0.04) | 0.77 (SE, 0.07) | -0.16 | ||
| Late-early | -0.12 (SE, 0.07) | 0.18 (SE, 0.10) | -0.30* | ||
. | Mean complexity . | . | . | ||
|---|---|---|---|---|---|
| Time point . | Progressors (n = 10) . | Nonprogressors (n = 10) . | Difference: progressor-nonprogressor . | ||
| Coinfected (n = 16) | |||||
| Early | 0.78 (SE, 0.02) | 0.54 (SE, 0.09) | 0.24 | ||
| Late | 0.65 (SE, 0.07) | 0.72 (SE, 0.07) | -0.07 | ||
| Monoinfected (n = 4) | |||||
| Early | 0.53 (SE, 0.03) | 0.81 (SE, 0.18) | -0.28 | ||
| Late | 0.48 (SE, 0.23) | 1.0 (SE, 0.0) | -0.52 | ||
| Total | |||||
| Early | 0.73 (SE, 0.04) | 0.59 (SE, 0.08) | 0.14 | ||
| Late | 0.61 (SE, 0.04) | 0.77 (SE, 0.07) | -0.16 | ||
| Late-early | -0.12 (SE, 0.07) | 0.18 (SE, 0.10) | -0.30* | ||
Frequencies were determined by the proportion of unique nucleotide sequences to the number of clones assessed for each patient at early and late time points.
P = .01 for difference in unique nucleotide sequence frequencies between progressors and nonprogressors over time (generalized estimating equation model with binomial distribution and logit link).
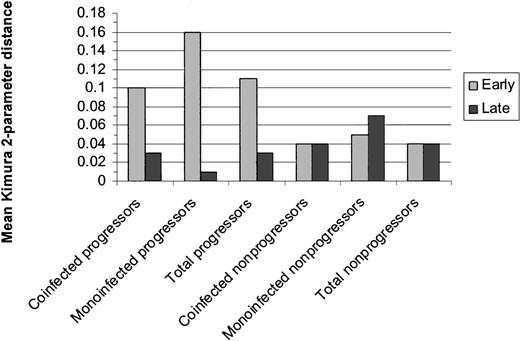
HCV quasispecies diversity. Diversity was assessed by comparing the mean Kimura 2-parameter distances between early and late sets of clones for matched cases/controls. Early and late distances are displayed in Figure 1. Regardless of HIV status, progressors as a group tended to have greater genetic diversity at the early time point than nonprogressors (P = .04) and demonstrated significant narrowing of genetic diversity over time (P = .04). Nonprogressors as a group maintained the same level of diversity over the study duration. MEGA was used to compute net change in intrasubject distances between early and late groups of clones by the formula dA = dXY – (dX – dY)/2 where dXY is the average distance between groups X (early clones) and Y (late clones), and dX and dY are the mean within-group distances. The mean intrasubject change in distance for progressors was 0.32 (SE, 0.11) versus 0.11 (SE, 0.03) for controls (P = .09). Phylogenetic trees illustrating the distances between clones for each patient are shown in Figure 2.
Mean genetic distances within the HVR1 between early and late clonal sets for progressors and nonprogressors. Distance was calculated via the mean Kimura 2-parameter pairwise distances for each patient. Regardless of HIV status, progressors as a group tended to have greater genetic diversity at the early time point than nonprogressors (P = .04) and demonstrated significant narrowing of genetic diversity over time (P = .04). Nonprogressors as a group maintained the same level of diversity over the study duration.
Mean genetic distances within the HVR1 between early and late clonal sets for progressors and nonprogressors. Distance was calculated via the mean Kimura 2-parameter pairwise distances for each patient. Regardless of HIV status, progressors as a group tended to have greater genetic diversity at the early time point than nonprogressors (P = .04) and demonstrated significant narrowing of genetic diversity over time (P = .04). Nonprogressors as a group maintained the same level of diversity over the study duration.
Phylogenetic trees from early/late E2/NS1 HVR1 clonal sets from ESLD progressors (cases) and nonprogressors (controls), constructed in MEGA2.1 using the neighbor-joining method of Saitou and Nei.23 Sequences from early time point clones are labeled with an “E” at the end of the sequence name. Late time point sequences are labeled with an “L.” One matched pair is not represented here due to a subtype shift in the progressor that precluded tree generation in MEGA. Progressors trended toward greater genetic diversity at the early time point than nonprogressors (P = .04) and demonstrated significant narrowing of genetic diversity over time, regardless of infection status (P = .04). Nonprogressors maintained the same level of diversity over the study duration.
Phylogenetic trees from early/late E2/NS1 HVR1 clonal sets from ESLD progressors (cases) and nonprogressors (controls), constructed in MEGA2.1 using the neighbor-joining method of Saitou and Nei.23 Sequences from early time point clones are labeled with an “E” at the end of the sequence name. Late time point sequences are labeled with an “L.” One matched pair is not represented here due to a subtype shift in the progressor that precluded tree generation in MEGA. Progressors trended toward greater genetic diversity at the early time point than nonprogressors (P = .04) and demonstrated significant narrowing of genetic diversity over time, regardless of infection status (P = .04). Nonprogressors maintained the same level of diversity over the study duration.
Mutational selection
The nonsynonymous-to-synonymous (dN/dS) mutation ratio, a marker of selective pressure, was compared between early and late time points for both cases and controls. One patient (pair 2 case), who was infected with 2 HCV subtypes, displayed a dramatically large dN/dS ratio at the later time point (39.02) due to a shift in the dominant subtype from 1a to 1b. With this outlying matched pair removed from the analysis, progressors demonstrated an overall statistically nonsignificant decrease in dN/dS ratio (from 2.02 to 0.62; P = .13) over time, whereas nonprogressors remained stable (Table 3).
Mean dN/dS ratio comparisons within the HVR1 for progressors (cases) and nonprogressors (controls)
. | dN/dS ratio . | . | . | ||
|---|---|---|---|---|---|
| Time point . | Progressors . | Nonprogressors . | P for difference between progressors and nonprogressors . | ||
| Early dN/dS ratio | 2.02 (SE, 0.70) | 0.98 (SE, 0.22) | .25 | ||
| Late dN/dS ratio | 0.62 (SE, 0.22) | 0.98 (SE, 0.39) | .52 | ||
| Mean difference (early to late) in dN/dS ratio | -1.40 (SE, 0.83) | -0.006 (SE, 0.55) | .31 | ||
| P for difference over time | .13 | .99 | |||
. | dN/dS ratio . | . | . | ||
|---|---|---|---|---|---|
| Time point . | Progressors . | Nonprogressors . | P for difference between progressors and nonprogressors . | ||
| Early dN/dS ratio | 2.02 (SE, 0.70) | 0.98 (SE, 0.22) | .25 | ||
| Late dN/dS ratio | 0.62 (SE, 0.22) | 0.98 (SE, 0.39) | .52 | ||
| Mean difference (early to late) in dN/dS ratio | -1.40 (SE, 0.83) | -0.006 (SE, 0.55) | .31 | ||
| P for difference over time | .13 | .99 | |||
The numbers of synonymous mutations (dS) and the numbers of nonsynonymous mutations (dN) were determined using the Nei-Gojobori method with the Jukes-Cantor correction for multiple substitutions.22 The mean dN/dS ratio, a measure of selective pressure, was calculated for each patient. One matched pair is excluded due to a shift in dominant subtype in the progressor. Progressors demonstrated an overall decrease in dN/dS ratio over time, whereas nonprogressors remain stable.
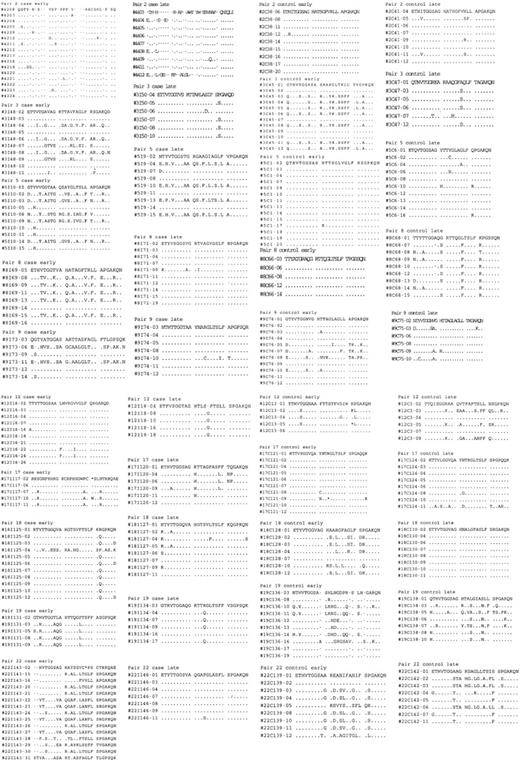
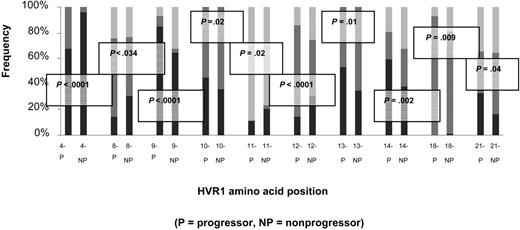
Amino acid sequences were predicted for the 81-bp HVR1 (amino acids 384-410) and aligned to determine the number of unique amino acids observed at each position. Sequences for each matched pair are shown in Figure 3. From these data, a profile of amino acid variation in HVR1 for cases and controls was created (Figure 4). The mean number of amino acid substitutions per site, including deletions, was 5.3 (SE, 0.54) for the progressors and 5 (SE, 0.61) for nonprogressors (P = .44). However, site-by-site analysis revealed statistically significant differences in the frequency of the most-conserved amino acid residues at various positions. A Thr residue is found at position 2 of the amplified sequence in all of the controls (151 of 151, 100%), but one progressor had a deletion at that location in 15 of 16 early time point clones so that the frequency in the case group was 136 of 150 (91%). At position 4, Val is predominant in both cases and controls, but in 94 of 150 cases (63%) versus 116 of 151 controls (77%; P = .03). Instead of Val, Thr occupies this position in 49 of 150 cases (33%) versus 6 of 151 controls (4%). An Ala residue is present at position 9 in 84 of 151 control clones (56%), but only in 59 of 150 cases (39%; P = .02). Sixty-seven of 150 cases (45%) have a Val residue occupying that position versus 13 of 151 controls (9%). Similar differences in frequencies of Thr and Asn are found at positions 13 and 27, respectively. Other positions are very highly conserved in both groups, including Gly at positions 6, 7, and 23, and Gln at position 2.
Predicted 81-bp HVR1 amino acid sequences for early and late clonal sets from progressors to ESLD (cases) and nonprogressors (controls). Sequence numbers are identified along the left column. Dots indicate no change from initial sequence (top line).
Predicted 81-bp HVR1 amino acid sequences for early and late clonal sets from progressors to ESLD (cases) and nonprogressors (controls). Sequence numbers are identified along the left column. Dots indicate no change from initial sequence (top line).
Number of amino acids, including deletions, represented at each site within the HVR1. Progressors (cases) are represented in gray; nonprogressors (controls), in black. The mean number of amino acid substitutions per site, including deletions, was 5.3 (SE- 0.54) for the progressors and 5 (SE- 0.61) for nonprogressors (P = .44). Certain sites (2, 6, 7, 23, and 26) were highly conserved for both cases and controls, but other sites (1, 9, 13, 27) had different numbers of substitutions between groups. Sites 2, 4, 9, 13, and 27 had significantly less of the predominant amino acid residue in the case group than in the control group (P =.002, .03, .02, .01, and .02, respectively). For site 2, this was due to one subject's early set of clones with a deletion. For the other sites, this was due to substitutions among several sets of clones.
Number of amino acids, including deletions, represented at each site within the HVR1. Progressors (cases) are represented in gray; nonprogressors (controls), in black. The mean number of amino acid substitutions per site, including deletions, was 5.3 (SE- 0.54) for the progressors and 5 (SE- 0.61) for nonprogressors (P = .44). Certain sites (2, 6, 7, 23, and 26) were highly conserved for both cases and controls, but other sites (1, 9, 13, 27) had different numbers of substitutions between groups. Sites 2, 4, 9, 13, and 27 had significantly less of the predominant amino acid residue in the case group than in the control group (P =.002, .03, .02, .01, and .02, respectively). For site 2, this was due to one subject's early set of clones with a deletion. For the other sites, this was due to substitutions among several sets of clones.
Because amino acid variation may alter hydropathic properties that are required for HVR1 structure and function, we assessed the proportions of hydrophilic, neutral, and hydrophobic residues at each site within the HVR1 using the scale of Black and Mould.25 Overall, sites 2, 6, 7, 18, and 23 were at least 80% neutral in sequences from both cases and controls. Sites 25, 26, and 27 were nearly 100% hydrophilic. Sites 16, 19, 20, and 24 were nearly 100% hydrophobic. The remaining sites (1, 3, 4, 8-15, 17, 21, and 22) appeared more tolerant of variability. Ten sites had more than 14% difference (P < .05) in frequency for the predominant hydropathic character of its residues between sequences from cases and controls. This subset of 10 sites is displayed in Figure 5.
Amino acid sites from the HVR1 with statistically significant variability in hydropathic character. Hydrophobicity was determined using the scale of Black and Mould.25 Hydrophobic residues include F, I, W, Y, L, V, M, P, C, and A. Neutral residues include G, S, and T. Hydrophilic residues include K, Q, N, H, E, D, and R. Proportion of residue hydropathic characteristics are presented as decreasing hydrophobicity from 0% up: hydrophobic residue proportions are represented in black, neutral in dark gray, and hydrophilic in light gray.
Amino acid sites from the HVR1 with statistically significant variability in hydropathic character. Hydrophobicity was determined using the scale of Black and Mould.25 Hydrophobic residues include F, I, W, Y, L, V, M, P, C, and A. Neutral residues include G, S, and T. Hydrophilic residues include K, Q, N, H, E, D, and R. Proportion of residue hydropathic characteristics are presented as decreasing hydrophobicity from 0% up: hydrophobic residue proportions are represented in black, neutral in dark gray, and hydrophilic in light gray.
Analysis of the core region
HCV quasispecies complexity. A total of 425 clones from the core protein-coding region of the 20 patient pairs were sequenced and analyzed. The mean time between early and late samples was 9.3 years (SE, 0.69 years). Mean numbers of clones per patient were 10.9 (SE, 0.42) for the early time point and 10.35 (SE, 0.29) for the follow-up time point. Complexity was assessed by the proportion of the number of unique core region nucleotide sequences to the number of clones from patients at early and late time points (Table 4). The total early time point mean of unique nucleotide sequences per total number of clones within the core region for progressors, regardless of infection status, was 0.54 (SE, 0.09). For nonprogressors, this mean was 0.76 (SE, 0.05; P = .06). At the late time point, the mean frequencies were 0.67 (SE, 0.09) and 0.75 (SE, 0.06) for progressors versus nonprogressors, respectively (P = .47). In contrast to the changes in quasispecies population complexity in the HVR1, where a narrowing of the population was seen in progressors compared to increased complexity for nonprogressors, no significant differences were apparent in the unique sequences of the core region for progressors versus nonprogressors.
Mean frequencies of unique core region nucleotide sequences for each patient subpopulation
. | Mean complexity . | . | . | ||
|---|---|---|---|---|---|
| Time point . | Progressors (n = 10) . | Nonprogressors (n = 10) . | Difference: progressor-nonprogressor* . | ||
| Coinfected (n = 16) | |||||
| Early | 0.56 (SE, 0.11) | 0.72 (SE, 0.06) | - 0.16 | ||
| Late | 0.61 (SE, 0.10) | 0.73 (SE, 0.07) | - 0.12 | ||
| Monoinfected (n = 4) | |||||
| Early | 0.47 (SE, 0.20) | 0.92 (SE, 0.08) | - 0.45 | ||
| Late | 0.89 (SE, 0.11) | 0.83 (SE, 0.08) | 0.06 | ||
| Total | |||||
| Early | 0.54 (SE, 0.09) | 0.76 (SE, 0.05) | - 0.22 | ||
| Late | 0.67 (SE, 0.09) | 0.75 (SE, 0.06) | - 0.08 | ||
| Late-Early | 0.13 (SE, 0.13) | - 0.01 (SE, 0.06) | 0.14 | ||
. | Mean complexity . | . | . | ||
|---|---|---|---|---|---|
| Time point . | Progressors (n = 10) . | Nonprogressors (n = 10) . | Difference: progressor-nonprogressor* . | ||
| Coinfected (n = 16) | |||||
| Early | 0.56 (SE, 0.11) | 0.72 (SE, 0.06) | - 0.16 | ||
| Late | 0.61 (SE, 0.10) | 0.73 (SE, 0.07) | - 0.12 | ||
| Monoinfected (n = 4) | |||||
| Early | 0.47 (SE, 0.20) | 0.92 (SE, 0.08) | - 0.45 | ||
| Late | 0.89 (SE, 0.11) | 0.83 (SE, 0.08) | 0.06 | ||
| Total | |||||
| Early | 0.54 (SE, 0.09) | 0.76 (SE, 0.05) | - 0.22 | ||
| Late | 0.67 (SE, 0.09) | 0.75 (SE, 0.06) | - 0.08 | ||
| Late-Early | 0.13 (SE, 0.13) | - 0.01 (SE, 0.06) | 0.14 | ||
Frequencies were determined by the proportion of unique nucleotide sequences to the number of clones assessed for each patient at early and late time points within the core-coding region.
P = NS for all comparisons.
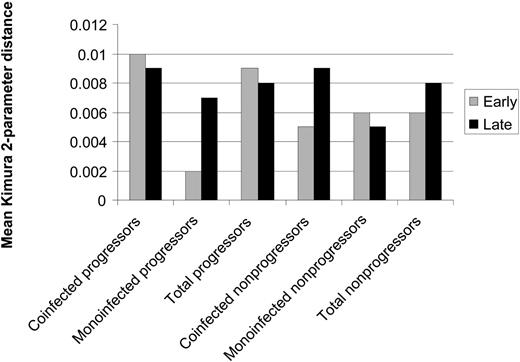
HCV quasispecies diversity. Diversity in the core region was assessed via Kimura 2-parameter pairwise distances between early and late sets of clones for the matched cases and controls. Differences in genetic distance are shown in Figure 6. The core region demonstrated less overall genetic diversity than the HVR1, as indicated by smaller Kimura 2-parameter distances. At baseline, there were no statistically significant differences in diversity between progressors and nonprogressors. Over time, 5 progressors and 1 nonprogressor showed a decrease in genetic distance within early and late sets of clones. Five progressors and 6 nonprogressors displayed an increase in diversity, and no progressors versus 3 nonprogressors demonstrated no net change. However, mean between-group genetic distances for early and late clonal sets were 0.04 (SE, 0.01) and 0.02 (SE, 0.003) for progressors versus nonprogressors, respectively (P = .02, data not shown). Eighty percent of the progressors had greater overall genetic distance than their matched controls.
Mean genetic distances within the core-coding region between early and late clonal sets for progressors and nonprogressors. Distance was calculated by the mean Kimura 2-parameter pairwise distances for each patient. At baseline, there were no statistically significant differences in diversity between progressors and nonprogressors. Over time, 5 progressors and 1 nonprogressor showed a decrease in genetic distance within early and late sets of clones. However, mean between-group genetic distances for early and late clonal sets were 0.04 (SE- 0.01) and 0.02 (SE- 0.003) for progressors versus nonprogressors, respectively (P = .02, data not shown); 80% of the progressors had greater overall genetic distance than their matched controls.
Mean genetic distances within the core-coding region between early and late clonal sets for progressors and nonprogressors. Distance was calculated by the mean Kimura 2-parameter pairwise distances for each patient. At baseline, there were no statistically significant differences in diversity between progressors and nonprogressors. Over time, 5 progressors and 1 nonprogressor showed a decrease in genetic distance within early and late sets of clones. However, mean between-group genetic distances for early and late clonal sets were 0.04 (SE- 0.01) and 0.02 (SE- 0.003) for progressors versus nonprogressors, respectively (P = .02, data not shown); 80% of the progressors had greater overall genetic distance than their matched controls.
Mutational selection
Mutational selection for the core region was determined by calculating mean dN/dS ratios for cases and controls over the early and late time points (Table 5). Although the progressor in pair 2 was clearly an outlier in the HVR1 dN/dS ratio analysis due to the significant nucleotide changes associated with a subtype shift, no outliers were evident in analysis of the core region. There was a statistically significant baseline difference in paired dN/dS ratio between progressors and nonprogressors. However, all dN/dS ratios were less than or equal to 1, indicating a lack of selective pressure driving nonsynonymous mutations. Mean dN/dS ratios of both progressors and nonprogressors decreased over time so that late time point differences were no longer significant.
Mean dN/dS ratios within the core-coding region for early and late time points for progressors (cases) versus nonprogressors (controls)
. | dN/dS ratio . | . | . | ||
|---|---|---|---|---|---|
| Time point . | Progressors (n = 10) . | Nonprogressors (n = 10) . | P for difference between progressors and nonprogressors . | ||
| Early dN/dS ratio | 0.15 (SE, 0.07) | 0.30 (SE, 0.10) | .05 | ||
| Late dN/dS ratio | 0.10 (SE, 0.03) | 0.16 (SE, 0.04) | .26 | ||
| Mean difference (early to late) in dN/dS ratio | - 0.05 (SE, 0.08) | - 0.13 (SE, 0.10) | .31 | ||
| P for difference over time | .54 | .21 | |||
. | dN/dS ratio . | . | . | ||
|---|---|---|---|---|---|
| Time point . | Progressors (n = 10) . | Nonprogressors (n = 10) . | P for difference between progressors and nonprogressors . | ||
| Early dN/dS ratio | 0.15 (SE, 0.07) | 0.30 (SE, 0.10) | .05 | ||
| Late dN/dS ratio | 0.10 (SE, 0.03) | 0.16 (SE, 0.04) | .26 | ||
| Mean difference (early to late) in dN/dS ratio | - 0.05 (SE, 0.08) | - 0.13 (SE, 0.10) | .31 | ||
| P for difference over time | .54 | .21 | |||
The numbers of dS and dN were determined using the Nei-Gojobori method with the Jukes-Cantor correction for multiple substitutions.22 The mean dN/dS ratio, a measure of selective pressure, was calculated for each patient.
Finally, no measures of quasispecies complexity, diversity, or mutational selection were found to be significantly associated with demographic characteristics, baseline CD4+/CD8+ counts, HCV viral load, or HIV infection status.
Discussion
Although it has been well documented that patients coinfected with HCV/HIV are at increased risk for progression to ESLD compared with monoinfected patients,2-4,26-28 the evolution of the HCV quasispecies during this process has not yet been clearly defined. HCV quasispecies complexity and diversity have been examined in relation to HIV-related immunosuppression,29-31 patient response to interferon-based treatment,8,32-37 response to liver transplantation,9,38-41 and in patients with alcoholism18,42 or hemophilia.43 These studies have generated conflicting results. Whereas some investigators have observed an association between complexity of the HVR1 and response to interferon,44,45 others have not.46-48 There is a paucity of longitudinal data regarding HCV quasispecies emergence and selection in patients progressing to ESLD. One report from Curran et al49 analyzed serum and biopsy specimens from 6 untreated HCV-infected patients with various stages of liver disease over approximately 5 years. Three patients with severe liver disease demonstrated a stable population, whereas 3 patients with mild liver disease demonstrated increasing viral complexity. However, not all groups have detected such a difference.50 This may be related to differences in sample size and duration of observation.
Studies in patients with inherited bleeding disorders have been similarly limited in scope and have yielded conflicting results. Many have focused on methods to detect the multiple genotypes, subtypes, and variants that may have existed in the donor pool rather than on the relationship between quasispecies heterogeneity and disease progression.51,52 Our study is the first to analyze HVR1 and core region variability in untreated HCV-infected and HCV/HIV-coinfected hemophilic patients with progression to ESLD, compared to matched, nonprogressing controls.
In this study, we demonstrated significant differences in changes in quasispecies complexity over time within the HVR1 for progressors compared with nonprogressors (P = .01). Progressors had greater quasispecies complexity at baseline (more unique nucleotide sequences per number of clones), but this complexity narrowed over the follow-up period of 9 years. Nonprogressors, in contrast, demonstrated an increase in complexity over time. These changes were evident regardless of HIV coinfection status. These results may initially appear to conflict with a prior cross-sectional study of coinfected and monoinfected patients that determined greater HVR1 sequence variability in coinfected patients, suggesting a possible accumulation of variants due to inability of those coinfected to clear the dominant type.30 However, although the duration of infection was not reported in that study, no patients had yet progressed to ESLD. Coinfected patients analyzed at a relatively early point of HCV infection would yield comparable findings.
We observed similar trends in quasispecies diversity within the HVR1, assessed via mean Kimura 2-parameter distances between early and late sets of clones for the matched progressors and nonprogressors. Progressors exhibited greater early time point genetic diversity than nonprogressors (P = .04) but that diversity narrowed significantly over time in progressors (P = .04) and remained constant in nonprogressors regardless of infection status. This is consistent with results from Tagariello et al43 who recently analyzed samples from 12 HCV-monoinfected hemophilic patients and found that those with progressive disease had high homogeneity as assessed by heteroduplex mobility assay, whereas nonprogressors had fairly static heterogeneity over time.
These differences within the HVR1 were confirmed in analysis of dN/dS ratios over time. After eliminating an outlying matched pair with a subtype shift in the progressor group, the progressors had slightly greater average dN/dS ratio at baseline than nonprogressors (P = .25). At the end point, however, the progressor group had a decreased average dN/dS ratio compared with no change for nonprogressors. Although the difference did not achieve statistical significance (P = .31), the trend is consistent with other measures of quasispecies evolution described. Moreover, site-by-site analysis of predicted amino acid sequences for both early and late time points revealed significant differences in conservation of amino acid residues and, consequently, differences in hydropathic properties between progressors and nonprogressors. Although we have not yet determined specific structural or functional significance of these changes, the differences suggest that nonprogressors are able to maintain an equilibrium of quasispecies variants, either through immune selection or through clearance of emerging mutants. Progressors may have an abrogated ability to clear new variants, resulting in early complexity and diversity but the eventual dominance of a variant with greater replicative fitness or greater pathogenic potential.
Because the HCV core protein-coding region has been associated with liver injury in vitro and in transgenic mouse models, we analyzed the same parameters of evolution over time in this portion of the HCV genome. Progressors demonstrated a slight increase in complexity over time compared with nonprogressors, although this change did not reach statistical significance. Likewise, although the early dN/dS ratio was significantly lower in progressors than in nonprogressors, both groups of patients had dN/dS ratios consistently less than 1, indicating a lack of selective pressure for this region. In contrast to the findings in the HVR1, genetic distances in the core region between early and late clonal sets were likely to be greater for progressors (P = .02), indicating greater divergence. The core region has been shown to regulate oncogenes,11,15,53 alter cytokine expression,12 and inhibit apoptosis,14 all of which are pathogenic mechanisms. Ray et al54 have postulated that specific mutations at core amino acid positions 9 and 11 help modulate nuclear factor-κB (NF-κB) activity. Our sequences displayed the RKT motif (positions 9-11) with 4 exceptions: T11 → P occurred in 3 clones, K10 → I occurred once, T11 → A once, and K10 → Qin 11 clones. All of these mutations occurred in progressors, and all generate increased hydrophobicity at those positions. This suggests that increased rates of specific mutation could lead to higher rates of progression to fibrosis, possibly through inhibition of inflammatory repressor activity, although the exact mechanism of this process remains unknown. Analysis of CD4+/CD8+ proliferative responses to HCV-specific epitopes may help elucidate the factors associated with clearance of specific epitopes.
In conclusion, we have demonstrated numerous and significant differences in HCV quasispecies complexity and diversity in the HVR1 as well as differences in genetic diversity in the core region among interferon-naive progressors and nonprogressors with inherited bleeding disorders. None of these differences were associated with HCV or HIV viral loads, demographic factors, or CD4+/CD8+ counts. Because patients infected through contaminated clotting factors were likely to have similar chances of exposure to multiple viral genotypes, subtypes, and variants through the large donor pool, these results suggest that multiple mechanisms, including viral fitness and host immune factors, drive quasispecies emergence and selection in patients with progressive versus nonprogressive disease. It is possible that more extensive cloning at each time point, and at interim time points, might clarify which mechanisms are operative during emergence and selection. Changes occurring in the HVR1 versus the core region in progressors versus nonprogressors are inconsistent. The HVR1 appears to exhibit the result of mutational selection; generally, the greater the immune selection process, the more variability will emerge over time. Patients progressing to ESLD, who likely exert less immune pressure on the virus, will demonstrate decreased variability in the HVR1 over time. The core region, however, undergoes stochastic rather than selectional mutation altering its hydropathic character. In progressors, the changes that occur in the core region appear to be associated more frequently with hydrophobic elements that may have important functional significance. Further understanding of these mechanisms may help identify factors related to rate of progression and to treatment response in HCV-infected patients with inherited bleeding disorders.
Appendix
Collaborating investigators (and institutions) in the Multicenter Hemophilia Cohort Study: J. J. Goedert, T. R. O'Brien, P. S. Rosenberg, C. S. Rabkin, E. A. Engels, M. H. Gail (National Cancer Institute, Rockville); M. Elaine Eyster (Division of Hematology and Oncology, Pennsylvania State University Medical Center, Hershey); B. Konkle (Cardeza Foundation Hemophilia Center, Philadelphia, PA); M. Manco-Johnson (Mountain States Regional Hemophilia and Thrombosis Program, University of Colorado, Aurora); D. DiMichele, M. W. Hilgartner (Hemophilia Treatment Center, New York Presbyterian Hospital, NY); Philip Blatt (Christiana Hospital, Newark, DE); L. M. Aledort, S. Seremetes (Hemophilia Center, Mount Sinai Medical Center, NY); K. Hoots (Gulf States Hemophilia Center, University of Texas at Houston); A. L. Angiolillo, N. L. C. Luban (Hemophilia Center, Children's Hospital National Medical Center, Washington, DC); A. Cohen, C. S. Manno (Hemophilia Center, Children's Hospital of Philadelphia, PA); C. Leissinger (Tulane University Medical School, New Orleans, LA); G. C. White II (Comprehensive Hemophilia Center, University of North Carolina, Chapel Hill); M. M. Lederman, S. Purvis, J. Salkowitz (Case Western Reserve University School of Medicine, Cleveland, OH); C. M. Kessler (Georgetown University Medical Center, Washington, DC); A. Karafoulidou, T. Mandalaki (Hemophilia Center, Second Regional Blood Transfusion Center, Laikon General Hospital, Athens, Greece); A. Hatzakis, G. Touloumi (National Retrovirus Reference Center, Athens University Medical School, Athens, Greece); W. Schramm, F. Rommel (Medizinische Klinik Innerstadt der Maximilian, Universitaet Muenchen, Munich, Germany); P. de Moerloose (Haemostasis Unit, Hôpital Cantonal Universitaire, Geneva, Switzerland); S. Eichinger (University of Vienna Medical School, Vienna, Austria); K. E. Sherman (University of Cincinnati Medical Center, Cincinnati, OH); D. Waters (Scientific Applications International Corporation, Frederick, MD); and B. L. Kroner (Research Triangle Institute, Rockville, MD).
Prepublished online as Blood First Edition Paper, September 16, 2004; DOI 10.1182/blood-2004-04-1452.
A complete list of the members of the Multicenter Hemophilia Cohort Study Group appears in the “Appendix.”
Supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH grant no. R01 AI49508), and the US Public Health Service General Clinical Research Centers Program, National Center for Research Resources, NIH (grant nos. M01 RR010732, M01 RR08084, and C06 RR016499).
Hongxing Qin died on March 9, 2004.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Angelos Hatzakis (Athens University Medical School, Athens, Greece) for critical review of this manuscript. This manuscript is dedicated to the memory of Hongxing Qin.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal