Abstract
B-cell chronic lymphocytic leukemia (CLL) consists of 2 prognostic entities where cases with mutated immunoglobulin VH genes have better outcome than unmutated cases. VH-mutated CLLs display longer telomeres compared with unmutated cases and telomere length has been indicated to predict outcome, although the prognostic value of telomere length has not been fully established in CLL. We analyzed telomere length, VH gene mutation status, and clinical parameters in a large series of CLL. Telomere length was assessed by quantitative polymerase chain reaction (PCR), giving a very good correlation to telomere length estimated by Southern blotting (P < .001). The prognostic information given by mutation status (n = 282) and telomere length (n = 246) was significant (P < .001, respectively). Telomere length was a prognostic factor for stage A (P = .021) and stage B/C (P = .018) patients, whereas mutation status predicted outcome only in stage A patients (P < .001). Furthermore, mutated CLLs were subdivided by telomere length into 2 groups with different prognoses (P = .003), a subdivision not seen for unmutated cases (P = .232). Interestingly, the VH-mutated group with short telomeres had an overall survival close to that of the unmutated cases. Thus, by combining VH mutation status and telomere length, an improved subclassification of CLL was achieved identifying previously unrecognized patient groups with different outcomes. (Blood. 2005;105:4807-4812)
Introduction
The end structures of the chromosomes, the telomeres, are composed of a repetitive DNA sequence, (TTAGGG)n, and specific proteins bound to the DNA.1,2 The telomeres protect the chromosomes from degradation, prevent end-to-end fusions, and are crucial for control of replicative senescence. In humans, the telomeres have an average length of 5 kilobases (kb) to 15 kb and in normal somatic cells the telomeric DNA shortens with 100 base pair (bp) to 200 bp every cell division due to the end-replication problem.3-5 The erosion of the telomeres can be counter-acted by telomerase, an enzyme that adds TTAGGG repeats to the chromosomal ends.6 Telomere maintenance, usually executed by telomerase, is a prerequisite for an extended or infinite cellular lifespan, and studies of telomere dynamics have been in focus ever since it was shown that telomerase activity is present in almost 90% of all malignant tumors.7-9 The nearly exclusive presence of telomerase in tumor cells has made the enzyme a theoretically attractive target for cancer therapy. In contrast, telomerase is restricted to a few normal cell types in humans, including germ cells, stem cells, activated lymphocytes, and epithelial cells with high reproductive demand.10-14 Additionally, during the germinal center (GC) reaction in lymph nodes a high level of telomerase activity is expressed in the GC B cells accompanied by a unique telomere lengthening process not observed in other cells in vivo.15-17
Telomere length reduction has been found with increasing age and was recently implicated as a predictive factor for overall survival, as high overall mortality was associated with short telomeres in peripheral blood cells in healthy individuals.18 Tumor cells often have shorter telomeres than cells from age-matched controls. Still, only a few studies have found a link between telomere length in malignant cells and prognosis. In myelodysplastic syndromes, short telomeres correlated with increased risk for transformation to acute leukemia and in chronic myeloid leukemia with a worse prognosis.19-22 In myeloma, a subgroup of cases with high telomerase activity and short telomeres had an increased mortality rate.23
B-cell chronic lymphocytic leukemia (CLL) is characterized by a monoclonal expansion of small B lymphocytes previously supposed to be derived from naive B cells of the mantle zone.24 However, analyses of the immunoglobulin heavy-chain variable region (IgVH) gene have shown that CLL can be divided into 2 subsets comprising cases with somatically mutated and unmutated VH genes.25-28 A more favorable prognosis has been found for CLL cases with mutated VH genes compared with unmutated cases.29-32 Somatic hypermutation of Ig genes is a normal process occurring in the GC reaction as part of the antigen-driven affinity maturation of antibodies.33 CLL cases with germ line VH genes are hence considered to originate from pre-GC B lymphocytes and the mutated from GC-experienced B lymphocytes. Interestingly, in a recent study a strong correlation existed between VH gene mutation status and telomere length, where mutated (post-GC) CLL cases had longer telomeres than germ line (pre-GC) cases.32 A similar pattern was also described in some cases of familial CLL after age adjustment of telomere lengths.34 The telomere length and mutation status in CLL thus appear to be linked, possibly reflecting the origin of the clonal cells with regard to the GC reaction. Furthermore, both telomere length and telomerase activity have been found to be associated with outcome in CLL, since a shorter survival was associated with short telomeres and high telomerase activity.32,35,36 However, in a recent study telomere length analyzed by flow-fluorescence in situ hybridization (FISH) was not a significant prognostic factor for CLL.37
In the present study, a large series of CLL cases have been analyzed for telomere length, using a recently described polymerase chain reaction (PCR) method38 and in a subset also by flow-FISH.39,40 The telomere data were analyzed in relation to VH gene mutation status, stage, and overall survival. The correlations between VH gene mutation status, telomere length, and prognosis previously shown were verified. Furthermore, we could show that telomere length could further subdivide the VH-mutated cases into 2 groups with significantly different outcomes, where the group with short telomeres had a prognosis similar to VH gene-unmutated cases. Thus, by combining Ig gene mutation analysis and telomere length evaluation, a better subclassification of CLL can be achieved.
Patients, materials, and methods
Patients
Tumor samples from 310 patients with CLL diagnosed between 1973 and 2002 were obtained from the archives of the Departments of Pathology at the University Hospitals in Umeå (n = 74), Uppsala (n = 156), and Linköping (n = 80), Sweden. The tumor material was acquired from peripheral blood (178 cases, 58%), bone marrow (91 cases, 29%), lymph node (32 cases, 10%), and spleen (9 cases, 3%). There were 216 male (70%) and 94 female (30%) patients with a male-female ratio of 2.3:1. Survival data were available for 296 patients from Swedish population and cancer registries. The median age at diagnosis was 66 years (range, 32-88 years) and the median survival time (estimated by Kaplan-Meier method) was 85 months. Patient follow-up extended from 1 month to 189 months with a median value of 61 months. Data on staging according to Binet were available for 201 patients (110 stage A and 91 stage B or C). Informed consent was provided according to the Declaration of Helsinki, and the study was approved by the ethics committee of Uppsala University (Ups 01-082).
Morphology and immunophenotyping
Besides morphologic classification of smears, sections, and imprints, immunophenotyping was performed on all samples using flow cytometry and a panel of monoclonal antibodies. Only cases fulfilling the criteria for CLL (CD5+, CD19+, CD23+, and a weak expression of surface Ig) in the Royal Marsden scoring system were included.24
Telomere length measurement by quantitative real-time PCR (Tel-PCR)
The method for relative telomere length measurement using real-time PCR has been described38 and is here called Tel-PCR. Briefly, genomic DNA samples diluted to approximately 1.75 ng/μL were incubated for 5 minutes at 95°C, cooled on ice for 5 minutes, and centrifuged briefly at 730g. Two separate PCR runs were performed for each sample, the first to determine the cycle threshold (Ct) value for telomere amplification (plate 1), and the second to determine the Ct value for control gene amplification (plate 2). A standard curve was generated in each run, consisting of reference DNA (CCRF-CEM cell line) diluted serially. Both reference and sample DNA were analyzed in triplicate (35 ng DNA/aliquot).
Two master mixes of PCR reagents were prepared, one for telomere amplification and one for control gene (β2-globin on chromosome 11) amplification. The final concentrations of the PCR reagents were 1.25 U AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), 150 nM 6-ROX and 0.2x Sybr Green I (Roche Diagnostics GmBH, Mannheim, Germany), 50 mM KCl, 2 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate (dNTP) (MBI Fermentas, Amherst, NY), 5 mM dithiothreitol (DTT), 1% dimethyl sulfoxide (DMSO), and 15 mM Tris-HCl pH 8.0. The final telomere primer concentrations were: tel 1, 270 nM; tel 2, 900 nM. In the control gene master mix primers HBG1 and HBG2 were used at the concentration of 400 nM. The primer sequences (written 5′‡3′) were as follows: tel 1, GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT; tel 2, TCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA; HBG1, GCTTCTGACACAACTGTGTTCACTAGC; HBG2, CACCAACTTCATCCACGTTCACC.
Telomere sequences were amplified in a Prism 7000 sequence Detection System (Applied Biosystems) using the following conditions: 95°C, 10 minutes, to activate Taq polymerase; 35 cycles of denaturation at 95°C, 15 seconds, and annealing/extension at 54°C, 2 minutes. The conditions for HBG gene amplification were: 95°C, 10 minutes; 40 cycles at 95°C, 15 seconds, and 58°C, 1 minute. ABI Prism 7000 SDS Software v 1.0 was used for analysis.
The Ct values generated in both runs were used to calculate relative telomere-single-copy-gene ratio (T/S) values for each sample: T/S = 2-ΔCt (where ΔCt = Ctcontrol-Cttelomere).
Telomere length estimated by Southern blotting
In order to verify the reliability of the Tel-PCR method, 43 CLL samples included in this study were also analyzed for telomere length by Southern blotting. Hybridization with a telomeric probe (TTAGGG)4 was performed and mean telomere restriction fragment (TRF) length was calculated as described.41 The lambda DNA/EcoI StyI/MluI Marker (MBI Fermentas) and the DNA molecular weight marker X (Boehringer Mannheim Gmbh, Mannheim, Germany) were used as molecular weight standards.
Fluorescence in situ hybridization and flow cytometry (flow-FISH)
A subset of the CLL cases (n = 87) was analyzed using a previously described flow cytometric assay for telomere length estimation.39 All samples tested had been DMSO frozen at the time of sampling and kept in liquid nitrogen until analysis. After in situ hybridization with a fluorescein-labeled PNA probe, (C3TA2)3, and DNA staining with propidium iodide the samples were run in a flow cytometer (FACS Calibur; Becton Dickinson, San Jose, CA). The tetraploid T-cell line 1301 with very long telomeres was used as an internal control. Telomere length was given as an arbitrary value derived from the ratio between the signal intensity of the internal control and the sample and where the 1301 signal was set to 1. Compensation for cellular DNA content was made as described.39
VH gene analysis
VH gene family-specific PCR amplification was performed on genomic DNA using 6 family-specific VH primers and one JH as previously described.42 Clonal PCR products were sequenced directly or using cloning of the PCR products as previously outlined.42,43 All sequence reactions were analyzed using an automated DNA sequencer (ABI 377 or ABI3700, Applied Biosystems; and MegaBACE 500 DNA Analysis System, Amersham Biosciences, Piscataway, NJ). The sequences were aligned to IgH sequences from the GenBank database (National Center for Biotechnology Information, Bethesda, MD), the V-BASE database (MRC, Centre for Protein Engineering, Cambridge, United Kingdom), and the Immunogenetics database (http://imgt.cines.fr:8104). A VH gene sequence deviating more than 2% from the corresponding germ line gene was defined as mutated.44
Statistical analyses
Kaplan-Meier survival curves and the log-rank test were performed to study the prognostic significance of VH gene mutation status and telomere length in CLL using SPSS (SPSS, Chicago, IL.) Survival was calculated from the date of diagnosis until the last follow-up or death. Nonparametric correlation was calculated according to Spearman rank correlation. Differences between groups were analyzed with Pearson Chi-square. Probabilities of less than .05 were accepted as a significant value. Cox multiple regression was used to analyze the independence of the potential prognostic parameters of age, stage according to Binet, mutation status (cut off at 98% homology), and telomere length (cut off at median T/S value).
Correlation between telomere length estimated by Southern blotting and Tel-PCR. A significant correlation was found (r2 = 0.818, P < .001), giving the mathematical relation: Y = 3.1989X + 3.1280. TRF indicates telomere restriction fragment.
Correlation between telomere length estimated by Southern blotting and Tel-PCR. A significant correlation was found (r2 = 0.818, P < .001), giving the mathematical relation: Y = 3.1989X + 3.1280. TRF indicates telomere restriction fragment.
Results
Telomere length determination
In 246 of the 310 CLL cases, DNA was available for telomere length analysis by the PCR method. For 43 cases telomere length data were also assessed by Southern blotting, showing a high correlation to the Tel-PCR results (P < .001, r2 = 0.818; Figure 1). The Tel-PCR technique provides a relative T/S value for each sample in our CLL cases with a range from 0.01 to 2.21 with a mean value of 0.373. T/S values can be converted to TRF (telomere restriction fragment) length values in kb using the data shown in Figure 1 and in the present CLL material the “transformed” T/S values varied from 3.16 to 9.97 kb. The TRF value at a Tel-PCR value equal to 0 gives an estimation of the average subtelomeric DNA included in the Southern blot evaluation. The value obtained in our material using Tel-PCR (∼ 3.1 kb, Figure 1) corresponds very well to previous estimations using Southern blotting after Bal31 digestions or using flow-FISH.4,39
The time from diagnosis to sampling varied in our CLL cohort. In the group analyzed by Tel-PCR, 148 cases were sampled within one year and 78 cases after more than a year (mean 50 months) after diagnosis. Since telomeres are lost by time, we studied if the time span between diagnosis and sample collection could be of importance. No statistical difference in telomere lengths was found when comparing different time spans from diagnosis to sample collection (0-1, 1-3, and > 3 years; P = .530), indicating that this time factor did not influence the further analysis. A subset of 87 CLL cases was also studied for telomere length using the flow-FISH approach. For these cases, PCR telomere length data, but not Southern blot results, were available. A statistically significant association was found between the Tel-PCR and flow-FISH data (P = .001, r2 = 0.145) (not shown).
Analysis of VH gene mutations
Ig gene mutation data were successfully achieved from 297 patients. A total of 172 cases demonstrated unmutated VH genes (> 98% homology to germ line) and 126 cases showed somatic VH gene mutations (< 98% homology). Among the mutated samples 68 cases displayed less than 95% and 58 cases 95% to 98% homology. In 13 samples no mutation status data could be obtained. Two clonal rearrangements were found in 48 cases, 10 of which showed 2 mutated VH genes (regarded as mutated), 28 with 2 unmutated genes (regarded as unmutated), and 8 with one mutated and one unmutated VH gene (regarded as mutated). One case had 3 mutated VH genes and one case with one mutated and 2 unmutated genes was considered as mutated.
Telomere length in relation to VH gene mutation status
A significant correlation was found for the CLL cases comparing telomere length estimation by Tel-PCR and VH gene mutation status, with longer telomeres in mutated cases (P < .001). The mean Tel-PCR value for mutated cases was 0.51 and for unmutated 0.26. In the VH gene-mutated group cases with less than 95% homology to germ line had significantly longer telomeres (mean T/S = 0.63) compared with cases with 95% to 98% homology (mean T/S = 0.39; P < .01). Furthermore, sample site seemed to be of relevance for this association. In blood and bone marrow samples a significant correlation existed between telomere length and VH gene mutation status (P < .001), but not in samples from lymph nodes (P = .119) (not shown). No significant difference existed in number of mutated and unmutated cases depending on sample site (P = .676, Table 1). However, when the mutated cases were divided into 2 groups (< 95% and 95%-98% homology, respectively), significantly more lymph node cases were found in the group with higher sequence homology than in blood and bone marrow samples (Pearson Chi-square P = .002; Table 2). No significant differences in telomere length, sex, or age distribution were observed with regard to sample site (data not shown).
VH gene mutation status in CLL samples according to sample site
. | Bone marrow, no. cases . | Blood, no. cases . | Lymph node, no. cases . | Spleen, no. cases . | Total, no. cases . |
|---|---|---|---|---|---|
| Unmutated cases | 45 | 103 | 17 | 7 | 172 |
| Mutated cases | 41 | 68 | 14 | 2 | 125 |
| All cases | 86 | 171 | 31 | 9 | 297 |
. | Bone marrow, no. cases . | Blood, no. cases . | Lymph node, no. cases . | Spleen, no. cases . | Total, no. cases . |
|---|---|---|---|---|---|
| Unmutated cases | 45 | 103 | 17 | 7 | 172 |
| Mutated cases | 41 | 68 | 14 | 2 | 125 |
| All cases | 86 | 171 | 31 | 9 | 297 |
No difference in the distribution of mutated and unmutated cases was found with regard to sample site (P = .676, Pearson Chi-square).
Distribution of CLL cases with mutated VH genes, by sample site and sequence homology
. | Bone marrow, no. cases . | Blood, no. cases . | Lymph node, no. cases . | Spleen, no. cases . | Total, no. cases . |
|---|---|---|---|---|---|
| Sequence homology 95%-98% | 19 | 24 | 12 | 2 | 57 |
| Sequence homology less than 95% | 22 | 44 | 2 | 0 | 68 |
| All cases | 41 | 68 | 14 | 2 | 125 |
. | Bone marrow, no. cases . | Blood, no. cases . | Lymph node, no. cases . | Spleen, no. cases . | Total, no. cases . |
|---|---|---|---|---|---|
| Sequence homology 95%-98% | 19 | 24 | 12 | 2 | 57 |
| Sequence homology less than 95% | 22 | 44 | 2 | 0 | 68 |
| All cases | 41 | 68 | 14 | 2 | 125 |
CLL cases with mutated VH genes were divided at 95% sequence homology (the median frequency of homology to germline for the mutated cases) with regard to sample site. The data show a significant difference between groups (P = .002, Pearson Chi-square). Lymph node/spleen samples demonstrated a lower mutation frequency than bone marrow/blood samples.
Survival analysis
When the patient material was divided into mutated (n = 121) and unmutated (n = 162) cases, a statistically significant difference in overall survival was found between the groups (P < .001) (Figure 2A). The median survival for patients with mutated VH genes was 120 months and for those without mutations it was 68 months, calculated from the date of diagnosis. Regarding telomere length, analysis was performed dividing the material at the median Tel-PCR value showing a highly significant difference in survival (P < .001, Figure 2B). The median survival for cases with longer telomeres (Tel-PCR T/S value > 0.3) was 121 months compared with 78 months in the group with lower telomere signal (Tel-PCR T/S value < 0.3).
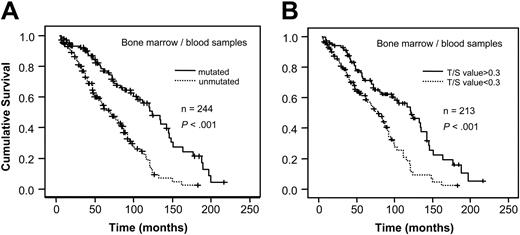
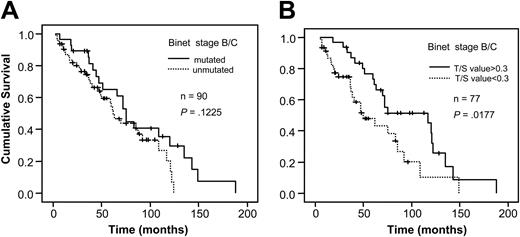
When analyzing VH gene mutation status and telomere length for survival in relation to sample type, a significant correlation to survival was found in bone marrow/blood samples for both mutation status and telomere length (P < .001, respectively; Figure 3A-B). However, no difference in survival existed among the lymph node samples (Figure 4A-B). For stage A patients the mutation status was a significant prognostic factor (P < .001) as was the Tel-PCR data (P = .0206; Figure 5A-B). Mutation status could not predict outcome for stage B/C patients (P = .1225; Figure 6A), in contrast to telomere length (P = .0177; Figure 6B).
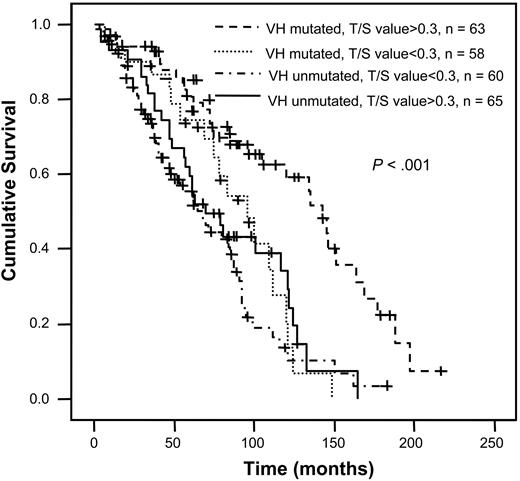
Furthermore, the prognostic value of telomere length was significant for VH gene-mutated cases but not for unmutated cases (P = .0031 and .2324, respectively). VH-mutated cases with short telomeres had a slightly better prognosis than unmutated cases (mean survival time 87 and 73 months, respectively; not significant) but significantly worse than a group with favorable outcome (mean survival time 127 months) defined as VH gene mutation homology less than 98% and telomere length more than median (Figure 7; P = .001).
CLL cases with a clonal rearrangement involving the VH3-21 gene are usually mutated but constitute a subgroup of CLL with an outcome comparable to unmutated cases.45 In the present material, VH3-21 cases were characterized by short telomeres (mean T/S value = 0.33) compared with mutated cases (mean T/S = 0.51), and analysis after excluding the VH3-21 cases still showed a significant difference in survival in the VH-mutated cases subgrouped after telomere length (P < .001).
Since CLLs with lymph node- and spleen-derived samples differed from the other CLLs with respect to several characteristics, they were excluded from the multiple regression analysis. Thus, in our CLL cases with bone marrow and peripheral blood samples the close association between mutation status and telomere length was demonstrated in the multivariate analysis with mutation status as the strongest prognostic indicator. When mutation status was excluded from the multivariate analysis telomere length (P = .003), Binet stage (P = .005), and age (P = .026), but not sex (P = .794) were independent prognostic factors. If only IgVH-mutated cases were analyzed, Binet stage was a significant prognostic factor (P = .005) but telomere length (P = .439) was not, indicating an association between short telomeres and higher stage and/or long telomeres and lower stage for this subgroup.
A subset of patients (n = 87) was also analyzed by flow-FISH but with this technique no difference in survival (P = .426) was achieved using a cut-off at the median flow-FISH value of 0.10639 (corresponding to about 5.5 kb in Southern blot TRF analysis) (not shown).
Survival analysis in relation to VH gene mutation status and telomere length determined by Tel-PCR. (A) VH gene mutation status; (B) telomere length by Tel-PCR.
Survival analysis in relation to VH gene mutation status and telomere length determined by Tel-PCR. (A) VH gene mutation status; (B) telomere length by Tel-PCR.
Discussion
Using Ig gene analysis, CLL can be divided into at least 2 groups, consisting of cases with somatically mutated or unmutated VH genes with a significantly better prognosis for the mutated, GC-experienced cases.25-32 In the present study, this subdivision according to VH gene mutation status was again confirmed. Furthermore, the clinical value of telomere length assessment in CLL was demonstrated using a recently described quantitative PCR method38 in our large cohort of CLL cases. Of special interest is that a combination of telomere length and VH gene mutation status could identify 2 previously unrecognized patient groups among the VH-mutated cases, one with longer telomeres and a favorable outcome and one with shorter telomeres and an unfavorable outcome.
During the GC reaction, telomerase is strongly up-regulated in the activated GC B cells, subsequently followed by a notable telomere elongation.15-17 This appears to be reflected in mature B-cell disorders in which GC-derived tumors have been found to exhibit longer telomeres than GC-inexperienced neoplasms.46 Thus, GC-derived follicular lymphomas, Burkitt lymphomas, and diffuse large B-cell lymphomas demonstrated longer telomeres than CLL and mantle cell lymphomas.46 For CLL the telomere length is strongly associated with mutation status32,37,46 and cases with long telomeres have a significantly better outcome compared with cases with shorter telomeres.32,35 These observations were further verified in the present study on a large cohort of CLL cases. Thus, GC-experienced CLL cells are characterized by hypermutated IgVH genes and longer telomeres.
Survival analysis in bone marrow/blood samples. Figure shows survival in relation to VH gene mutation status (A) and telomere length determined by Tel-PCR (B).
Survival analysis in bone marrow/blood samples. Figure shows survival in relation to VH gene mutation status (A) and telomere length determined by Tel-PCR (B).
The biologic background to the connection between telomere length and outcome can only be speculated upon. Since there is a very strong association between VH gene mutation status and telomere length in normal as well as in malignant samples,15-17,32,46 it can be argued that telomere length is merely a “pseudo marker” for GC-experienced B cells. However, further analysis of the relationship between telomere length and VH gene mutation status demonstrated that 2 patient subgroups with a significantly different outcome could be identified among the VH gene-mutated cases. These patient groups have previously been unrecognized and are important to identify, especially since the outcome for cases with mutated VH genes and short telomeres did not differ significantly from the unmutated group. Previously it has been demonstrated that extensive telomere shortening promotes genetic instability (reviewed in Hacket and Greider47 ) and that CLL unmutated cases show more genetic aberrations than the VH gene-mutated cases.48 These data indicate that in CLL, short telomeres can be a significant contributing factor to genetic instability and thereby also be an important prognostic parameter. An interesting future task will be to characterize VH-mutated CLL cases regarding genetic alterations in relation to telomere status. For clinical stage there seemed to be a difference between mutation status and telomere length regarding prognostication strength, since both parameters were highly significant for stage A patients, whereas only telomere length showed this power for stage B/C patients. In a recent French study, mutational status and stage were independent and complementary prognostic indicators in CLL,49 indicating to us unknown differences between the 2 patient groups. In IgVH-mutated CLL there seemed to be an association between stage and telomere length, suggesting that for this subgroup shorter telomeres and higher stage indicated a worse prognosis. This further substantiates the need to characterize, in detail, mutated CLL regarding genetic features at the chromosomal and molecular level.
Survival analysis in lymph node samples. Figure shows survival in relation to VH gene mutation status (A) and telomere length determined by Tel-PCR (B).
Survival analysis in lymph node samples. Figure shows survival in relation to VH gene mutation status (A) and telomere length determined by Tel-PCR (B).
In the nodal samples neither mutation status nor telomere length seemed to be a factor of prognostic relevance. It cannot be ruled out that CLL preferentially affecting lymphoid tissue could differ in some respects from the “common” CLL in blood and bone marrow, rendering mutation status or telomere length less important for the outcome. However, the nodal cases were few in the present study and a larger study cohort is needed to verify the difference indicated.
Survival analysis in stage A patients. Figure shows survival in relation to VH gene mutation status (A) and telomere length determined by Tel-PCR (B).
Survival analysis in stage A patients. Figure shows survival in relation to VH gene mutation status (A) and telomere length determined by Tel-PCR (B).
In CLL a biased usage of the VH3-21 and VH1-69 genes has been reported.45,50,51 VH1-69 cases are preferentially unmutated whereas VH3-21 cases often are mutated. However, irrespective of the mutation status the VH3-21 cases showed a similar clinical course to unmutated cases.45,51 Therefore, a connection between VH3-21 usage and mutated cases with short telomeres could be suspected, which was confirmed in this study. Accordingly, after excluding the VH3-21 cases from the analysis, mutated cases still could be subdivided into 2 groups with different outcomes, depending on the telomere length.
Survival analysis in stage B/C patients. Figure shows survival in relation to VH gene mutation status (A) and telomere length determined by Tel-PCR (B).
Survival analysis in stage B/C patients. Figure shows survival in relation to VH gene mutation status (A) and telomere length determined by Tel-PCR (B).
The application of the Tel-PCR methodology for telomere length analysis was shown to be highly consistent with Southern blot results, confirming its usefulness for telomere measurements. Compared with Southern blotting, the PCR approach is more convenient, since it is rapid and requires considerably less DNA. One possibly important feature of the PCR method is that only telomere repeats are amplified and recorded, excluding the subtelomeric parts included in the Southern blots. In the PCR method the telomere value has to be related to the value obtained for a single copy gene and for this purpose we have used the β-globin gene on chromosome 11. An alternative in the literature is 36B4 on chromosome 12,38 but since trisomy 12 is common in CLL we wanted to exclude the possibility of a bias due to a DNA imbalance. We did, however, compare the effect of trisomy 12 using both single-copy genes as loading controls in a few cases with known trisomy 12 and no obvious difference was shown, indicating that a disomy versus trisomy DNA difference is of minor importance after completion of the amplification rounds (P.G., unpublished data, January 2005).
Survival analysis in CLL after subdivision according to mutation status and telomere length determined by Tel-PCR as indicated in the figure.
Survival analysis in CLL after subdivision according to mutation status and telomere length determined by Tel-PCR as indicated in the figure.
In a recent study,37 no significant prognostic relevance of telomere length in CLL was found using flow-FISH. These data are in accordance with our present results using flow-FISH on a subset of the CLL samples. Our impression is that the flow technique is less reliable in the analysis of samples kept frozen in DMSO for many years. The correlation between our flow-FISH data and Southern blotting or Tel-PCR is indeed better using fresh cells compared with DMSO frozen samples (M.H., unpublished data, April 2004). Thus, for telomere length analysis of stored material, Tel-PCR seems to be the method of choice.
In conclusion, telomere length is a clinically significant factor in CLL and in combination with Ig gene-mutation analysis a better subclassification of CLL can be achieved, identifying 2 previously unrecognized patient groups with different outcomes.
Prepublished online as Blood First Edition Paper, March 3, 2005; DOI 10.1182/blood-2004-11-4394.
Supported by grants from the Swedish Cancer Society, the Medical Faculty, Umeå University, Lion's Cancer Research Foundation at Umeå University and Uppsala University, and by grant QLG1-1999-01341 from the European Union (G.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are grateful to Elisabeth Grönlund, Eleonor Lindström-Eriksson, and Kent Persson for skillful technical assistance. Richard Palmqvist is appreciated for statistical help.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal