Abstract
Omenn syndrome (OS) is characterized by severe combined immunodeficiency (SCID) associated with erythrodermia, hepatosplenomegaly, lymphadenopathy, and alopecia. In patients with OS, B cells are mostly absent, T-cell counts are normal to elevated, and T cells are frequently activated and express a restricted T-cell receptor (TCR) repertoire. Thus far, inherited hypomorphic mutations of the recombination activating genes 1 and 2 (RAG1/2) have been described in OS. We report on a first patient with clinical and immunologic features of OS caused by hypomorphic ARTEMIS mutations. The patient's T cells expressed α/β receptors with an oligoclonal repertoire but normal V(D)J recombination coding joints. Sequencing of the ARTEMIS gene revealed a compound heterozygosity in this nonhomologous end-joining (NHEJ) factor, explaining the enhanced radiosensitivity of the patient's primary dermal fibroblasts. The maternal allele contained a null mutation within the active center, whereas the expression of the paternal allele with a start codon (AUG to ACG) mutation partially restored V(D)J recombination and ARTEMIS function in vivo and in vitro.
Introduction
Omenn syndrome (OS), a rare autosomal recessive disease (OMIM 603 554), is characterized by symptoms of severe combined immunodeficiency (SCID) associated with erythrodermia, hepatosplenomegaly, lymphadenopathy, and alopecia. Since the first description by Omenn in 1965,1 about 70 patients have been reported.2 Laboratory findings of OS display elevated or normal patient T-cell counts with a restricted, oligoclonal T-cell receptor (TCR) repertoire.3-5 These T cells are often activated and skewed toward a Th2 phenotype, secreting predominantly Th2-type cytokines.6,7 B cells are typically largely absent. Almost uniformly, eosinophilia and high immunoglobulin E (IgE) levels are found that are most likely the result of the Th2-type cytokine secretion.8,9 Serum levels of other immunoglobulin classes are decreased or not detectable. Natural killer (NK) cell functions and absolute numbers are unaffected in OS; thus the majority of patients may be classified as T+B-NK+ SCID. The clinical course of OS is almost always fatal and bone marrow transplantation is successful only in half of the cases.8
Mutations of the recombination activating genes 1 and 2 (RAG1/2) with residual activity (ie, “hypomorphic” mutants) have so far been described in the majority of patients with OS.10-13
The RAG proteins are indispensable for V(D)J recombination, an essential process during lymphocyte development and thus for the implementation of the diversity of T- and B-cell receptors. At the genomic level, exons coding for immunoglobulin or TCR variable domains are recombined from distinct subgenic elements called variable (V), diversity (D), and joining (J) elements. The rearrangement of the DNA is guided by the recombination signal sequences (RSSs) that flank the individual V, (D), or J gene segments. The RAG proteins specifically recognize the RSSs and as a hetero-multimeric endonuclease initially introduce a nick in the DNA double-strand between a coding V, (D), or J element and the heptamer of its flanking RSS.14-17 After the generation of this initial cut, the resulting free 3′-OH attacks the phosphodiester bond of the opposite strand, leading to the formation of covalently sealed coding end hairpins at the ends of the V, (D), and J segments. Thereafter, the corresponding coding end hairpins are reopened and joined by the ubiquitously expressed factors of the nonhomologous end joining (NHEJ) DNA repair pathway.
Recently, it was demonstrated that the protein ARTEMIS is responsible for opening coding end hairpins.18 ARTEMIS associated with and phosphorylated by the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) possesses overhang endonucleolytic and DNA hairpin-opening activities in vitro.18 Without the activation by DNA-PKcs, ARTEMIS only exhibits its intrinsic DNA single-strand specific 5′ to 3′ exonuclease activity in vitro.
A lack of ARTEMIS in humans leads to the phenotype of B-cell–negative, T-cell–negative severe combined immunodeficiency (B-T-SCID) with a block of B-cell development in bone marrow.19,20 Since fibroblasts and hematopoietic progenitor cells deficient in ARTEMIS showed increased sensitivity to ionizing radiation as compared with wild-type cells, ARTEMIS, like several other components of the NHEJ pathway, seems to play a role in the general DNA double-strand break (DSB) repair pathway. Therefore, the type of severe combined immunodeficiency caused by a lack of ARTEMIS activity is called radio-sensitive SCID (RS-SCID).21
Some patients with OS without mutations of the RAG genes have been reported.8 It has been speculated that these patients may bear mutations of ARTEMIS or any other enzyme involved in the V(D)J recombination or NHEJ pathway. To date, however, no genetic defects other than RAG mutations have been identified as a molecular cause of OS.2
In this report, we present a case with classical clinical and immunologic features of OS; genetic, biochemical, and functional analyses pinpoint ARTEMIS mutations as the cause of the patient's OS.
Material and methods
Cells
Patient peripheral blood mononuclear cells (PBMCs) were obtained prior to bone marrow transplantation and were cryopreserved. In order to avoid potential T-cell contamination, for genomic studies skin-derived primary fibroblasts from the patient were also analyzed. For comparison of HLA-type and short tandem repeat (STR) analysis as well as genetic analysis, blood cells of the patient's mother, father, and the second brother were used for DNA preparation. Informed consent was provided according to the Declaration of Helsinki.
Immunofluorescence flow cytometry
The detection of T-, B-, and NK cells was performed with peripheral blood by direct immunofluorescence staining as described elsewhere.22 Briefly, triple-staining combinations of the following monoclonal antibodies (mAbs) were used (from Coulter/Immunotech, Marseilles, France, or Dako, Glostrup, Denmark): CD3/CD4/CD8; CD5/CD19; CD16/CD56/CD3; CD45RA/CD45RO/CD4, directly conjugated with fluorescein isothiocyanate (FITC), phycoerythrin (PE), or PE-Cy5, respectively.
For TCR expression studies, 2 × 105 thawed PBMCs were stained with CD3-FITC, CD4-Cy5, or CD8-Cy5 (Dako, Hamburg, Germany), and TCRαβ-PE or TCRγδ-PE (Immunotech) after blocking with Fc-blocking reagent (Miltenyi Biotec, Bergisch Gladbach, Germany). Isotype controls (Dako) were used to determine background fluorescence. Cells were analyzed on a Coulter Epics flow cytometer.23,24
Isolation of CD3+ cells
To enrich T cells, 2 × 106 PBMCs were resuspended in 300 μL magnetic activated cell sorting (MACS) buffer (phosphate-buffered saline [PBS] with 0.5% human albumin and 5 mM EDTA [ethylenediaminetetraacetic acid; Delta Pharma, Pfullingen, Germany]). The cells were incubated with 100 μL Fc-blocking reagent (Miltenyi Biotec) for 10 minutes at 4°C. Subsequently, 100 μL CD3-labeled immunomagnetic beads (Miltenyi Biotec) were added. After an incubation for 15 minutes at 4°C cells were washed once in MACS buffer, resuspended, and passed through a positive selection column (MS; Miltenyi Biotec) according to the manufacturer's instructions. Purity was checked immediately after selection by immunofluorescence flow cytometry (see “Immunofluorescence flow cytometry”).
NK cytotoxicity assay
Cytotoxic activity of NK cells was tested by a flow cytometric nonradioactive assay with the use of DiOC318-stained K562 cells as described previously.25,26 In brief, fluorescent DiOC318-stained K562 cells were gated and propidium iodide–stained cells emitting red fluorescence were evaluated for percentage of lysis.
Immunoglobulin serum levels
IgA, IgM, IgG, and IgE immunoglobulin serum levels were measured using a turbidimetric method (Dade Behring, Marburg, Germany).
T-cell functions
In vitro T-cell functions were determined by measuring 3H-TdR uptake after stimulation of PBMCs with phytohemagglutinin (PHA-P) at 20 μg/mL (Difco Laboratories, Detroit, MI), poke weed mitogen (PWM; Sigma-Aldrich, Steinheim, Germany), and concanavalinA (ConA; Sigma-Aldrich) as described previously.24,27
Isolation of DNA and RNA
For TCR studies, total RNA was extracted from cells by the guanidinium thiocyanate-phenol-chloroform method using RNAzol B (Biotecx Laboratories, Houston, TX) or TriStar (AGS, Heidelberg, Germany). After RNA extraction, DNA was recovered from the remaining phases of the reagent according to the manufacturer's instructions. When larger amounts of patient and control cells were available and higher amounts of DNA or RNA were required, the Qiagen DNA or RNA extraction kits were used (Qiagen, Hilden, Germany).
TCR repertoire
The experimental strategy of quantitative analysis of rearranged T-cell receptor β-chain sequences (TRBV) and CDR3 length imaging (“spectratyping”) was carried out as described previously.28 In brief, first-strand cDNA was copied from 1 μg total RNA by MMuLV reverse transcriptase (PE Applied Biosystems, Weiterstadt, Germany) following oligo (dT) priming. For CDR3 length imaging, 26 aliquots of 0.2 μL to 1.0 μL cDNA were subjected to 26 to 29 cycles of PCR coamplification with an exogenous standard and incorporation of a fluorescence-labeled constant-region primer. Aliquots (3 μL) of the fluorescence-labeled polymerase chain reaction (PCR) products were heat-denatured in 50% formamide and loaded onto a 6% denaturing polyacrylamide gel. Electrophoresis was performed on a DNA sequencer (PE Applied Biosystems) at constant current. Quantitative determination of DNA fragments was performed by measurement of laser light–induced fluorescence. Typically, each of the 24 Vβ-specific amplicons contains transcripts of 7 to 10 different lengths. For data analysis, the GeneScan software (PE Applied Biosystems) was used. The fluorescence intensity of an amplicon is expressed as area under the curve (AUC) in relative fluorescence units. The relative expression of sequences of one TRBVβ-specific amplification in a T-cell population was calculated by normalization with the semihomologous standard. For assessment of the clonal contingent, an algorithm similar to that described earlier by Gorochov and coworkers was used.29 The lengths of CDR3 regions were defined by the criteria described by Moss and Bell.30,31
Irradiation experiments
To check their radiosensitivity, viable fibroblasts were suspended at a concentration of 4 × 104 cells/mL and irradiated at 0, 1, 3, and 6 Gy in 2 independent experiments. After irradiation, the cells were seeded at a density of 1 × 104 cells/mL in T75 flasks in triplicate. Ten days later the adherent cells were trypsinized and counted. The mean of the triplicates was calculated; the resulting values were set in relation to the non-irradiated cells.
Analysis of ARTEMIS mRNA
Expression of ARTEMIS mRNA was investigated by reverse transcriptase (RT)–PCR. RNA was transcribed by using the Superscript II RT reverse transcriptase (Gibco Life Technologies, Gathersburg, MD) and a poly-T-primer (5′-TTT TTT TTT TTT TTT TTT TV-3′; Thermo Electron, Ulm, Germany). Samples without addition of reverse transcriptase were used as controls. cDNA was amplified either by the Expand High Fidelity PCR System (Roche, Mannheim, Germany) or the Taq Polymerase System (Qiagen) in combination with DNA polymerization mix (Amersham, Piscataway, NJ). Primers (Thermo Electron) were designed to amplify the complete ARTEMIS mRNA or a 300-bp fragment of the 5′ part of the ARTEMIS mRNA including the start codon: ARTcDNA-F (5′-TAT GAG TTC TTT CGA GGG GC-3′), ARTcDNA360M (5′-CAC AAC AAT CTC TTC CTT CTC TCC-3′), and ARTcDNA1200M (5′-CAA AGA GAT AGT CAT CTT CCT CC-3′). The transcribed cDNA was checked by amplification of a fragment of the human hypoxanthine phosphoribosyltransferase (HPRT) mRNA. ARTEMIS cDNA was cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA). Subsequently, the cDNA sequences were analyzed using the BigDye Terminator v1.1 Cycle Sequencing Kit (PE Applied Biosystems) and primers M13 Reverse (5′-CAGGAAACAGCTATGAC-3′) and M13 Forward (5′-GTAAAACGACGGCCAG-3′).
Analysis of ARTEMIS and RAG1/2 genes on genomic DNA
Coding sequences and the exon/intron boundaries of the ARTEMIS and RAG1/2 genes were amplified using the Taq polymerase system (Qiagen). PCR products were sequenced directly or cloned into the pCR2.1 vector (Invitrogen) and sequenced. Primers used for PCR and sequencing are available on request.
Construction of ARTEMIS-myc-His and mutated ARTEMIS-myc-His expression plasmids
Full-length human ARTEMIS cDNA was amplified by recombinant Pfu DNA polymerase (Stratagene, La Jolla, CA) from a human thymus matchmaker cDNA library (BD Biosciences, Palo Alto, CA). ARTEMIS cDNA was amplified using primers Kpn1ARTcDNA5′N (5′-GGGTACCGCTATGAGTTCTTTCGAGGG-3′) and Not1ARTcDNA3′w/oSTOP (5′-ATAAGAATGCGGCCGCCAGGTATCTAAGAGTGAG-C-3′). A plasmid encoding the fusion protein ARTEMIS-myc-His (pcDNA6/ArtWT-myc-His) was constructed by cloning the wild-type ARTEMIS cDNA (ART-WT) into the pcDNA6/myc-His vector Version A (Invitrogen) after a KpnI/NotI digest. The point mutated ARTEMIS-expressing vectors pcDNA6/ArtH35A-myc-His (ARM12), pcDNA6/ArtH115A-myc-His (ARM14), pcDNA6/ArtH35D-myc-His (ARM27), pcDNA6/ArtAA8-692-myc-His (ARM29), and pcDNA6/ArtM1T-myc-His (ARM32) were generated using the Quick-Change Site-Directed Mutagenesis Kit (Stratagene). Primers for mutagenesis are available on request. Subsequent to cloning, the wild-type and mutant ARTEMIS open reading frames (ORF) of the corresponding plasmids were confirmed by sequencing.
Immunoblotting
To investigate the expression of mutated ARTEMIS proteins, HEK293 and HEK293T cells were transfected by the calcium phosphate precipitation method32 with the pcDNA6/myc-His Version A plasmids, coding for either wild-type or mutated versions of ARTEMIS fused to the myc-His tag. After 48 hours cells were harvested and lysed in the lysis buffer (50 mM Tris-Cl pH 8.0, 62.5 mM EDTA, 1% (wt/vol) NP-40, 0.4% (wt/vol) sodium deoxycholat). Lysates were spun down at 14 000g. Protein concentrations of the supernatants were determined by the Bio-Rad Dc Protein assay (Bio-Rad, Hercules, CA). A quantity of 10 μg of each lysate was run on a 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel. The proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA) and the blots were developed using mouse anti-myc18 and mouse anti-SV40 large T (Santa Cruz Biotechnology) antibodies.
Immunostaining of HEK293T cells
HEK293T cells placed on sterile cover slips were transfected by the calcium phosphate precipitation method32 with pcDNA6/myc-His Version A, coding for wild-type or mutant forms of ARTEMIS. After 48 hours, cells were stained as previously published.33 Subsequently, cells were analyzed by confocal fluorescence microscopy.
Protein purification, DNA-PKcs kinase assay, and in vitro nuclease assay
Protein purification and the DNA-PKcs kinase and in vitro nuclease assays were carried out as described.18
V(D)J recombination assay in fibroblasts
V(D)J recombination assays were performed as described previously.33 In short, human primary ARTEMIS-negative or ARTEMIS-positive dermal fibroblasts were transfected using the AMAXA NHDF-Neo Nucleofector Kit (AMAXA Biosystems, Cologne, Germany). Transfections were performed with 1.2 μg pcWTRAG1, 1.8 μg pcWTRAG2, 8.0 μg pMACS11-19VDJ, or pMACS11-19Flip and either 1.0 μg pcDNA6/myc-His Version A or pcDNA6/ArtWT-myc-His or pcDNA6 vectors coding for mutant ARTEMIS proteins. After 48 hours, cells were harvested and analyzed by immunofluorescence cytometry.
Transfected fibroblasts were detected with biotin–anti-mouse H-2KK antibody (BD Biosciences) against the truncated major histocompatibility complex (MHC) class I protein H-2KK encoded by the assay plasmids. The biotin-labeled antibodies were developed by streptavidin–peridin chlorophyll protein (PerCP) staining (BD Biosciences). The excitation wavelength for the FACS analysis was 488 nm. In the first analysis step, healthy fibroblasts were grouped according to their morphology by a forward/sideward scatter measurement. The fibroblasts transfected with either pMACS11-19VDJ or pMACS11-19Flip (PerCP-positive cells) were detected in fluorescence channel 3 (650 nm longpass) and were distinguishable from fibroblasts showing only autofluorescence when a fluorescence channel 2 (FL2; 585 nm ± 21 nm) versus fluorescence channel 3 (PerCP) analysis was performed. In the third step, enhanced green fluorescent protein (EGFP)–positive (ie, recombination-positive) fibroblasts were specified in channel 1 (EGFP; 530 nm ± 15 nm) out of the subpopulation of PerCP-positive cells by a fluorescence channel 1 (EGFP) versus fluorescence channel 2 (FL2) analysis.
Results
Patient history and immunophenotype
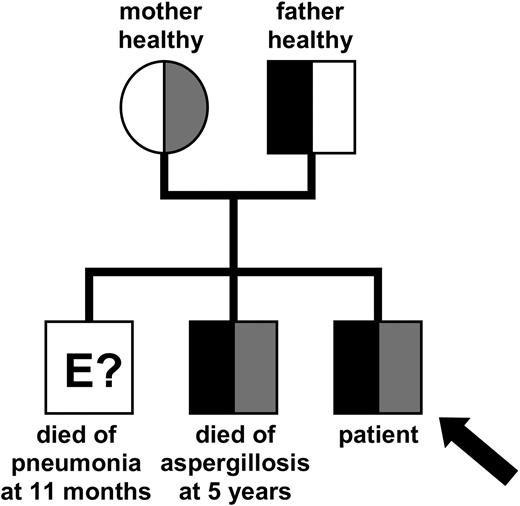
We report a male patient affected by a T+B-NK+ SCID with an Omenn syndrome phenotype. This patient was the third male child of nonconsanguinous healthy parents (Figure 1). The first boy of the family presented with an erythrodermatitis and died at the age of 11 months of an atypical pneumonia. The second boy died at the age of 5 years of aspergillosis. He suffered from thrombocytopenia and autoimmune hemolytic anemia, which was considered an Evans syndrome.
The third boy, who we focus on in this report, was admitted to a children's hospital at the age of 5 months for septicemia, failure to thrive (body mass and height < third percentiles), generalized lymphadenopathy, hepatomegaly, splenomegaly, and erythrodermatitis. Skin lesions consisted of large ichtyotic scales and scattered erythematous papules. In addition, he exhibited alopecia of the scalp, eyebrows, and lashes.
The blood culture revealed the presence of Staphylococcus aureus; Pseudomonas aeruginosa was isolated from the feces. At the site of Bacille-Calmette-Guérin (BCG) vaccination ulceration with lymph node involvement was observed. A lymph node histology was taken from the left axilla, the draining site of the vaccination. Numerous eosinophils, a marked proliferation of HLA-DR+ CD45R0+ T cells, and a complete absence of B lymphocytes, were noted. No other clinical changes due to the BCG infection were observed.
Immunosuppressive treatment led to an almost complete resolution of the dermatitis, alopecia, hepatosplenomegaly, and lymphadenopathy. Significant weight gain was observed. At the age of 10 months, peripheral blood stem cell transplantation after T-cell depletion and myeloablative conditioning from the HLA-haploidentical mother was performed. This therapy resulted in a complete donor chimerism and normal immune functions.
The patient's first immunologic assessment was initiated at the age of 5 months. Lymphocyte counts were elevated (Table 1). The most important finding was the complete absence of B cells in the peripheral blood, as well as in a lymph node biopsy. An inverse CD4/CD8 ratio was noted. NK cell counts were low (0.53 × 109/L), but consistently present, NK cell function was normal.
Cell counts and immune functions at first diagnostic work-up
. | Value . |
|---|---|
| Cell counts | |
| Leukocytes | 31.00 × 109/L |
| Lymphocytes | 15.50 × 109/L |
| T cells | |
| CD3+ | 14.57 × 109/L |
| CD3+ CD4+ | 4.39 × 109/L |
| CD3+ CD8+ | 8.91 × 109/L |
| CD3+ DR+ | 7.60 × 109/L |
| B cells | |
| CD19+ | 0.00 × 109/L |
| NK cells | |
| CD56+ | 0.53 × 109/L |
| T-cell functions, stimulation index* | |
| PHA | 14.1% |
| PWM | 36.2% |
| ConA | 17.4% |
| Serum immunoglobulin levels† | |
| IgA | 0.22 g/L (0.08-0.50 g/L) |
| IgG | 0.16 g/L (1.80-8.00 g/L) |
| IgM | 0.00 g/L (0.20-1.00 g/L) |
| IgE | 123 kU/L (2-24 kU/L) |
. | Value . |
|---|---|
| Cell counts | |
| Leukocytes | 31.00 × 109/L |
| Lymphocytes | 15.50 × 109/L |
| T cells | |
| CD3+ | 14.57 × 109/L |
| CD3+ CD4+ | 4.39 × 109/L |
| CD3+ CD8+ | 8.91 × 109/L |
| CD3+ DR+ | 7.60 × 109/L |
| B cells | |
| CD19+ | 0.00 × 109/L |
| NK cells | |
| CD56+ | 0.53 × 109/L |
| T-cell functions, stimulation index* | |
| PHA | 14.1% |
| PWM | 36.2% |
| ConA | 17.4% |
| Serum immunoglobulin levels† | |
| IgA | 0.22 g/L (0.08-0.50 g/L) |
| IgG | 0.16 g/L (1.80-8.00 g/L) |
| IgM | 0.00 g/L (0.20-1.00 g/L) |
| IgE | 123 kU/L (2-24 kU/L) |
Stimulation index as accounted in percent of internal healthy control
A functional analysis of the immune system revealed that in vitro T-cell functions were very low, but still present. Unfortunately, T-cell reactivity to tuberculin purified protein derivative (PPD) was not assessed. Immunoglobulin levels, namely IgA, IgG, and IgM levels, were severely decreased or absent. In contrast, IgE levels were elevated (123 kU/L, Table 1).
Taken together, the patient presented with all clinical signs of Omenn syndrome, a T+B-NK+ SCID immunophenotype, and with elevated IgE levels and eosinophilia.
Origin and characteristics of T cells and TCR repertoire
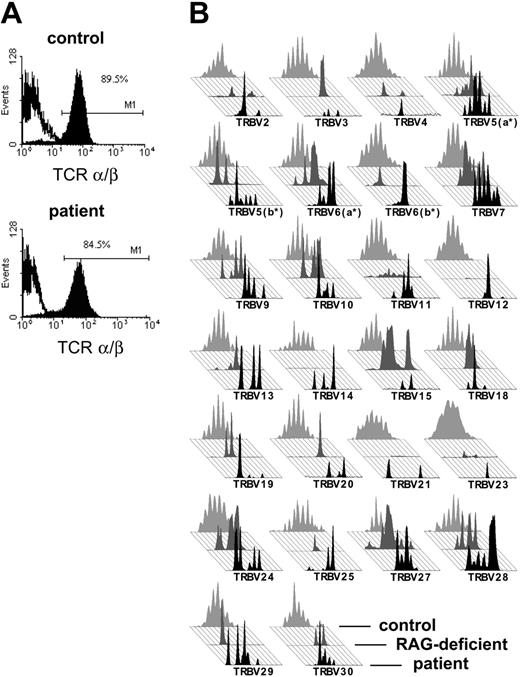
The patient's T cells expressed almost exclusively an α/β T-cell receptor in normal quantity and stained positive for CD3 (Figure 2A). To exclude materno-fetal transfusion, we checked the origin of T cells in a peripheral blood sample prior to transplantation. T cells were positively selected with CD3-immunomagnetic beads and were 93% pure after selection as judged by immunofluorescence cytometric analysis (data not shown).
Inheritance of the mutated ARTEMIS alleles. Pedigree of the patient's family indicating the inheritance of the mutated ARTEMIS alleles. The genetic status of the eldest brother could not be determined at this point. Gray shaded areas indicate the affected maternal allele; black areas indicate the affected paternal allele.
Inheritance of the mutated ARTEMIS alleles. Pedigree of the patient's family indicating the inheritance of the mutated ARTEMIS alleles. The genetic status of the eldest brother could not be determined at this point. Gray shaded areas indicate the affected maternal allele; black areas indicate the affected paternal allele.
Materno-fetal transfusion was excluded by comparing the patient's DNA from the CD3+ fraction, from the CD3-, fraction and from fibroblasts to DNA from the mother, father, and the second sibling by genetic HLA-typing and STR analysis (Supplemental Table 1 and Supplemental Figure 1; see the Supplemental Materials link at the top of the online article on the Blood website).
The fact that the patient had his own T cells supported the OS diagnosis. In addition, we tested the patient's TCR repertoire, because patients with OS exhibit a severely restricted TCR repertoire. Indeed, in a spectratyping analysis all TRBV families of the patient showed a restricted repertoire that was not discernable from a RAG-deficient patient with OS (Figure 2B).
The analysis of the few TCR sequences recovered displayed a normal usage of V, D, and J elements (Table 2). Of 28 different TCR sequences, 2 sequences appeared 4 times and 4 sequences appeared twice, of the other sequences only single copies were obtained. These sequences displayed the usage of 11 different Vβ elements, both Dβ elements, and 9 different Jβ elements. Trimming of 1 to 7 nucleotides was found at the border of the V elements and up to 14 nucleotides were cut off from the J elements. Eight sequences contained N-regions and 6 sequences displayed P nucleotides adjacent to the D or J elements.
Sequences of TCRβ rearrangements of the ARTEMIS deficient Omenn syndrome patient
TRBV . | TRBD . | TRBJ . | .TRBV---------------------|P . | N . | P|------TRBD------|P . | N . | P|-------------------TRBJ. . | No. of clones . |
|---|---|---|---|---|---|---|---|---|
| V5-1 | J2.7 | .CTTTATCTTTGCGCCAGCAGCTTGG| | |--CCTACGAGCAGTACTTCGGGC. | 1× | ||||
| V5-1 | J2.7 | .CTTTATCTTTGCGCCAGCAGCTTG-| | |-TCCTACGAGCAGTACTTCGGGC. | 2× | ||||
| V5-1 | D1/2 | J2.7 | .CTTTATCTTTGCGCCAGCAGCTTGG| | |G-----------| | |---CTACGAGCAGTACTTCGGGC. | 1× | ||
| V5-1 | D2 | J2.7 | .CTTTATCTTTGCGCCAGC-------| | TCC | |-------CG-------| | T | |-TCCTACGAGCAGTACTTCGGGC. | 1× |
| V5-2 | J1.1 | .GTGTATCTCTGTGCCAGCAGCTT--| | |----CACTGAAGCTTTCTTTGGA. | 2× | ||||
| V5-2 | D1 | J1.1 | .CTGTATCTCTGTGCCAGCAGCTTG-| | |---ACAG-----| | TGTTCA|TGAACACTGAAGCTTTCTTTGGA. | 1× | ||
| V5-2 | D1 | J1.5 | .CTGTATCTCTGTGCCAGCAGC----| | |---ACAGGGGGC| | TA|TAGCAATCAGCCCCAGCATTTTG. | 1× | ||
| V5-2 | D1/2 | J1.2 | .CTGTATCTCTGTGCCAGCA------| | |-----AG-----| | |--------------ACCTTCGGT. | 1× | ||
| V7-2 | D1 | J1.5 | .GTGTATCTCTGTGCCAGCAGCTT--| | |---ACAGGG---| | |------TCAGCCCCAGCATTTTG. | 1× | ||
| V7-2 | D1/2 | J1.2 | .GTGTATCTCTGTGCCAGCAGCTT--| | A | |---ACAG-----| | |CTAACTATGGCTACACCTTCGGT. | 1× | |
| V9 | D1 | J1.3 | .TTGTATTTCTGTGCCAGCAGCGTA-| | |----CAG-----| | |----GGAAACACCATATATTTTG. | 1× | ||
| V9 | D1 | J1.3 | .TTGTATTTCTGTGCCAGCAGCG---| | |---ACAGG----| | |----GGAAACACCMTATATTTTG. | 1× | ||
| V9 | D1/2 | J1.2 | .TTGTATTTCTGTGCCAGCAGCG---| | |-GGACAG-----| | |CTAACTATGGCTACACCTTCGGT. | 1× | ||
| V9 | D1/2 | J2.7 | .TTGTATTTCTGTGCCAGCAGCG---| | |---A--------| | |CTCCTACGAGCAGTACTTCGGGC. | 1× | ||
| V12-5 | D2 | J2.6 | .TGTATTTTTGTGCTAGTGGTTT---| | |-----TAG--------| | |CTCTGGGGCCAACGTCCTGACTT. | 4× | ||
| V14 | D1 | J1.5 | .TTTATTTCTGTGCCAGCAGCCAAGA| | |----CA------| | A|TAGCAATCAGCCCCAGCATTTTG. | 1× | ||
| V14 | D1 | J2.7 | .TTTATTTCTGTGCCAGCAGCCAAGA| | |-------GGGGC| G | |------CGAGCAGTACTTCGGGC. | 1× | ||
| V15 | D2 | J2.5 | .TGTACCTGAGTGCCACCAGCAGA--| | |----CTAGCG------| | |--CAAGAGACCCAGTACTTCGGG. | 1× | ||
| V20-1 | D1 | J1.2 | .GCTTCTACATCTGCAGTGCTAGAG-| | |-GGACAGGG---| | |-----TATGGCTACACCTTCGGT. | 1× | ||
| V20-1 | D1/2 | J1.2 | .GCTTCTACATCTGCAGTGCTAG---| | |------GGGG--| | |-----TATGGCTACACCTTCGGT. | 2× | ||
| V20-1 | D1/2 | J1.2 | .GCTTCTACATCTGCAGTGCTAGAG-| | |GGGAC-------| | |--AACTATGGCTACACCTTCGGT. | 1× | ||
| V20-1 | D2 | J2.7 | .GCTTCTACATCTGCAGTGCTAG---| | |GGGACTAGCGGGGGG-| | T | |----TACGAGCAGTACTTCGGGC. | 1× | |
| V27 | D1 | J1.1 | .CTCTGTACTTCTGTGCCAGCAG---| | TTT | |--GACAGG----| | |-GAACACTGAAGCTTTCTTTGGA. | 1× | |
| V27 | D1 | J2.7 | .TGTACTTCTGTGCCAGCAGT-----| | TT | |-----AGGGGGC| GCCC | |--CCTACGAKCAGTACTTCGGGC. | 1× | |
| V27 | D1/2 | J2.2 | .TGTACTTCTGTGCCAGCAG------| | TT | |G-----------| | |-GAACACCGGGGAGCTGTTTTTT. | 1× | |
| V28 | D1 | J2.7 | .TGTACCTCTGTGCCAGCAG------| | CCCTG | CC|GGGACAGGGGG-| | |---CTACGAGCAGTACTTCGGGC. | 4× | |
| V29-1 | J1.4 | .GCATATATCTCTGCAGCGTT-----| | |--------GAAAAACTGTTTTTT. | 1× | ||||
| V29-1 | D1 | J2.7 | .GCATATATCTCTGCAGCGTT-----| | C | |---ACAGGG---| | |--CCTACGAGCAGTACTTCGGGC. | 2× |
TRBV . | TRBD . | TRBJ . | .TRBV---------------------|P . | N . | P|------TRBD------|P . | N . | P|-------------------TRBJ. . | No. of clones . |
|---|---|---|---|---|---|---|---|---|
| V5-1 | J2.7 | .CTTTATCTTTGCGCCAGCAGCTTGG| | |--CCTACGAGCAGTACTTCGGGC. | 1× | ||||
| V5-1 | J2.7 | .CTTTATCTTTGCGCCAGCAGCTTG-| | |-TCCTACGAGCAGTACTTCGGGC. | 2× | ||||
| V5-1 | D1/2 | J2.7 | .CTTTATCTTTGCGCCAGCAGCTTGG| | |G-----------| | |---CTACGAGCAGTACTTCGGGC. | 1× | ||
| V5-1 | D2 | J2.7 | .CTTTATCTTTGCGCCAGC-------| | TCC | |-------CG-------| | T | |-TCCTACGAGCAGTACTTCGGGC. | 1× |
| V5-2 | J1.1 | .GTGTATCTCTGTGCCAGCAGCTT--| | |----CACTGAAGCTTTCTTTGGA. | 2× | ||||
| V5-2 | D1 | J1.1 | .CTGTATCTCTGTGCCAGCAGCTTG-| | |---ACAG-----| | TGTTCA|TGAACACTGAAGCTTTCTTTGGA. | 1× | ||
| V5-2 | D1 | J1.5 | .CTGTATCTCTGTGCCAGCAGC----| | |---ACAGGGGGC| | TA|TAGCAATCAGCCCCAGCATTTTG. | 1× | ||
| V5-2 | D1/2 | J1.2 | .CTGTATCTCTGTGCCAGCA------| | |-----AG-----| | |--------------ACCTTCGGT. | 1× | ||
| V7-2 | D1 | J1.5 | .GTGTATCTCTGTGCCAGCAGCTT--| | |---ACAGGG---| | |------TCAGCCCCAGCATTTTG. | 1× | ||
| V7-2 | D1/2 | J1.2 | .GTGTATCTCTGTGCCAGCAGCTT--| | A | |---ACAG-----| | |CTAACTATGGCTACACCTTCGGT. | 1× | |
| V9 | D1 | J1.3 | .TTGTATTTCTGTGCCAGCAGCGTA-| | |----CAG-----| | |----GGAAACACCATATATTTTG. | 1× | ||
| V9 | D1 | J1.3 | .TTGTATTTCTGTGCCAGCAGCG---| | |---ACAGG----| | |----GGAAACACCMTATATTTTG. | 1× | ||
| V9 | D1/2 | J1.2 | .TTGTATTTCTGTGCCAGCAGCG---| | |-GGACAG-----| | |CTAACTATGGCTACACCTTCGGT. | 1× | ||
| V9 | D1/2 | J2.7 | .TTGTATTTCTGTGCCAGCAGCG---| | |---A--------| | |CTCCTACGAGCAGTACTTCGGGC. | 1× | ||
| V12-5 | D2 | J2.6 | .TGTATTTTTGTGCTAGTGGTTT---| | |-----TAG--------| | |CTCTGGGGCCAACGTCCTGACTT. | 4× | ||
| V14 | D1 | J1.5 | .TTTATTTCTGTGCCAGCAGCCAAGA| | |----CA------| | A|TAGCAATCAGCCCCAGCATTTTG. | 1× | ||
| V14 | D1 | J2.7 | .TTTATTTCTGTGCCAGCAGCCAAGA| | |-------GGGGC| G | |------CGAGCAGTACTTCGGGC. | 1× | ||
| V15 | D2 | J2.5 | .TGTACCTGAGTGCCACCAGCAGA--| | |----CTAGCG------| | |--CAAGAGACCCAGTACTTCGGG. | 1× | ||
| V20-1 | D1 | J1.2 | .GCTTCTACATCTGCAGTGCTAGAG-| | |-GGACAGGG---| | |-----TATGGCTACACCTTCGGT. | 1× | ||
| V20-1 | D1/2 | J1.2 | .GCTTCTACATCTGCAGTGCTAG---| | |------GGGG--| | |-----TATGGCTACACCTTCGGT. | 2× | ||
| V20-1 | D1/2 | J1.2 | .GCTTCTACATCTGCAGTGCTAGAG-| | |GGGAC-------| | |--AACTATGGCTACACCTTCGGT. | 1× | ||
| V20-1 | D2 | J2.7 | .GCTTCTACATCTGCAGTGCTAG---| | |GGGACTAGCGGGGGG-| | T | |----TACGAGCAGTACTTCGGGC. | 1× | |
| V27 | D1 | J1.1 | .CTCTGTACTTCTGTGCCAGCAG---| | TTT | |--GACAGG----| | |-GAACACTGAAGCTTTCTTTGGA. | 1× | |
| V27 | D1 | J2.7 | .TGTACTTCTGTGCCAGCAGT-----| | TT | |-----AGGGGGC| GCCC | |--CCTACGAKCAGTACTTCGGGC. | 1× | |
| V27 | D1/2 | J2.2 | .TGTACTTCTGTGCCAGCAG------| | TT | |G-----------| | |-GAACACCGGGGAGCTGTTTTTT. | 1× | |
| V28 | D1 | J2.7 | .TGTACCTCTGTGCCAGCAG------| | CCCTG | CC|GGGACAGGGGG-| | |---CTACGAGCAGTACTTCGGGC. | 4× | |
| V29-1 | J1.4 | .GCATATATCTCTGCAGCGTT-----| | |--------GAAAAACTGTTTTTT. | 1× | ||||
| V29-1 | D1 | J2.7 | .GCATATATCTCTGCAGCGTT-----| | C | |---ACAGGG---| | |--CCTACGAGCAGTACTTCGGGC. | 2× |
The left region indicates subgenic elements used for recombination. The middle region exposes the CDR3 regions of the recombinations sequenced (P denotes palindromic sequences; N denotes matrix independently added nucleotides). On the right, the number of identified clones of an individual recombination is depicted.
Radiosensitivity of primary skin fibroblasts
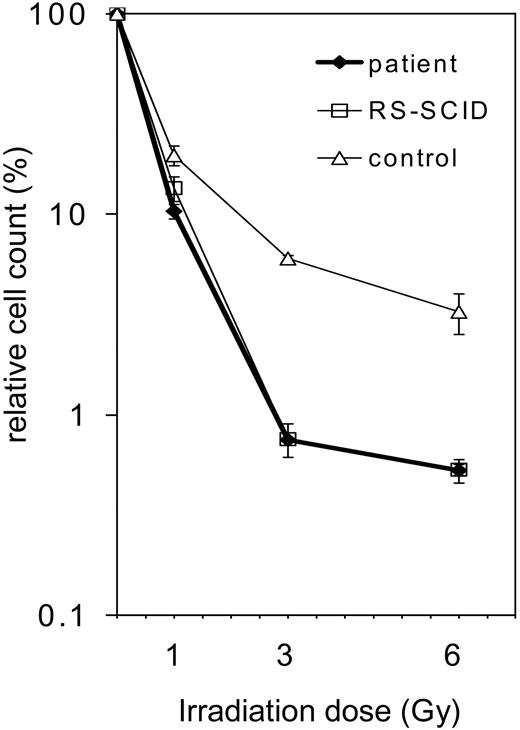
After the exclusion of RAG1 and RAG2 mutations (data not shown) and given the restriction of the TCR repertoire of our patient, we presumed that an underlying gene defect might be located somewhere else in the V(D)J recombination pathway. A general test for the function of NHEJ enzymes is radiosensitivity analysis. Defects in DNA-PKcs, KU70, KU80, ARTEMIS, Ligase IV, and XRCC4 can lead to a reduced tolerance of gamma irradiation. Therefore, patient and control primary skin fibroblasts were irradiated with 0, 1, 3, and 6 Gy, and cell survival after irradiation was determined (Figure 3). The survival of the cells from the patient with OS was identical to those of a complete ARTEMIS-deficient SCID patient. In addition, the OS cells exhibited reduced survival when compared with control cells, indicating their higher irradiation sensitivity.
TCR expression of the patient with OS. (A) Cell surface staining of the TCRαβ on CD3+ T cells of the patient and a healthy control in immunofluorescence flow cytometry. The percentage of TCRαβ cells in PBMCs as well as the intensity of TCRαβ expressed is comparable between the patient's T cells and a healthy control. (B) Comparison of the TCR repertoire of our patient (in black, foreground) with a patient with OS with a RAG-deficient genotype (in dark gray, middle) and an age-matched healthy control (in light gray, background). The TCR repertoire of the patient is severely restricted and matches the pattern of a RAG-deficient patient with OS, while the repertoire of the healthy control displays a Gaussian distribution.
TCR expression of the patient with OS. (A) Cell surface staining of the TCRαβ on CD3+ T cells of the patient and a healthy control in immunofluorescence flow cytometry. The percentage of TCRαβ cells in PBMCs as well as the intensity of TCRαβ expressed is comparable between the patient's T cells and a healthy control. (B) Comparison of the TCR repertoire of our patient (in black, foreground) with a patient with OS with a RAG-deficient genotype (in dark gray, middle) and an age-matched healthy control (in light gray, background). The TCR repertoire of the patient is severely restricted and matches the pattern of a RAG-deficient patient with OS, while the repertoire of the healthy control displays a Gaussian distribution.
Radiosensitivity of the fibroblasts of the patient with OS. Survival of fibroblasts after irradiation with 1, 3, or 6 Gy. Survival is calculated as percentage of cell count without irradiation (0 Gy). The patient's fibroblasts (♦) are compared with an age-matched healthy control (▵) and an ARTEMIS-deficient patient (RS-SCID; □). The OS patient's fibroblasts display the same irradiation sensitivity as the ARTEMIS-deficient control. Error bars indicate standard deviation.
Radiosensitivity of the fibroblasts of the patient with OS. Survival of fibroblasts after irradiation with 1, 3, or 6 Gy. Survival is calculated as percentage of cell count without irradiation (0 Gy). The patient's fibroblasts (♦) are compared with an age-matched healthy control (▵) and an ARTEMIS-deficient patient (RS-SCID; □). The OS patient's fibroblasts display the same irradiation sensitivity as the ARTEMIS-deficient control. Error bars indicate standard deviation.
ARTEMIS RNA expression and mutation analysis
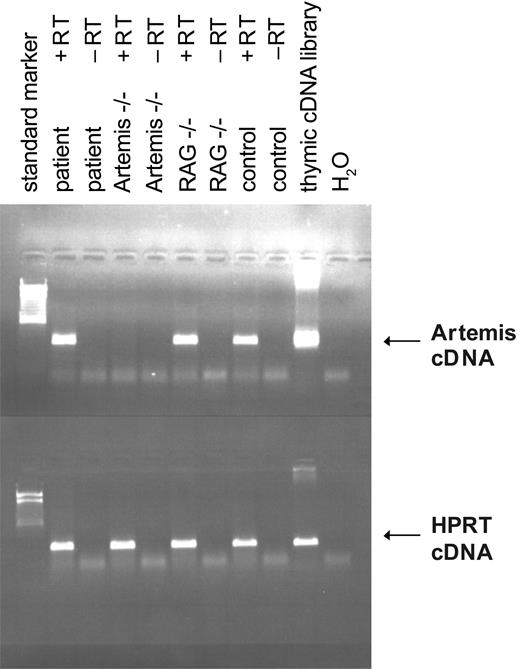
Since irradiation sensitivity was confirmed and RAG defects were excluded and since the phenotype of the patient was comparable to that of hypomorphic RAG-deficient OS, we tested whether ARTEMIS RNA was present in the patient's fibroblasts. We used an RT-PCR for a 300-bp fragment of the 5′ end of the ARTEMIS RNA including the start codon (Figure 4). The patient's cells expressed ARTEMIS RNA in levels comparable to control and RAG-deficient cells. The lack of RNA expression in cells from a known ARTEMIS-deficient SCID patient with a homozygous deletion of exons 1 to 3 served as a specificity control. A similar observation was made when complete ARTEMIS RNA was analyzed by RT-PCR (data not shown).
ARTEMIS RNA expression of the fibroblasts of the patient with OS. Reverse transcriptase–PCR of ARTEMIS RNA derived from fibroblasts of the patient with OS, an ARTEMIS-deficient patient, a RAG-deficient patient, and an age-matched healthy control. Amplification from a thymic cDNA library and H2O served as positive and negative controls, respectively. Transcription was performed with (+RT) and without (-RT) reverse transcriptase. Equal usage of cDNA was tested by amplifying HPRT cDNA.
ARTEMIS RNA expression of the fibroblasts of the patient with OS. Reverse transcriptase–PCR of ARTEMIS RNA derived from fibroblasts of the patient with OS, an ARTEMIS-deficient patient, a RAG-deficient patient, and an age-matched healthy control. Amplification from a thymic cDNA library and H2O served as positive and negative controls, respectively. Transcription was performed with (+RT) and without (-RT) reverse transcriptase. Equal usage of cDNA was tested by amplifying HPRT cDNA.
We concluded that at least 1 ARTEMIS allele was transcribed. To check whether the RNA might contain a mutation that could affect the function of the encoded protein, we sequenced the cDNA. This analysis revealed a compound heterozygosity (Table 3). The paternal mutation consisted of the replacement of thymidine by cytidine at the site of the protein translation initiation codon, resulting theoretically in an alteration of methionine to threonine at the protein level. The maternal mutation was found at the site of the putative active center of the protein: the codon 35 GAC (aspartatic acid) was present instead of the wild-type codon CAC (histidine).
Mutated codons of the patient's ARTEMIS alleles and predicted amino acid changes
. | Codon 1 . | Codon 35 . |
|---|---|---|
| Wild-type | ATG → Met | CAC → His |
| Patient | ||
| Paternal allele | ACG → Thr | Wild type |
| Maternal allele | Wild type | GAC → Asp |
. | Codon 1 . | Codon 35 . |
|---|---|---|
| Wild-type | ATG → Met | CAC → His |
| Patient | ||
| Paternal allele | ACG → Thr | Wild type |
| Maternal allele | Wild type | GAC → Asp |
Sequencing the genomic DNA of the patient and his relatives and of the CD3- and CD3+ fractions of the patient's mononuclear cells confirmed the compound heterozygosity for mutations in the ARTEMIS gene found in the patient's cDNA. Both mutated alleles were also identified in the genomic DNA of the deceased second brother (Figure 1).
Function of the mutated ARTEMIS proteins
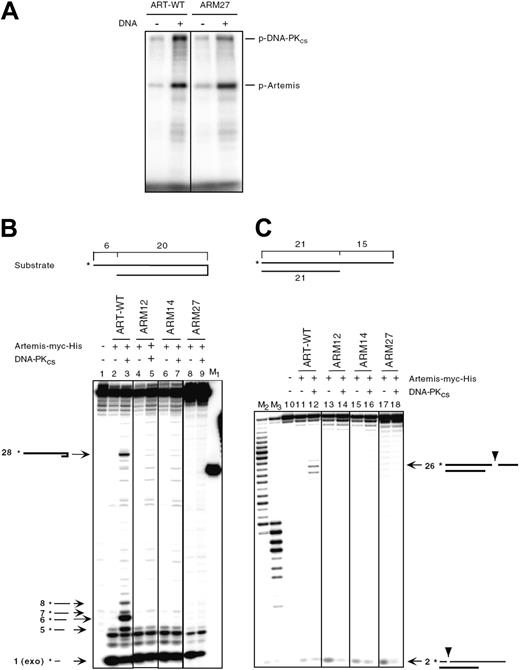
To examine the function of the ARTEMIS proteins encoded by the 2 patient alleles, we generated expression plasmids encoding WT-ARTEMIS and mutated cDNA (ARM27 encodes the mutant H35D, ARM32 encodes M1T) as well as 1 ARTEMIS expression plasmid starting with the next possible initiation codon at codon 8 (ARM29). All 3 ARTEMIS variants were overexpressed in HEK293T cells and were detected in the nucleus (Supplemental Figure 2B). Furthermore, ARM27 was phosphorylated in vitro to levels comparable to wild-type ARTEMIS (Figure 5A).
Two different substrates were used to test whether the mutant ARM27 protein still exhibited both exonuclease and endonuclease activities in an in vitro DNA cleavage assay. The first substrate was a DNA hairpin of 20-bp length with a 6 nt 5′ overhang (Figure 5B). The second substrate was a 21 bp double-strand DNA with a 15 nt 3′ overhang (Figure 5C). Only wild-type ARTEMIS (ART-WT) showed endonucleolytic activities. ARM27 (H35D), the ARTEMIS mutant protein encoded by the patient's maternal allele, did not reveal any endonucleolytic activity. Neither of the negative controls, ARM12 (H35A) and ARM14 (H115A), resembling active-site mutants, exhibited detectable hairpin opening or endonucleolytic activities, as was previously shown.33 ARTEMIS exonuclease activity was not impaired by the H35D mutation.
ARTEMIS mutant ARM32 (M1T) is expected to be expressed as wild-type ARTEMIS although at lower efficiencies, and therefore may be phosphorylated and may function like WT-ARTEMIS (see “Discussion”).33
The capability of ARTEMIS proteins to perform in the process of V(D)J rearrangement was investigated in a plasmid-based system that tests for inversional V(D)J recombination in vivo on an extrachromosomal substrate in primary dermal ARTEMIS-negative fibroblasts (Table 4).33 Furthermore, the OS fibroblasts were also analyzed for their ability to perform V(D)J recombination. Transfection efficiencies for the ARTEMIS-positive, ARTEMIS-negative, and OS fibroblasts ranged from 10% to 35%. In ARTEMIS-deficient cells V(D)J recombination was RAG2 as well as ARTEMIS dependent. Without the addition of the ARTEMIS expression plasmid, the ARTEMIS-deficient cells did not carry out significant numbers of detectable recombination events (up to 0.1% of transfected cells exhibited green fluorescence), whereas ARTEMIS rescued V(D)J recombination to a level of 4.8% to 6.5% in transfected cells. The OS fibroblasts showed also a strict dependence on the RAG2 expression plasmid but were able to perform V(D)J recombination even in the absence of a WT-ARTEMIS expression vector with 0.5% to 0.7% of transfected cells being positive, amounts that were intermediate between the ARTEMIS-negative (0.0%-0.1%) and ARTEMIS-positive fibroblasts (2.0%-2.8%). When expression plasmids for mutant ARTEMIS proteins (ARM27 encodes mutant H35D, ARM32 encodes mutant M1T, and ARM29 encodes mutant Del aa 1-7) were introduced into the ARTEMIS-negative fibroblasts, ARM27 and ARM29 were unable to complement the defect in V(D)J recombination at all. In contrast, the addition of an expression vector encoding for the paternal ARTEMIS variant (M1T) resulted in a V(D)J recombination efficiency of 2.1% to 2.7%. This corresponds to a recombination efficiency of between 37% and 48% when compared with the assay with WT-ARTEMIS.
V(D)J recombination assays in human primary dermal fibroblasts
. | Positive control . | -RAG2 . | -ART . | +ART . | +ARM27 (H35D) . | +ARM32 (M1T) . | +ARM29 (aa 8-692) . |
|---|---|---|---|---|---|---|---|
| ARTEMIS-positive fibroblasts | |||||||
| Experiment 1 | 54.09 | 0.02 | 1.71 | 1.99 | |||
| Experiment 2 | 65.52 | 0.08 | 2.61 | 2.51 | |||
| Experiment 3 | 67.43 | 0.06 | 2.48 | 2.84 | |||
| Omenn syndrome fibroblasts | |||||||
| Experiment 1 | 61.97 | 0.05 | 0.66 | 5.07 | |||
| Experiment 2 | ND | ND | ND | ND | |||
| Experiment 3 | 70.88/72.47* | 0.02/0.04* | 0.68/0.50* | 5.43/5.10* | |||
| ARTEMIS-negative fibroblasts | |||||||
| Experiment 1 | 54.15 | 0.05 | 0.01 | 5.66 | 0.14 | 2.09 | 0.04 |
| Experiment 2 | 64.35 | 0.06 | 0.06 | 4.77 | 0.07 | 2.30 | 0.10 |
| Experiment 3 | 65.22 | 0.05 | 0.02 | 6.50 | 0.05 | 2.65 | 0.09 |
. | Positive control . | -RAG2 . | -ART . | +ART . | +ARM27 (H35D) . | +ARM32 (M1T) . | +ARM29 (aa 8-692) . |
|---|---|---|---|---|---|---|---|
| ARTEMIS-positive fibroblasts | |||||||
| Experiment 1 | 54.09 | 0.02 | 1.71 | 1.99 | |||
| Experiment 2 | 65.52 | 0.08 | 2.61 | 2.51 | |||
| Experiment 3 | 67.43 | 0.06 | 2.48 | 2.84 | |||
| Omenn syndrome fibroblasts | |||||||
| Experiment 1 | 61.97 | 0.05 | 0.66 | 5.07 | |||
| Experiment 2 | ND | ND | ND | ND | |||
| Experiment 3 | 70.88/72.47* | 0.02/0.04* | 0.68/0.50* | 5.43/5.10* | |||
| ARTEMIS-negative fibroblasts | |||||||
| Experiment 1 | 54.15 | 0.05 | 0.01 | 5.66 | 0.14 | 2.09 | 0.04 |
| Experiment 2 | 64.35 | 0.06 | 0.06 | 4.77 | 0.07 | 2.30 | 0.10 |
| Experiment 3 | 65.22 | 0.05 | 0.02 | 6.50 | 0.05 | 2.65 | 0.09 |
ARTEMIS-positive fibroblasts, ARTEMIS-negative fibroblasts, and Omenn syndrome fibroblasts were tested in 3 independent experiments. Human primary dermal fibroblasts were cotransfected with V(D)J recombination substrate vector and expression plasmids coding for RAG1, RAG2, wild-type ARTEMIS (+ART), or mutant ARTEMIS proteins (+ARM27, 32, 29), respectively. As controls the same cotransfections without the expression plasmids coding for RAG2 (-RAG2) or wild-type ARTEMIS (-ART) were performed. As a positive control, a pre-recombined substrate vector was transfected. In each assay 5 × 104 fibroblasts were analysed by FACS. The percentages of recombination-positive cells out of the subpopulation of transfected fibroblasts are depicted.
ND indicates not determined.
Two independent values were ascertained
In vitro function of ARTEMIS mutants. (A) DNA-PKcs assay of ARTEMIS and ARTEMIS mutants. Wild-type ARTEMIS and mutant ARTEMIS ARM27 were subjected to a DNA-PKcs phosphorylation assay in the absence and presence of exogenous 35 bp DNA. The positions of phosphorylated DNA-PKcs (p-DNA-PKcs) and phosphorylated ARTEMIS (p-ARTEMIS) are indicated. The ARTEMIS mutant ARM27 is indistinguishable from wild-type ARTEMIS in the ability of being phosphorylated by DNA-PKcs. (B) In vitro nuclease assay of ARTEMIS and ARTEMIS mutants. To analyze 5′ overhang processing and hairpin opening activities of ART-WT, ARM27, ARM12, and ARM14, a 20-bp hairpin with a 6 nt 5′ overhang was used as the substrate. The asterisks indicate the positions of the radioactive label. Wild-type ARTEMIS and mutant ARTEMIS proteins were incubated with the substrate indicated in the absence and presence of DNA-PKcs. Autoradiographs of sequencing gels are shown. M1 marks the position of the hairpin opening product if the hairpin was opened at the tip. Positions and sizes of the major hairpin opening and overhang processing products are indicated by arrows and numbers (in nt). The exonucleolytic product by ARTEMIS (1 nt) is indicated by “exo” adjacent to the size of the product. Diagrams adjacent to the arrows are depicted to show the cleavage positions in the substrate that result in the corresponding products. (C) A 21-bp DNA with a 15 nt 3′ overhang was used as the substrate. The asterisks indicate the positions of the radioactive label. Diagrams adjacent to the arrows are depicted to show the cleavage positions in the substrate that result in the corresponding products. M2 and M3 were generated by incubating the labeled strand of the 3′ overhang substrate with Klenow enzyme at room temperature for 30 and 60 minutes, respectively.
In vitro function of ARTEMIS mutants. (A) DNA-PKcs assay of ARTEMIS and ARTEMIS mutants. Wild-type ARTEMIS and mutant ARTEMIS ARM27 were subjected to a DNA-PKcs phosphorylation assay in the absence and presence of exogenous 35 bp DNA. The positions of phosphorylated DNA-PKcs (p-DNA-PKcs) and phosphorylated ARTEMIS (p-ARTEMIS) are indicated. The ARTEMIS mutant ARM27 is indistinguishable from wild-type ARTEMIS in the ability of being phosphorylated by DNA-PKcs. (B) In vitro nuclease assay of ARTEMIS and ARTEMIS mutants. To analyze 5′ overhang processing and hairpin opening activities of ART-WT, ARM27, ARM12, and ARM14, a 20-bp hairpin with a 6 nt 5′ overhang was used as the substrate. The asterisks indicate the positions of the radioactive label. Wild-type ARTEMIS and mutant ARTEMIS proteins were incubated with the substrate indicated in the absence and presence of DNA-PKcs. Autoradiographs of sequencing gels are shown. M1 marks the position of the hairpin opening product if the hairpin was opened at the tip. Positions and sizes of the major hairpin opening and overhang processing products are indicated by arrows and numbers (in nt). The exonucleolytic product by ARTEMIS (1 nt) is indicated by “exo” adjacent to the size of the product. Diagrams adjacent to the arrows are depicted to show the cleavage positions in the substrate that result in the corresponding products. (C) A 21-bp DNA with a 15 nt 3′ overhang was used as the substrate. The asterisks indicate the positions of the radioactive label. Diagrams adjacent to the arrows are depicted to show the cleavage positions in the substrate that result in the corresponding products. M2 and M3 were generated by incubating the labeled strand of the 3′ overhang substrate with Klenow enzyme at room temperature for 30 and 60 minutes, respectively.
Discussion
Our results pinpoint a hypomorphic ARTEMIS allele as a novel genetic basis for Omenn syndrome. The clinical presentation as well as the immunophenotype of the patient was indistinguishable from that caused by hypomorphic RAG1/2 mutations.10,11,36,37 As with RAG-deficient OS, the patient had no peripheral B cells, but the presence of serum IgE suggests that Ig-producing plasma cells must be present. Previously, a thorough search for precursor B cells in the bone marrow of RAG OS patients failed completely to detect them, most likely due to experimental limitations caused by the high frequency of T cells within the bone marrow.37
The T cells of our patient expressed mostly an α/β TCR with wild-type intensity levels. An oligoclonal repertoire was assessed by spectratyping. Sequencing of the CDR3 regions of rearranged Vβ genes revealed no obvious conspicuous recombination events, similar to RAG OS. The differential diagnosis of a SCID with materno-fetal transfusion of T cells was ruled out by HLA and STR analyses.
Sequencing of the ARTEMIS gene showed a compound heterozygosity at both the RNA and genomic levels; the heterozygosity was confirmed by family analyses. The maternally mutated allele leads to an H35D substitution at the protein level. When investigated, the corresponding ARTEMIS mutant protein exhibited neither endonuclease nor hairpin opening activity in biochemical assays in vitro nor was the protein functional in an in vivo V(D)J recombination assay. Thus, we assume the defective maternal allele to be a null mutant. These results correspond closely to previous observations that amino acid 35 is an essential residue of the active center of ARTEMIS.33,38 By modeling the active center of the metallo-β-lactamase/β-CASP domain of ARTEMIS, we and others had found H35 to be involved in the coordination of 1 of the 2 divalent cations of the reactive site.33,38
The allele inherited from the father bears an alteration of the protein translation initiation codon (AUG to ACG). In our extrachromosomal V(D)J recombination assay, we scored residual function (about 40%) of the protein(s) translated from an expression plasmid carrying this paternal mutation.As was shown for RAG1 hypomorphic alleles,12 we considered the possibility that internal methionines may be used as translation start codons. This possibility was ruled out, since the V(D)J recombination efficiency of ARTEMIS expression vectors starting at 8M (this study), 39M, 121M, and 147M (data not shown) was as low as the negative control. Therefore, most likely, the ACG mutant of the first codon may be expressed as a wild-type protein in low levels. Unfortunately, with the available anti–ARTEMIS antibodies we were unable to achieve by immunoblotting a reproducible detection of ARTEMIS proteins either in control fibroblasts or in fibroblasts from the patient (data not shown). The phenomenon that an altered start codon initiates translation has already been described for other genes of mammalian cells and their viruses.39 The efficiency of protein synthesis of an ACG start codon was determined to be about 10% when compared with the AUG initiation codon.39 Homozygous start codon mutations may lead to severely affected phenotypes, as was shown for the desert hedgehog (DHH) gene,40 which result in a partial gonadal dysgenesis (PGD) accompanied by minifascicular neuropathy. Heterozygous mutations of the initiation codon of the beta-globin gene can result in β-thalassemia.41,42
V(D)J recombination efficiency in primary dermal patient's fibroblasts was present to a level of about 25% of wild-type efficiency, supporting the notion that residual ARTEMIS function is present in cells of the patient.
Most interestingly, while the endogenous ARTEMIS activity supported V(D)J recombination, the OS fibroblasts were as sensitive to irradiation as were cells from an ARTEMIS SCID patient. We speculate that the low amounts of functional ARTEMIS in the OS cells are insufficient to repair the multiple DNA double-strand breaks occurring in irradiated cells, but they can in part handle the number of breaks introduced on the test plasmids of the extrachromosomal V(D)J assay. In addition, it is presently unknown whether ARTEMIS binds with different affinities to respective DNA intermediates of V(D)J recombination or DNA repair.
In conclusion, we demonstrate that a patient with OS can result from an ARTEMIS defect with 1 null and 1 hypomorphic mutation, the latter most likely causing low expression of a wild-type protein. In vivo, ARTEMIS protein levels may become rate-limiting in V(D)J recombination and the absolute amount of ARTEMIS protein per cell needs tight regulation for the development of a diverse adaptive immune system.
Prepublished online as Blood First Edition Paper, February 24, 2005; DOI 10.1182/blood-2004-12-4861.
Supported by a grant from the Interdisciplinary Center for Clinical Research (IZKF) of the University Hospital Ulm (M.E.).
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors like to thank the members of the Schwarz and Lieber laboratories for helpful discussions. We thank S. Koerper for very kind help in confocal microscopy. We are grateful to I. Brackmann, I. Janz, and E.-M. Rump for excellent technical support.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal