Abstract
We have investigated the effects of coexpression of protein 4.2 and three protein-4.2 variants with band 3 in the Xenopus oocyte expression system. Normal protein 4.2 increased band-3–specific chloride transport in the oocytes. Protein 4.2 also coimmunoprecipitated with band 3 and colocalized with band 3 at the oocyte plasma membrane. The increase in band-3–mediated chloride transport and coimmunoprecipitation of protein 4.2 required the presence of the N-terminal cytoplasmic domain of band 3. Protein 4.2 also localized to the oocyte plasma membrane in the absence of band 3. The protein-4.2 variants 4.2 Tozeur (R310Q) and 4.2 Komatsu (D175Y) had impaired ability to bind to band 3 and these variants did not localize to the oocyte plasma membrane when expressed on their own or when coexpressed with band 3. Unexpectedly, 4.2 Nippon (A142T) behaved similarly to normal protein 4.2. In the absence of a crystal structure of protein 4.2, we propose a homology model of protein 4.2 based on the structure of the sequence-related protein transglutaminase. Using our results in oocytes and this homology model we speculate how these mutations affect protein 4.2 and result in hereditary spherocytosis.
Introduction
Protein 4.2 is a major constituent of the red blood cell (RBC) membrane skeletal network, present at about 200 000 copies per RBC.1 The protein-4.2 gene EPB42 contains 13 exons.2,3 There are two isoforms of protein 4.2, a minor 74-kDa isoform obtained when all these exons of the gene are expressed and a 72-kDa major isoform of protein 4.2 that lacks 30 of the 33 amino acids that are encoded by exon 1.4,5
The exact role of protein 4.2 in RBCs has not been elucidated, but protein 4.2 binds to the N-terminal cytoplasmic domain of the band-3 anion exchanger (AE1) and also interacts with ankyrin in RBCs.6,7 The presence of band 3 is critical for the stable incorporation of protein 4.2 into the RBC membrane, since human,8 mouse,9 and cow10 RBCs deficient in band 3 are also completely deficient in protein 4.2. The functional significance of the association of protein 4.2 with band 3 in RBCs is currently unclear. In liposomes containing reconstituted band 3, anion transport activity decreased in the presence of increasing amounts of protein 4.2,11 which suggested that protein 4.2 was a negative modulator of band-3 anion exchange activity. However, band-3–mediated anion transport activity has been reported to be unaffected12 or increased13 in human RBCs with protein-4.2 deficiency. In contrast, the absence of protein 4.2 slightly decreased anion transport activity in protein-4.2–null mouse RBCs.14 Protein 4.2 also associates with spectrin,15,16 ankyrin,7 and protein 4.17 in solution, although the significance of these associations in vivo has yet to be demonstrated. Recently, protein 4.2 has been shown to interact with CD47, which probably contributes to the anchoring of the Rh complex to the RBC skeleton.17,18 Protein 4.2 can probably interact with the inner leaflet of the lipid bilayer since it is fatty acylated with myristoyl and palmitoyl chains.19,20
The complete or nearly complete absence of protein 4.2 is associated with an atypical form of hereditary spherocytosis (HS), highlighting the important role 4.2 plays in maintaining the stability and flexibility of RBCs. To date, 9 protein-4.2 mutations have been found associated with HS. Five lead to the premature termination of translation.17,21-24 Other protein-4.2 variants result from missense mutations that yield amino acid substitutions: protein 4.2 Nippon (GCT>ACT; A142T25 ; occurs sporadically in the Japanese population and has been encountered once in whites),26 protein 4.2 Tozeur (CGA>CAA; R310Q),27 protein 4.2 Shiga (CGC>TGC; R317C),28 and protein 4.2 Komatsu (GAT>TAT; D175Y).29 Although there is a strict correlation between the occurrence of HS and the presence of these mutations (in homozygous or compound heterozygous states), the fact that these mutations are the direct cause of the absence of protein 4.2 has not been formally established. One possibility is that these point mutations affect the binding of protein 4.2 to band 3 and therefore its function. This has proved difficult to study using biochemical methods because of the tendency of purified protein 4.2 and protein-4.2 fragments to aggregate.
We have used the Xenopus oocyte expression system to investigate the effects of coexpression of protein 4.2 and 3 protein-4.2 variants with band 3. Normal protein 4.2 increased band-3–specific chloride transport induced in the oocytes, coimmunoprecipitated with band 3, and colocalized with band 3 at the oocyte plasma membrane and this was dependent on the presence of the N-terminal cytoplasmic domain of band 3. We used these properties of protein 4.2 displayed in oocytes to examine 3 protein-4.2 point mutations associated with HS (4.2 Nippon, A142T; 4.2 Komatsu, D175Y; 4.2 Tozeur, R310Q). Each protein-4.2 mutation had strikingly different effects on protein-4.2 function in this assay system. Protein 4.2 Tozeur had markedly reduced band-3 binding, whereas protein 4.2 Komatsu did not bind to band 3. Neither mutant protein had an effect on band-3 chloride transport nor did they colocalize with band 3 at the plasma membrane. Unexpectedly, protein 4.2 Nippon behaved in a similar way to normal 4.2 in oocytes, suggesting that the HS induced by this mutation is not directly due to the loss of band-3 binding. We have constructed a homology model for protein 4.2 using its amino acid sequence similarity with the transglutaminase family.1,2 We speculate how the locations of the different protein-4.2 HS mutations result in the different properties observed when expressed in oocytes or patient's RBCs.
Materials and methods
DNA constructs
The RBC band 3 Xenopus expression constructs BSXG1.B3 and BSXG.B3Mem (which encodes residues 360-911 of band 3) have been described previously.30 BSXG.4.2 was constructed by amplification of human protein 4.2 cDNA using complementary primers incorporating BamHI sites, cut with BamHI, and ligated into BglII-cut BSXG plasmid. Protein-4.2 mutations Nippon (A142T), Tozeur (R310Q), and Komatsu (D175Y) were constructed using BSXG4.2 and Quick Change Site-Directed Mutagenesis Kit (Stratagene, Amsterdam-Zuidoost, The Netherlands). BSXGAnk-R89 containing the 89-kDa band-3–binding domain (residues 1 to 827) of human RBC ankyrin (Ank1) was prepared using 3 overlapping clones provided by Dr K. John and Dr S. E. Lux (Harvard Medical School, Boston, MA). All constructs were confirmed by DNA sequencing.
Preparation of cRNAs and expression in Xenopus oocytes
The methods used for the synthesis of cRNA, cell-free translation, isolation of Xenopus oocytes, injection with cRNA, and chloride transport assay were as previously described.30 [35Cl]-chloride uptake into oocytes was measured over a 20-minute period. Band-3–specific chloride transport induced in the oocytes was estimated from the difference in chloride uptake in the absence and in the presence of 2 mM DNDS (4,4′-dinitro-2,2-stillbene disulfonate).
Coimmunoprecipitation of band 3 and protein 4.2
Xenopus oocytes were injected with 5 ng of the cRNAs for band 3, band-3 membrane domain, or protein 4.2 alone, or 5 ng cRNA of band 3 or band-3 membrane domain were coinjected with 5 ng cRNA of protein 4.2 or protein 4.2 Nippon, protein 4.2 Tozeur, or protein 4.2 Komatsu. The oocytes were labeled using Easy Tag [35S]-L-methionine (NEN, London, United Kingdom) for 48 hours and chased for 4 hours as described previously.30 Groups of 10 oocytes were homogenized in 100 μL immunoprecipitation buffer (IP buffer; 150 mM NaCl, 6 mM EDTA [ethylenediaminetetraacetic acid], 50 mM Tris-HCl [pH 7.4], 200 μM PMSF [phenylmethylsulfonyl fluoride], 10 μg/mL aprotinin, and 5 μg/mL each of leupeptin, pepstatin A, and antipain) before the addition of 400 μL IP buffer containing 2.5% Triton X-100 and 1% bovine serum albumin (BSA). The lysate was precleared with protein A–agarose beads (Bio-Rad, Hemel Hempstead, United Kingdom) and then incubated overnight at 4°C with protein A–agarose beads preloaded with anti–band-3 C-terminal murine monoclonal antibody BRIC155.31 The beads were washed with IP buffer containing 1% Triton X-100, the immune complexes were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and radiolabeled proteins were detected by fluorography.
Immunofluorescence and confocal microscopy
Xenopus oocytes were injected with band-3 cRNA, protein-4.2 cRNA alone, band-3 cRNA together with cRNA for protein 4.2, or a protein-4.2 variant and incubated at for 48 hours at 18°C. Individual oocytes were placed inside pieces of chicken liver (an inert support for sectioning), overlayed with embedding medium (optimum cutting temperature compound; BDH, Poole, United Kingdom), and quick-frozen in liquid nitrogen–cooled isopentane. The cells were frozen sectioned and air dried and sections were fixed and permeabilized with acetone at -20°C. After phosphate-buffered saline (PBS) washes, the sections were blocked for 1 hour using 10% BSA at room temperature. The sections were incubated with primary antibodies (undiluted hybridoma culture supernatant of mouse monoclonal anti–band-3 antibody BRIC155 or rabbit anti–protein 4.2 antibody27 diluted 1:500 in PBS containing 4% BSA) for 1 hour at room temperature. After PBS washes, sections were incubated for 1 hour with the appropriate secondary antibodies. After further washes, the preparations were mounted in Vectashield (Vector Labs, Burlingame, CA). Fluorescence imaging was performed using a Confocal Zeiss Axiovert 135 microscope (Carl Zeiss, Welwyn Garden City, United Kingdom) with a 40×/1.2 water immersion objective, and images were acquired with LSM 410 confocal system using an Ar/He-Ne laser or using a Leica TCS-NT confocal laser scanning microscope (Leica-Microsystems, Milton Keynes, United Kingdom) using a 63×/1.32 oil immersion objective equipped with a Kr/Ar laser. Images were processed with Abode Photoshop 6.0 software, and figures were assembled with Adobe Illustrator 10 software (Adobe, San Jose, CA).
Molecular modeling
The sequence of protein 4.2 was used to search the database of known protein 3-dimensional structures (pdb) using Blastp.32 This identified 2 candidate template structures for homology modeling, a transglutaminase from sea bream33 (1G0D; (resolution, 2.5 A; sequence identity, 32%) and from human34 (1KV3; resolution, 2.8 A; sequence identity, 34%). The sea bream structure had a more complete set of residues structurally defined so this was chosen as the template. A homology model was constructed by standard methods using InsightII and Discover (Accelrys, Cambridge, United Kingdom). The model had an appropriate disposition of charged and polar residues on the surface and hydrophobic residues in the core, indicating that the sequence is compatible with the template structure.
Results
Expression of protein 4.2 and protein-4.2 variants with band 3 in Xenopus oocytes
We examined the effects of expressing normal protein 4.2 and protein 4.2 carrying the Nippon, Komatsu, or Tozeur mutations on the anion transport function of band 3 using intact band 3 (B3) or the membrane domain alone of band 3 (BM). Purified in vitro–transcribed cRNAs for either band or BM were co-injected with cRNA for protein 4.2 or the protein-4.2 variants into Xenopus oocytes, and the band-3–specific anion transport was estimated from the stilbene-sensitive chloride uptake of chloride induced in the oocytes.
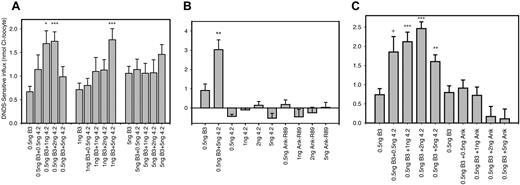
Coexpression of band 3 with protein 4.2 or ankyrin band-3–binding domain in Xenopus oocytes. Band-3 (B3) cRNA was coinjected into oocytes with the indicated concentration of protein 4.2 or Ank-R89 (Ank) cRNA. DNDS-sensitive chloride influx (20 min) was measured 24 hours after injection with cRNA, using groups of 10 to 15 oocytes as described previously.30 Results are shown as means ± SEM for comparisons with the relevant B3 alone sample by Student t test: ***P < .0005, **P < .001, *.002 <P < .005, and + .005 < P < .01. Panel A shows the effects of varying the B3 concentration from 0.5 ng to 5 ng using 0.5 ng to 5 ng protein 4.2. This suggests that the best combination of cRNA to use for observation of the effects of protein 4.2 on B3-specific transport is 0.5 ng B3 cRNA and 1 to 2 ng protein-4.2 cRNA. Panel B demonstrates that neither protein 4.2 nor Ank-R89 have a significant effect on endogenous chloride transport when 0.5 ng to 5 ng of cRNA is injected into oocytes. In comparison, the enhancement effect can be clearly observed when 0.5 ng of band 3 is co-injected with 5 ng protein 4.2. Panel C shows a comparison of the effects of protein 4.2 and Ank-R89 at 0.5 to 5 ng with B3 cRNA concentration held constant at 0.5 ng. Under these conditions, protein 4.2 enhanced B3-specific chloride, whereas Ank-R89 did not increase B3-specific chloride transport. This suggests that the protein-4.2 enhancement effect was due specifically to 4.2 binding to B3 and not simply a response to protein binding at the N-terminus of band 3.
Coexpression of band 3 with protein 4.2 or ankyrin band-3–binding domain in Xenopus oocytes. Band-3 (B3) cRNA was coinjected into oocytes with the indicated concentration of protein 4.2 or Ank-R89 (Ank) cRNA. DNDS-sensitive chloride influx (20 min) was measured 24 hours after injection with cRNA, using groups of 10 to 15 oocytes as described previously.30 Results are shown as means ± SEM for comparisons with the relevant B3 alone sample by Student t test: ***P < .0005, **P < .001, *.002 <P < .005, and + .005 < P < .01. Panel A shows the effects of varying the B3 concentration from 0.5 ng to 5 ng using 0.5 ng to 5 ng protein 4.2. This suggests that the best combination of cRNA to use for observation of the effects of protein 4.2 on B3-specific transport is 0.5 ng B3 cRNA and 1 to 2 ng protein-4.2 cRNA. Panel B demonstrates that neither protein 4.2 nor Ank-R89 have a significant effect on endogenous chloride transport when 0.5 ng to 5 ng of cRNA is injected into oocytes. In comparison, the enhancement effect can be clearly observed when 0.5 ng of band 3 is co-injected with 5 ng protein 4.2. Panel C shows a comparison of the effects of protein 4.2 and Ank-R89 at 0.5 to 5 ng with B3 cRNA concentration held constant at 0.5 ng. Under these conditions, protein 4.2 enhanced B3-specific chloride, whereas Ank-R89 did not increase B3-specific chloride transport. This suggests that the protein-4.2 enhancement effect was due specifically to 4.2 binding to B3 and not simply a response to protein binding at the N-terminus of band 3.
Figure 1A-C shows that chloride uptake by intact B3 was consistently increased 2- to 3-fold in the presence of normal protein 4.2. Figure 1A shows that this enhancement of chloride uptake by band 3 is dependent on coexpression of protein 4.2 with a low band-3 cRNA concentration (0.5 ng per oocyte) and with protein-4.2 concentrations at 1 to 2 ng/oocyte. Interestingly, when protein-4.2 cRNA was coinjected at 5 ng with 0.5 ng of band-3 cRNA, the chloride uptake was reduced (Figure 1A,C). We suggest that this is due to the 10-fold higher concentration of protein-4.2 cRNA competing with the low concentration of band-3 cRNA for the oocyte protein translation machinery. Similar effects were seen on band-3–dependent chloride uptake when low concentrations of band 3 are coinjected with high levels of other purified cRNAs (see next paragraph for Ank-R89). It can also be observed from Figure 1A that after a relatively small increment in band-3 concentration from 0.5 ng to 1 ng, only the 5 ng/oocyte protein-4.2 concentration had a significant effect on chloride uptake. Furthermore, when the band-3 concentration was increased further to 5 ng, no statistically significant enhancement of band-3–specific chloride transport occurred. Figure 1B shows that in the absence of band 3, protein 4.2 has no effect on endogenous oocyte chloride influx over the range of the protein-4.2 concentrations used in our experiments. Therefore, protein 4.2 can enhance the chloride transport of band 3 in oocytes, but this enhancement is most evident when a low concentration of band-3 cRNA is coinjected with protein 4.2.
We coexpressed the band-3–binding domain of erythroid ankyrin (Ank-R89) with band 3 to investigate whether enhancement of band-3–specific chloride transport is a general response to proteins binding to the N-terminus of band 3. Figure 1B shows that Ank-R89 alone has no significant effect on endogenous oocyte chloride uptake. Figure 1C shows that the coexpression of band 3 with Ank-R89 did not enhance band-3 chloride uptake under conditions where protein 4.2 does enhance band-3 transport. The presence of Ank-R89 appeared to reduce band-3 transport when injected at high cRNA concentrations (2 and 5 ng), suggesting that at these concentrations the Ank-R89 cRNA is competing for the oocyte protein machinery with the low B3 cRNA concentration (0.5 ng).
Figure 2A shows that there is no enhancement effect on band-3 chloride uptake when protein 4.2 is coexpressed with BM (which lacks the protein-4.2 binding site present on N-terminal cytoplasmic of B3). When 2 ng protein 4.2 was injected, the transport by BM was reduced, again suggesting that the coexpressed cRNA is competing with the low concentration (0.3 ng) of BM cRNA for the oocyte protein translation machinery. Taken together, these results suggest that the enhancement of band-3 anion transport activity by protein 4.2 requires the presence of the protein-4.2 binding site on the N-terminus of band 3 and does not appear to be a general response to the binding of proteins to the N-terminus of band 3.
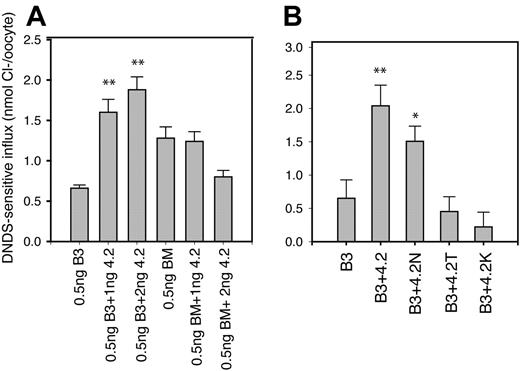
The effect of removal of band-3 N-terminus and of protein-4.2 HS mutations on the enhancement of B3-specific chloride transport by protein 4.2. The cRNAs encoding either band 3 (B3) or band-3 membrane domain (BM) were injected into oocytes either alone or with protein 4.2 or protein-4.2 variants. DNDS-sensitive chloride influx (20 min) was measured 24 hours after injection with cRNA using groups of 12 to 15 oocytes as described previously.30 Results are shown as means ± SEM for comparisons with the relevant B3 alone sample by Student t test: **P < .005, *P < .05. Similar results to those shown in panels A and B have been observed in at least 3 separate experiments. Panel A shows the effects of the removal of the B3 N-terminus (construct BM) on the 4.2 enhancement effect. B3 (0.5ng) or BM (0.3 ng) cRNAs were coinjected with 1 and 2 ng protein 4.2. Protein 4.2 only enhanced B3-specific chloride transport and not that of BM. This result is consistent with the protein-4.2 binding site being on the B3 N-terminus. Panel B shows the effect of 3 different HS mutations (4.2 Nippon, 4.2N; 4.2Tozeur, 4.2T; 4.2Komatsu, 4.2K) on the ability of protein 4.2 to enhance band-3 chloride transport. In panel B, 0.5 ng B3 cRNA was coinjected with 1.5 ng protein 4.2 or protein-4.2 variant cRNA. Protein 4.2 Nippon increased the chloride uptake induced by intact B3 nearly as much as normal protein 4.2. Neither protein 4.2 Tozeur nor protein 4.2 Komatsu had an effect on chloride uptake when coexpressed with intact B3.
The effect of removal of band-3 N-terminus and of protein-4.2 HS mutations on the enhancement of B3-specific chloride transport by protein 4.2. The cRNAs encoding either band 3 (B3) or band-3 membrane domain (BM) were injected into oocytes either alone or with protein 4.2 or protein-4.2 variants. DNDS-sensitive chloride influx (20 min) was measured 24 hours after injection with cRNA using groups of 12 to 15 oocytes as described previously.30 Results are shown as means ± SEM for comparisons with the relevant B3 alone sample by Student t test: **P < .005, *P < .05. Similar results to those shown in panels A and B have been observed in at least 3 separate experiments. Panel A shows the effects of the removal of the B3 N-terminus (construct BM) on the 4.2 enhancement effect. B3 (0.5ng) or BM (0.3 ng) cRNAs were coinjected with 1 and 2 ng protein 4.2. Protein 4.2 only enhanced B3-specific chloride transport and not that of BM. This result is consistent with the protein-4.2 binding site being on the B3 N-terminus. Panel B shows the effect of 3 different HS mutations (4.2 Nippon, 4.2N; 4.2Tozeur, 4.2T; 4.2Komatsu, 4.2K) on the ability of protein 4.2 to enhance band-3 chloride transport. In panel B, 0.5 ng B3 cRNA was coinjected with 1.5 ng protein 4.2 or protein-4.2 variant cRNA. Protein 4.2 Nippon increased the chloride uptake induced by intact B3 nearly as much as normal protein 4.2. Neither protein 4.2 Tozeur nor protein 4.2 Komatsu had an effect on chloride uptake when coexpressed with intact B3.
We next examined the effects of the protein-4.2 mutations associated with HS on band-3 anion transport activity under the conditions where normal protein 4.2 enhances band-3 chloride influx into oocytes (coexpressing 0.5 ng/oocyte B3 cRNA with 1.5 ng/oocyte protein-4.2 cRNA). Figure 2B shows the effects of coexpressing protein-4.2 variants on band-3–specific chloride uptake induced in Xenopus oocytes in comparison with normal 4.2. Protein 4.2 Nippon increased the chloride uptake induced by intact band 3 nearly as much as normal protein 4.2 (Figure 2B). Importantly, no significant change in the band-3 chloride uptake was obtained by coexpression of cRNAs for either protein 4.2 Tozeur or protein 4.2 Komatsu (Figure 2B-C), suggesting that these mutations impair protein-4.2 interaction with band 3.
It is possible that protein 4.2 enhances band-3–induced anion transport in oocytes by increasing the amount of band 3 at the cell surface. A cell-surface proteolysis assay has previously been used to estimate the amount of band 3 at the oocyte cell surface.30 However, we were unsuccessful in obtaining reproducible results using 0.5 ng band-3 cRNA (despite several attempts) in this assay, probably due to the low amounts of band 3 present at the surface35 (Supplemental Figure S1A-B; see the Supplemental Figure link at the top of the online article, available on the Blood website).
Since coexpression of glycophorin A (GPA) also enhances band-3–induced anion transport in oocytes,30 we also examined the effects of coexpression of both GPA and protein 4.2 on band-3–induced anion transport. The coexpression of both glycophorin cRNA and protein-4.2 cRNA did not further enhance band-3–induced anion transport above that observed when glycophorin A cRNA or protein-4.2 cRNA were injected alone with band 3 (results not shown).
Protein 4.2 coimmunoprecipitates with band 3
We investigated the ability of protein 4.2 and the protein-4.2 variants expressed in oocytes to bind to band 3. Although the enhancement effect on band-3 chloride transport was observed using 0.5 ng cRNA, this represents the synthesis of a very low amount of band-3 protein that would be difficult to detect immunologically. Therefore, for practical reasons we coinjected oocytes with 5 ng of purified cRNA of both band 3 and protein 4.2, assuming that although no significant effect on band-3 chloride transport is observed, both proteins would be synthesized at detectable levels and an association would still occur. Oocytes coexpressing band 3 or BM, either alone or with protein 4.2 or protein-4.2 variants, were metabolically labeled and C-terminal BRIC155 anti–band-3 antibody was used to immunoprecipitate band 3 under nondenaturing conditions. The immunoprecipitate from oocytes expressing band 3 alone contained only labeled bands corresponding to intact band 3 (Figure 3A, lane 3). When protein 4.2 was coexpressed with band 3, the band-3 immunoprecipitate contained an additional labeled band (Figure 3A, lane 4) that had the same mobility as labeled protein 4.2 prepared by cell-free translation of protein-4.2 cRNA (Figure 3A, lane 2). Protein 4.2 was not immunoprecipitated by anti–band-3 from oocytes coexpressing BM and protein 4.2 (Figure 3B, lane 4). No band with mobility corresponding to protein 4.2 was immunoprecipitated by anti–band-3 from oocytes expressing protein 4.2 alone or water-injected oocytes (Figure 3A-B, lanes 8-9). These data show that protein 4.2 coimmunoprecipitates with intact band 3 and this interaction is dependent on the presence of the N-terminal cytoplasmic domain of band 3. Importantly, Figure 3A (lane 5) protein 4.2 Nippon was immunoprecipitated from oocytes coexpressing band 3 and protein 4.2 Nippon but not from oocytes coexpressing BM and protein 4.2 Nippon (Figure 3B, lane 5). Similar coimmunoprecipitation experiments using protein 4.2 Tozeur and 4.2 Komatsu showed that less protein 4.2 Tozeur (24% ± 6% compared with normal protein 4.2; SEM n = 4) and no protein 4.2 Komatsu was coimmunoprecipitated with intact band 3 under these conditions (Figure 3A, lanes 6 and 7, respectively). As expected, none of the protein-4.2 variants was coimmunoprecipitated with BM (Figure 3B, lanes 5-7).
Coimmunoprecipitation of band 3 and protein 4.2 from metabolically labeled oocytes. Oocytes were injected with either 5 ng of B3 cRNA or 5 ng BM cRNA alone, or 5 ng B3 cRNA or BM cRNA were coinjected with 5 ng of protein 4.2, protein 4.2 Nippon (4.2N), protein 4.2 Tozeur (4.2T), or protein 4.2 Komatsu (4.2K) cRNA. Control oocytes were either injected with 5 ng protein-4.2 cRNA alone or left uninjected. Oocytes were incubated with 35[S]-labeled methionine for 48 hours and then chased with unlabeled amino acids for 4 hours. 35S-methionine–labeled proteins were immunoprecipitated using the B3 C-terminal antibody BRIC155 from homogenates derived from the equivalent of 5 oocytes. In each case 35S-methionine–labeled cell-free–translated (CFT) B3, BM, or protein 4.2 were also separated on duplicate SDS-PAGE gels to provide an indication of size. Panel A shows the results of the coimmunoprecipitation when using B3 run on an 8% SDS-PAGE gel. This shows that both protein 4.2 and protein 4.2 Nippon coimmunoprecipitate with band 3, but markedly less protein 4.2 Tozeur and no protein 4.2 Komatsu was coimmunoprecipitated under the same conditions. Panel B shows the results of coimmunoprecipitation using BM on a 10% SDS-PAGE gel. This shows that the ability to coimmunoprecipitate protein 4.2 with band 3 required the N-terminal cytoplasmic domain of band 3. Similar results to those shown in panel A and panel B have been observed in at least 3 independent experiments and the immunoprecipitations were carried out in duplicate for each experiment.
Coimmunoprecipitation of band 3 and protein 4.2 from metabolically labeled oocytes. Oocytes were injected with either 5 ng of B3 cRNA or 5 ng BM cRNA alone, or 5 ng B3 cRNA or BM cRNA were coinjected with 5 ng of protein 4.2, protein 4.2 Nippon (4.2N), protein 4.2 Tozeur (4.2T), or protein 4.2 Komatsu (4.2K) cRNA. Control oocytes were either injected with 5 ng protein-4.2 cRNA alone or left uninjected. Oocytes were incubated with 35[S]-labeled methionine for 48 hours and then chased with unlabeled amino acids for 4 hours. 35S-methionine–labeled proteins were immunoprecipitated using the B3 C-terminal antibody BRIC155 from homogenates derived from the equivalent of 5 oocytes. In each case 35S-methionine–labeled cell-free–translated (CFT) B3, BM, or protein 4.2 were also separated on duplicate SDS-PAGE gels to provide an indication of size. Panel A shows the results of the coimmunoprecipitation when using B3 run on an 8% SDS-PAGE gel. This shows that both protein 4.2 and protein 4.2 Nippon coimmunoprecipitate with band 3, but markedly less protein 4.2 Tozeur and no protein 4.2 Komatsu was coimmunoprecipitated under the same conditions. Panel B shows the results of coimmunoprecipitation using BM on a 10% SDS-PAGE gel. This shows that the ability to coimmunoprecipitate protein 4.2 with band 3 required the N-terminal cytoplasmic domain of band 3. Similar results to those shown in panel A and panel B have been observed in at least 3 independent experiments and the immunoprecipitations were carried out in duplicate for each experiment.
It was interesting that only a small proportion of protein 4.2 associated with band 3 (protein 4.2–band-3 ratio of approximately 1:25) in our immunoprecipitations. This suggests less protein 4.2 than band 3 may be present in the oocytes. Alternatively, only the mature population of band 3 and protein 4.2 in the oocytes may associate, as suggested by the cell-free translation results described in the next paragraph, or a proportion of band 3 may be located in oocyte compartments inaccessible to protein 4.2 (and vice versa). To investigate these possibilities, we attempted reciprocal immunoprecipitation studies using our anti–protein-4.2 antibody, but this antibody was unsuitable for this use (results not shown). We also attempted Western blotting of both proteins in oocytes injected with 5 ng of both band 3 and 4.2 cRNA. Although band-3 protein was successfully detected by Western blotting in our oocyte lysates (but only when injected at 5 ng cRNA or more), protein 4.2 was not detected with the anti–protein 4.2 antibody, even when 10 ng protein-4.2 cRNA was injected per oocyte and the amount of oocyte lysate used in the Western blot was doubled (10 oocytes; results not shown).
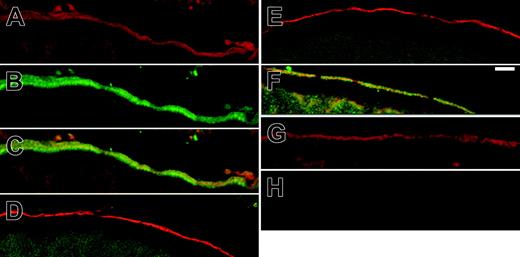
Colocalization of band 3 and 4.2 in Xenopus oocytes. Band-3 (0.5 ng) cRNA alone or B3 with 1.5 ng protein 4.2 or protein-4.2 variant cRNA were injected into Xenopus oocytes; these oocytes were processed for confocal imaging as described in “Materials and methods.” The sections were incubated sequentially with mouse monoclonal anti–band-3 antibody BRIC155, then rabbit anti–protein-4.2 antibody diluted 1:500, and these primary antibodies were detected using a goat antirabbit antibody conjugated to Alexa488 and a goat antimouse antibody conjugated to Alexa546. The micrographs in panels A-C were taken from the same oocyte coexpressing band-3 and 4.2 with (A) band-3 imaged with Alexa546 (red; B), 4.2 Alexa488 fluorescence (green; C), merged image of B3 (A) and 4.2 (B) showing colocalization at the plasma membrane (yellow). The images in panels D-F and H are the corresponding merged images for (D) band-3 and 4.2 Tozeur, (E) band-3 and 4.2 Komatsu, (F) band-3 and 4.2 Nippon, and (H) uninjected oocyte control. The image in panel G shows a section of an oocyte injected with band-3 alone, incubated with BRIC155, and then detected with anAlexa546 secondary. Only protein 4.2 (C) and protein 4.2 Nippon (F) colocalized with B3 at the plasma membrane. Scale bar = 10 μM.
Colocalization of band 3 and 4.2 in Xenopus oocytes. Band-3 (0.5 ng) cRNA alone or B3 with 1.5 ng protein 4.2 or protein-4.2 variant cRNA were injected into Xenopus oocytes; these oocytes were processed for confocal imaging as described in “Materials and methods.” The sections were incubated sequentially with mouse monoclonal anti–band-3 antibody BRIC155, then rabbit anti–protein-4.2 antibody diluted 1:500, and these primary antibodies were detected using a goat antirabbit antibody conjugated to Alexa488 and a goat antimouse antibody conjugated to Alexa546. The micrographs in panels A-C were taken from the same oocyte coexpressing band-3 and 4.2 with (A) band-3 imaged with Alexa546 (red; B), 4.2 Alexa488 fluorescence (green; C), merged image of B3 (A) and 4.2 (B) showing colocalization at the plasma membrane (yellow). The images in panels D-F and H are the corresponding merged images for (D) band-3 and 4.2 Tozeur, (E) band-3 and 4.2 Komatsu, (F) band-3 and 4.2 Nippon, and (H) uninjected oocyte control. The image in panel G shows a section of an oocyte injected with band-3 alone, incubated with BRIC155, and then detected with anAlexa546 secondary. Only protein 4.2 (C) and protein 4.2 Nippon (F) colocalized with B3 at the plasma membrane. Scale bar = 10 μM.
In contrast to the results obtained using the proteins expressed in oocytes, we were unsuccessful in coimmunoprecipitation of labeled protein 4.2 and band-3 proteins coexpressed from the cRNAs using the rabbit reticulocyte cell-free translation system and microsomes. Ank-R89 did immunoprecipitate with band 3 under these conditions (results not shown). This suggests that in order to become competent to interact with each other, either protein 4.2 and/or band 3 require posttranslational modifications that occur outside the endoplasmic reticulum or the cell-free translation system with microsomes is not competent to carry out these modifications.
Confocal imaging of protein 4.2 in oocytes
We next investigated the localization of band 3 and protein 4.2 and protein-4.2 variants in oocytes. Oocytes injected with band-3 cRNA alone, cryosectioned, and then incubated with band-3–specific antibody revealed that the band 3 was located at the plasma membrane (Figure 4G). In oocytes coexpressing band 3 and normal protein 4.2, confocal imaging using anti–band-3 and anti–protein-4.2 antibodies revealed an overlapping localization of the 2 proteins at the plasma membrane (Figure 4A-B and merged image 5C). This is as expected from much data that indicate that protein 4.2 associates with band 3 at the RBC plasma membrane (reviewed in Cohen et al1 ). Band 3 and protein 4.2 Nippon also colocalized at the oocyte plasma membrane (Figure 4F), but neither protein 4.2 Tozeur nor protein 4.2 Komatsu colocalized with band 3 at the plasma membrane (Figure 4D-E). This is consistent with these proteins having no effect on band-3 anion transport activity and exhibiting markedly reduced (4.2 Tozeur) or no binding (4.2 Komatsu) to band 3.
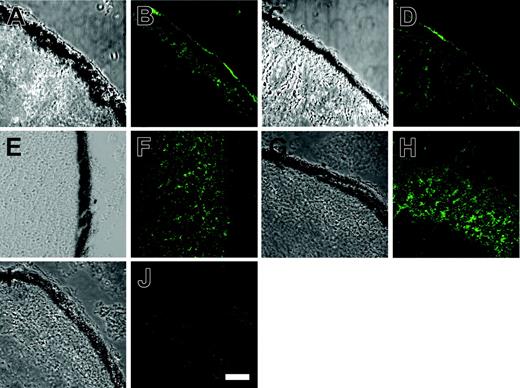
We investigated whether the plasma membrane association of protein 4.2 was dependent on its interaction with band 3 by expressing the protein 4.2 and protein-4.2 mutants alone in Xenopus oocytes. When normal protein 4.2 or protein 4.2 Nippon were expressed alone in oocytes some protein 4.2 was localized to the plasma membrane (Figure 5B,D). In contrast, both protein 4.2 Tozeur (Figure 5F) and protein 4.2 Komatsu (Figure 5H) had some intracellular staining, but very little immunostaining was observed at the oocyte plasma membrane. Therefore, at least when expressed in oocytes, protein-4.2 association with the plasma membrane is not dependent on the presence of band 3 and the 4.2 Tozeur and protein 4.2 Komatsu mutations appear to affect this association.
Discussion
To avoid the complications associated with using protein-4.2–deficient RBCs and the isolation of the proteins from RBC membranes in pure and native form, we have investigated the interaction of band 3 and protein 4.2 using the Xenopus oocyte expression system. Our results clearly show that when the proteins are coexpressed in Xenopus oocytes, protein 4.2 coimmunoprecipitated with band 3 and colocalized with band 3 at the oocyte plasma membrane. Importantly, protein 4.2 also enhanced band-3–mediated chloride transport in oocytes, and this was most evident when using a low band-3 cRNA concentration (0.5 ng/oocyte).
The reasons why the enhancement effect is less evident at higher band-3 cRNA concentrations remain unclear. Although protein 4.2 was detected by coimmunoprecipitation with band 3 in metabolically labeled oocytes or by using indirect immunofluorescence, our inability to detect protein 4.2 by Western blotting suggests that only low levels of protein 4.2 may be expressed in oocytes. Therefore, when large amounts of band 3 are synthesized, the amount of protein 4.2 may be limiting, resulting in a much-reduced enhancement of band-3 transport. In the presence of low levels of band 3 (eg, after injecting 0.5 ng cRNA) the 4.2 binding sites may become saturated, maximizing the enhancement effect in the transport assay. In addition, at high band-3–high protein-4.2 concentrations, competition between the 2 purified cRNAs for the oocyte's protein translation machinery may decrease net expression levels of band 3 relative to oocytes injected with just band-3 cRNA alone, effectively masking the 4.2 enhancement effect.
Protein 4.2 localizes to the plasma membrane in Xenopus oocytes. The cRNAs corresponding to normal protein 4.2 and protein 4.2 Nippon were injected alone into Xenopus oocytes and then processed for confocal imaging as described in “Immunofluorescence and confocal microscopy.” The oocyte sections were incubated with the rabbit anti–protein-4.2 antibody (diluted 1:500) and detected using a goat antirabbit antibody conjugated to Alexa488. Panels A, C, E, G, and I are bright field images and panels B, D, F, H, and J are corresponding fluorescent images. (A-B) Injected with 1.5 ng normal protein-4.2 cRNA, (C-D) injected with 1.5 ng protein 4.2 Nippon cRNA, (E-F) injected with 1.5 ng protein 4.2 Tozeur cRNA, (G-H) injected with 1.5 ng protein 4.2 Komatsu cRNA, and (I-J) uninjected oocyte control. Both protein 4.2 and protein 4.2 Nippon had immunoreactive protein localized to the plasma membrane in oocytes. The majority of protein 4.2 Tozeur and protein 4.2 Komatsu immunostaining was intracellular, with very little present at the oocyte plasma membrane. No protein-4.2 staining was observed in control oocytes. Scale bar in panel K = 30 μM.
Protein 4.2 localizes to the plasma membrane in Xenopus oocytes. The cRNAs corresponding to normal protein 4.2 and protein 4.2 Nippon were injected alone into Xenopus oocytes and then processed for confocal imaging as described in “Immunofluorescence and confocal microscopy.” The oocyte sections were incubated with the rabbit anti–protein-4.2 antibody (diluted 1:500) and detected using a goat antirabbit antibody conjugated to Alexa488. Panels A, C, E, G, and I are bright field images and panels B, D, F, H, and J are corresponding fluorescent images. (A-B) Injected with 1.5 ng normal protein-4.2 cRNA, (C-D) injected with 1.5 ng protein 4.2 Nippon cRNA, (E-F) injected with 1.5 ng protein 4.2 Tozeur cRNA, (G-H) injected with 1.5 ng protein 4.2 Komatsu cRNA, and (I-J) uninjected oocyte control. Both protein 4.2 and protein 4.2 Nippon had immunoreactive protein localized to the plasma membrane in oocytes. The majority of protein 4.2 Tozeur and protein 4.2 Komatsu immunostaining was intracellular, with very little present at the oocyte plasma membrane. No protein-4.2 staining was observed in control oocytes. Scale bar in panel K = 30 μM.
The enhancement of band-3 chloride transport by protein 4.2 and the ability to immunoprecipitate band 3 with protein 4.2 requires the presence of the N-terminal cytoplasmic domain of band 3, the known site of association of protein 4.2 on band 3.6 The effect of protein 4.2 on band-3 anion transport activity was not mimicked by coexpression of the 89-kDa band-3–binding domain of ankyrin-R with band 3, suggesting that the effect of protein 4.2 on band 3 is not a general effect of ligand binding to the N-terminal domain of band 3 but is specific to protein 4.2. Information on the binding site of protein 4.2 on the N-terminus of band 3 is limited, but natural mutations in the N-terminal domain of band 3 at Glu40,36 Gly130,37 and Pro32738 lead to protein-4.2 deficiency and HS. However, these mutations do not localize to a common region of the 3-dimensional structure of the cytoplasmic domain of band 3.39
Further work is necessary to demonstrate how protein 4.2 enhances anion transport induced by band 3 in oocytes. Native band 3 is a dimeric protein.40,41 One possibility is that at low concentrations of band-3 protein, the binding of protein 4.2 to the N-terminus of band 3 may promote the formation of band-3 dimers, since protein 4.2 is itself oligomeric.1 Interestingly, ankyrin is known to only interact with band-3 tetramers,42 which may explain the lack of a enhancement effect when band 3 was coinjected with the Ank-R89 cRNA. Other possible explanations are that protein-4.2 binding to the band-3 N-terminus may favor a more transport-active conformation of the band-3 molecule or that binding of protein 4.2 to the N-terminal domain of band 3 may perturb the structure of the band-3 dimer so that it becomes more open to the anion transport substrates.
An interesting observation was that protein 4.2 localized to the plasma membrane when expressed alone in Xenopus oocytes. Protein 4.2 was found to associate with the plasma membrane in Sf9 cells.20 Protein 4.2 is fatty acylated with myristoyl and palmitoyl chains,19,20 which could mediate the protein's association with the plasma membrane. This result is surprising in the context of the RBC, since the presence of band 3 is critical for the stable incorporation of protein 4.2 into the RBC membrane.8-10 In addition, natural mutations that occur in the N-terminal domain of band 3,36-38 which are assumed to affect the 4.2 binding site,39 lead to protein-4.2 deficiency. This suggests that although band 3 may not be required for an initial association of 4.2 with the plasma membrane, the interaction with band 3 is required for protein-4.2 retention at the RBC plasma membrane. Another possibility is that oocytes and Sf9 cells contain plasma membrane protein-4.2 binding sites that are absent in RBCs.
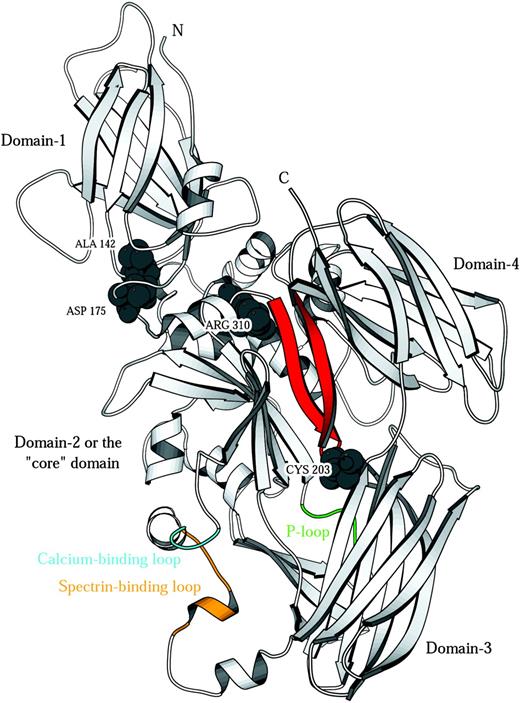
We studied the effects of 3 protein-4.2 point mutations associated with HS, 4.2 Nippon (A142T),25 4.2 Komatsu (D175Y),29 and 4.2 Tozeur (R310Q),27 on binding to band 3 and band-3–mediated anion transport in oocytes. These missense mutations cause an almost complete absence (in the case of protein 4.2 Nippon25 ) or total absence of RBC protein 4.2 (4.2 Tozeur and Komatsu).27,29 We have examined the effect of these different HS mutations by expression in Xenopus oocytes and can now compare this with our knowledge of their predicted location in the homology model of protein 4.2 (Figure 6).
Protein-4.2 homology model. Ribbon representation of the model of protein 4.2. Space-filled residues are the 3 mutation sites (Nippon, A142T; Komatsu, D175Y; Tozeur, R310Q) and the palmitoylation site (C203). The proposed band-3–binding site on protein 4.243 is located on a hairpin region (red) that also contains the known palmitoylation site of protein 4.243 (C203 in the hairpin reverse turn). Transglutaminases such as 1G0D,33 1KV3,34 and 1EVU44 (factor XIII) have a catalytic triad comprising H332, C333, and D355 (1G0D numbering). The corresponding residues in the model of protein 4.2 are T, Q, and D, hence the catalytic triad is missing and no transglutaminase activity is expected. It has been shown that protein 4.2 binds adenosine triphosphate and that binding is associated with a P-loop type sequence GEGQRGR (residues 346-352; green).45 The protein-4.2 model has 2 conserved glutamate residues whose side chains coordinate a calcium ion in other family members (protein 4.2: E469, E474; 1L9N: E443, E44846 ; 1EVU: E485, E49044 ). This putative calcium-binding site (cyan) is at the N-terminus of a 3-turn helix corresponding to helix-14 in the 1G0D template. The template structure has a stretch of 10 residues that do not appear in the crystal structure (due to disorder) connecting the C-terminal end of this helix (G0D1 residue) to the first strand of domain 3. Protein 4.2 also has a sequence insertion of 9 residues with respect to the template in this region. Hence the protein-4.2 model required a 19-residue linker between domains 2 and 3 to be modeled ab initio and must be considered as the least reliable part of the model with respect to structural detail. Interestingly, this region (colored orange) contains the sequence (482 EKEKMEREKD 491) that is responsible for the binding of spectrin by protein 4.2.16 The proximity of the calcium-binding loop to the spectrin-binding loop is suggestive of a role for calcium in controlling spectrin binding.
Protein-4.2 homology model. Ribbon representation of the model of protein 4.2. Space-filled residues are the 3 mutation sites (Nippon, A142T; Komatsu, D175Y; Tozeur, R310Q) and the palmitoylation site (C203). The proposed band-3–binding site on protein 4.243 is located on a hairpin region (red) that also contains the known palmitoylation site of protein 4.243 (C203 in the hairpin reverse turn). Transglutaminases such as 1G0D,33 1KV3,34 and 1EVU44 (factor XIII) have a catalytic triad comprising H332, C333, and D355 (1G0D numbering). The corresponding residues in the model of protein 4.2 are T, Q, and D, hence the catalytic triad is missing and no transglutaminase activity is expected. It has been shown that protein 4.2 binds adenosine triphosphate and that binding is associated with a P-loop type sequence GEGQRGR (residues 346-352; green).45 The protein-4.2 model has 2 conserved glutamate residues whose side chains coordinate a calcium ion in other family members (protein 4.2: E469, E474; 1L9N: E443, E44846 ; 1EVU: E485, E49044 ). This putative calcium-binding site (cyan) is at the N-terminus of a 3-turn helix corresponding to helix-14 in the 1G0D template. The template structure has a stretch of 10 residues that do not appear in the crystal structure (due to disorder) connecting the C-terminal end of this helix (G0D1 residue) to the first strand of domain 3. Protein 4.2 also has a sequence insertion of 9 residues with respect to the template in this region. Hence the protein-4.2 model required a 19-residue linker between domains 2 and 3 to be modeled ab initio and must be considered as the least reliable part of the model with respect to structural detail. Interestingly, this region (colored orange) contains the sequence (482 EKEKMEREKD 491) that is responsible for the binding of spectrin by protein 4.2.16 The proximity of the calcium-binding loop to the spectrin-binding loop is suggestive of a role for calcium in controlling spectrin binding.
Surprisingly, protein 4.2 Nippon had similar properties to normal protein 4.2. Protein 4.2 Nippon coimmunoprecipitated with band 3, colocalized with band 3 at the plasma membrane, and also enhanced band-3–mediated chloride transport. This suggests that the HS induced by this mutation is probably not directly due to defective binding to band 3. This mutant also retained the ability to associate with the plasma membrane in the absence of band 3. We speculate that the deficiency of protein 4.2 Nippon in the patient's RBCs may occur because its mRNA is unstable in RBCs, the protein is degraded before it associates with band 3, or this mutant protein 4.2 has an altered interaction with other cytoskeletal partner proteins. An examination of our model of protein 4.2 (Figure 6) shows that the residue position 142 is on a tight loop connecting strands on each face of the β-sandwich (domain 1) in the N-terminal domain of 4.2. The residue side chain points toward the hydrophobic core of the domain but is quite loosely packed. The model has space to readily accommodate the change from A to T, however the replacement of a rather hydrophobic side chain with a hydrophilic one is likely to destabilize the fold of the N-terminal domain and shorten the lifetime of the Nippon mutant.
Previous work showed that recombinant protein 4.2 Tozeur had normal binding to inside-out RBC membrane vesicles but was more susceptible to proteolysis than normal protein 4.2.27 It was proposed that the disappearance of protein 4.2 in mature RBCs was due to an imbalance between the destruction and synthesis of protein 4.2 Tozeur prior to reaching the plasma membrane.27 Our work supports this hypothesis, since protein 4.2 Tozeur coexpressed with band 3 in oocytes retained the ability to associate with the N-terminal cytoplasmic domain of band 3 (albeit at reduced levels), but no protein 4.2 Tozeur was observed to reach the oocyte plasma membrane and this mutant did not enhance band-3–mediated chloride transport. Residue position 310 is in a highly conserved stretch of sequence CTVLRCLG across known members of this fold family. The arginine is partly buried and its tip has electrostatic interactions with main chain carbonyls of residues E187 to N193, which are located on a turn between the C-terminus of a short helix (180-190) at the start of this domain (Figure 6, domain 2) and a hairpin (193-213). This hairpin region contains the proposed band-3–binding site43 and also contains C203, the palmitoylation site of protein 4.2.43 Although the mutation is quite conservative in a steric sense, it is expected that replacing the charged residue R310 with the neutral glutamine will disrupt the native hydrogen bonding and electrostatic interactions in this area. Such perturbations are likely to disrupt the band-3–binding site and also to reduce the stability of the protein.
The missense mutation in protein 4.2 Komatsu appeared to have the most severe effect on protein-4.2 function. This protein-4.2 variant did not coimmunoprecipitate with band 3 from oocytes, did not associate with the plasma membrane when expressed alone, did not colocalize with band 3 at the oocyte plasma membrane, and had no effect on band-3 chloride transport. Therefore, 4.2 Komatsu mutation may cause HS through its inability to bind the band-3 cytoplasmic domain, leading to its premature degradation. A similar dependence of protein 4.2 on the presence of band 3 is reported in human,8 mouse,9 and cow10 RBCs lacking band 3. Another possibility is that the protein 4.2 Komatsu mutation affects the stability of protein 4.2; this would result in the destruction of protein 4.2 before any interaction with band 3 can occur. Residue position 175 is on the linker strand between domains 1 and 2 in our model (Figure 6). The side chain of D175 is tightly packed into the hydrophilic interface between domain 1 and domain 2, forming a salt bridge to R65. Replacement of D175 by the larger, neutral tyrosine residue would cause severe steric clashes and break the salt bridge. It is expected that this will disrupt the domain 1/domain 2 interface. Since this interface is adjacent to the site of R301 and the band-3–binding region (as described in the previous paragraph for Tozeur 4.2), this mutation would result in a severe destabilization of 4.2 Komatsu and a likely abrogation of band-3 binding.
In summary, this work has demonstrated that the Xenopus oocyte expression system may be successfully used to investigate the effects of mutations in protein 4.2 on the interaction of protein 4.2 with band 3. An examination of the effects of three 4.2 HS mutations suggests that each mutation has a distinct effect on the function of protein 4.2. Protein 4.2 Nippon had properties similar to the normal protein, suggesting that the HS associated with this mutation is not due to altered binding with band 3 or membrane association. A reduced amount of protein 4.2 Tozeur was found to bind band 3 and did not associate with band 3 at the oocyte surface and had no affect on anion transport, consistent with this mutant being unstable. The protein 4.2 Komatsu variant affects a more important residue for maintaining the cellular function of protein 4.2 since this mutant had none of the properties observed for normal 4.2 expressed in Xenopus oocytes.
Prepublished online as Blood First Edition Paper, February 3, 2005; DOI 10.1182/blood-2004-05-1895.
Supported by grants from the Wellcome Trust, the Indo-French Center for the Promotion of Advanced Research (IFCPAR Project 1903.1), the Institut de la Santé et de la Recherche Médicale (Unité 473), the Institut National de la Santé et de la Recherche Médicale jointly with the Association Française contre les Myopathies (`Réseaux de Recherche sur les Maladies Rares'), the Assistance Publique-Hôpitaux de Paris, and the Faculté deMédecine Paris-Sud.
Presented in part at the 43rd Annual Meeting of the American Society of Hematology, Orlando, FL, December 7-11, 2001.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Prof D. Anstee for antibodies, R. Mushens for culturing antibodies, and Drs K. John and S. E. Lux for providing the overlapping ankyrin clones that were used to construct Ank-R89. We also thank the Medical Research Council for providing an infrastructure award to establish the School of Medical Sciences Imaging Facility and Dr M. Jepson and A. Leard for their assistance with the imaging studies.

![Figure 3. Coimmunoprecipitation of band 3 and protein 4.2 from metabolically labeled oocytes. Oocytes were injected with either 5 ng of B3 cRNA or 5 ng BM cRNA alone, or 5 ng B3 cRNA or BM cRNA were coinjected with 5 ng of protein 4.2, protein 4.2 Nippon (4.2N), protein 4.2 Tozeur (4.2T), or protein 4.2 Komatsu (4.2K) cRNA. Control oocytes were either injected with 5 ng protein-4.2 cRNA alone or left uninjected. Oocytes were incubated with 35[S]-labeled methionine for 48 hours and then chased with unlabeled amino acids for 4 hours. 35S-methionine–labeled proteins were immunoprecipitated using the B3 C-terminal antibody BRIC155 from homogenates derived from the equivalent of 5 oocytes. In each case 35S-methionine–labeled cell-free–translated (CFT) B3, BM, or protein 4.2 were also separated on duplicate SDS-PAGE gels to provide an indication of size. Panel A shows the results of the coimmunoprecipitation when using B3 run on an 8% SDS-PAGE gel. This shows that both protein 4.2 and protein 4.2 Nippon coimmunoprecipitate with band 3, but markedly less protein 4.2 Tozeur and no protein 4.2 Komatsu was coimmunoprecipitated under the same conditions. Panel B shows the results of coimmunoprecipitation using BM on a 10% SDS-PAGE gel. This shows that the ability to coimmunoprecipitate protein 4.2 with band 3 required the N-terminal cytoplasmic domain of band 3. Similar results to those shown in panel A and panel B have been observed in at least 3 independent experiments and the immunoprecipitations were carried out in duplicate for each experiment.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/10/10.1182_blood-2004-05-1895/6/m_zh80100578570003.jpeg?Expires=1769111507&Signature=WxYCsfopgNhzuYpzYCmFus-CE17uyiGiunsC14zylW~tXiUZmotBrL-YrboYa9OWVCHRDdVw7BnG8SaQ9ghVEgghHY3NXX-nsXDQe3rcZnQrL988l7kOZ0EQczf2JcHLtxzaUMjqus5Y3PeMvXKdTx7-hgi--x50kAHT-SnBOMrVH-KHuFsu0BxXaMOUVdBMgA8V6AHBZeLSE7KRII-wxACY61hQe5Mg6IRHvCFnGTmSaXovT7etJb043XWlaGAsF44CkN18FFdjFtoDNeW3x~tQ5lQ70wBZgho6z86kzPwVIw0N~k2dHPFScUNSbjlcQVc~XUXZz7ErZ5uJGU9kWw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal