Abstract
Type IV hemochromatosis is associated with dominant mutations in the SLC40A1 gene encoding ferroportin (FPN). Known as the “ferroportin disease,” this condition is typically characterized by high serum ferritin, reduced transferrin saturation, and macrophage iron loading. Previously FPN expression in vitro has been shown to cause iron deficiency in human cell lines and mediate iron export from Xenopus oocytes. We confirm these findings by showing that expression of human FPN in a human cell line results in an iron deficiency because of a 3-fold increased export of iron. We show that FPN mutations A77D, V162Δ, and G490D that are associated with a typical pattern of disease in vivo cause a loss of iron export function in vitro but do not physically or functionally impede wild-type FPN. These mutants may, therefore, lead to disease by haploinsufficiency. By contrast the variants Y64N, N144D, N144H, Q248H, and C326Y, which can be associated with greater transferrin saturation and more prominent iron deposition in liver parenchyma in vivo, retained iron export function in vitro. Because FPN is a target for negative feedback in iron homeostasis, we postulate that the latter group of mutants may resist inhibition, resulting in a permanently “turned on” iron exporter.
Introduction
Hemochromatosis is an iron overload disease characterized by excessive iron uptake through the enterocytes of the gut and subsequent deposition in the liver, spleen, and heart, leading to tissue damage. Currently 4 subtypes of hemochromatosis are recognized. In Caucasian populations disease is predominantly associated with mutations in the HFE gene, discovered in 19961 ; HFE-linked hemochromatosis is designated type I. A more severe form of the disease, juvenile hemochromatosis (type II hemochromatosis), is linked to mutations in either the recently identified hemojuvelin2 or the antimicrobial peptide hepcidin.3,4 Hepcidin is normally up-regulated in response to high serum iron, but it is unexpectedly low in patients with hemochromatosis because of mutations in HFE,5 hemojuvelin,2 and transferrin receptor 2 (TfR2).6,7 TfR2, which is expressed by hepatocytes,8,9 is mutated in hemochromatosis type III. The iron exporter ferroportin/iron-regulated transporter 1/metal transporter protein 1 (FPN/IREG-1/MTP-1; gene symbol SLC40A1) was discovered simultaneously by 3 groups.10-12 Since that time, numerous mutations in the gene have been implicated in patients from diverse ethnic origins with previously unexplained hemochromatosis. Iron overload disease because of a mutation in FPN is referred to as type IV hemochromatosis or ferroportin disease.13
FPN is expressed on basolateral membranes of mature intestinal enterocytes and the basal membrane of the placental syncytiotrophoblast.10-12 Another site of high expression of FPN is in macrophages, including Kupffer cells in the liver and in the red pulp of the spleen.12,14 These sites of expression are consistent with a role for FPN in transport of iron from the gut to the serum, from the mother to the fetus, and in iron recycling from effete red blood cells back into the serum.
HEK293T cells transfected with mouse Slc40a1 were shown to increase binding of iron-regulated protein-1 to an iron response element (IRE), indicating a low cellular iron state, and the same cells were shown to be low in the iron storage protein ferritin.12 Studies expressing FPN in Xenopus oocytes have shown that it mediates iron export, which may be enhanced in the presence of the multicopper-oxidase, ceruloplasmin.10
One of the groups that discovered FPN found that the weh zebra fish, which exhibits severe microcytic anemia, had an L167F mutation in FPN12 ; the human homologue of this substitution is L170F. In humans, disease-associated mutations of the SLC40A1 gene discovered to date are Y64N,15 A77D,16 N144D,17 N144H,18 N144T,19 V162Δ,20-22 Q248H,23,24 C326Y,25 and G490D26 in patients with iron overload and N174I, Q182H, and G323V in patients with hyperferritinaemia.27 Patients with ferroportin disease often have high serum ferritin despite normal transferrin saturation (at least in early stages of disease) and iron deposits principally in the liver macrophages but also in some cases in the hepatocytes.15-23,25,26,28 Patients often have a borderline anemia, and these cases do not respond well to phlebotomy.26,29
FPN mutations exert their disease phenotype in an autosomal dominant fashion, but it is unclear how mutations in FPN lead to hemochromatosis. The expression of the protein on macrophages and predominant loading of iron in these cells in patients with A77D and V162Δ mutations suggest a loss of function and haploinsufficiency model.16,30 In this scenario, one wild-type (wt) and one dysfunctional-mutated FPN are able to mediate iron absorption through the enterocyte but cannot meet the much greater demands of iron recycling from red blood cells by macrophages. Consequently, iron trapped in the macrophages leads to insufficient iron availability for erythropoiesis, and the bone marrow signals to the intestine to up-regulate absorption. An alternative possibility is a dominant-negative effect, in which the incorporation of mutant proteins in a membrane could disrupt the function of the wild type.
A second model for how mutations could cause disease is a gain of function model, whereby overactive FPN leads to inappropriately high transport of iron from the diet through the gut enterocytes, resulting in an iron overload phenotype, particularly observed in patients who have mutations at N14417,18 and C326Y.25 In contrast to the aforementioned patients with loss of function, these patients tend to have high transferrin saturation and high serum ferritin, and on liver biopsy iron is found throughout the tissue, not just confined to the macrophages17,18 (V.V., A.T.M.-C., Y.C., Chanin Lim Wongse, and K.I.H.R., manuscript in preparation: A novel ferroportin mutation (C326Y) causes a dominant form of inherited iron overload in the far east).
We report that transfection of human wtFPN into a human epithelial cell line results in surface expression of the protein and causes an up-regulation of transferrin receptor 1 (TfR1), reduced ferritin, lower iron accumulation, and increased cellular iron export. FPN mutants A77D, V162Δ, L170F, and G490D all lose this function and are retained inside the cell. However, mutants Y64N, N144H, N144D, Q248H, and C326Y locate to the cell surface and function comparably to wtFPN to deplete the cell of iron. We find no evidence for multimerization of FPN, and no evidence of physical or functional interaction between wild-type and mutant forms of the protein. We postulate that FPN mutations fall into 2 categories, distinguishable by disease phenotype, one causing disease by haploinsufficiency because of loss of function in 1 allele, while the other retains function and so could cause disease by resisting normal negative feedback signals.
Materials and methods
Antibodies
Fluorescein isothiocyanate (FITC)–conjugated anti–c-Myc antibody (clone 9E10) was obtained from Santa Cruz Biotechnologies (Santa Cruz, CA), biotinylated anti–c-Myc was obtained from Covance (Berkeley, CA), biotinylated anti-TfR1 and FITC-conjugated anti-CD8 were obtained from BD Biosciences (Oxford, United Kingdom). Unconjugated anti–c-Myc, biotinylated anti-FLAG (peptide marker sequence DYKDDDDY), anti-FLAG–horseradish peroxidase (HRP) conjugate, and goat anti–mouse immunoglobulin G (IgG)–phycoerythrin (PE) conjugate were obtained from Sigma (Poole, United Kingdom). Streptavidin-phycoerythrin and streptavidin-AlexaFluor568 conjugates were obtained from Molecular Probes (Invitrogen, Paisley, United Kingdom). MR12 was obtained from DAKO (Ely, Cambs, United Kingdom). An anti–FPN polyclonal rabbit sera was produced against the C-terminal amino acid sequence CGPDAKEVRKENQUANTSVV as used by Donovan et al,12 anti-FPNct.
Expression vectors
pcDNA3.1myc-His containing the gene encoding human FPN was a kind gift from Dr A. McKie (King's College, London, United Kingdom). Naturally occurring mutations of the gene in humans and the L170F analogous to the L167F weh zebra fish mutation were introduced using the Quikchange mutagenesis kit from Stratagene (La Jolla, CA) and oligonucleotides designed containing the required sequence, synthesized by Sigma-Genosys (Haverhill, United Kingdom). The wtFPN-FLAG construct was created by removing c-Myc tag with XbaI and ClaI (ClaI had been introduced after the stop codon using site-directed mutagenesis) and ligating with oligonucleotides 5′-ctagattacaaggatgacgacgataagtgat-3′ and 5′-ggatcacttatcgtcgtcatccttgtaat-3′ encoding the FLAG sequence. Plasmid containing CD8 was a kind gift from Dr George Gao (Oxford, United Kingdom) and was used as a control in transfection experiments, as CD8 has no known role in iron metabolism.
Maintenance and transfection of 293T cells
The human kidney epithelial line 293T was maintained in Dulbecco Modified Eagle medium containing 10% fetal calf serum (Sigma). The day before transfection flasks/plates were seeded at 50% confluency. Cells were transfected using Effectene (Qiagen, Crawley, United Kingdom) following the manufacturer's standard protocol. This gave 80% to 100% efficiency with a plasmid encoding green fluorescent protein.
Ferritin ELISA
Total cellular ferritin was measured from cell lysates using ferritin enzyme-linked immunosorbent assay (ELISA) kits obtained from Laguna Scientific (Laguna Niguel, CA). Cells were transfected as described in “Maintenance and transfection of 293T cells,” in the presence or absence of 1 mg/mL human holo transferrin (hTf) (Sigma). After culturing for 3 days, the cells were lysed in 0.5% NP-40 buffer (containing 50 mM trizma base, 5 mM EDTA (ethylenediaminetetraacetic acid), and 150 mM sodium chloride) at 2 × 107 cells/mL. Protein content of lysates was measured using the Bradford assay (Bio-Rad, Hercules, CA) and normalized by diluting with lysis buffer as necessary. Doubling dilutions were made, and 20 μL of these were used per well in the ferritin enzyme-linked immunosorbent assay ELISA. Each dilution was tested in triplicate and compared against ferritin standards provided at 0, 15, 80, 250, 500, and 1000 ng/mL. These gave a linear range between 15 and 250 ng/mL, and so dilutions resulting in values within this range were used to calculate nanograms of ferritin per 107 cells. It was generally found that hTf was required in the media to increase cellular ferritin levels to the linear range, as 293T cells naturally express low levels of ferritin.
Fluorescence-activated cell sorting (FACS) staining
Cells were tested for surface expression of TfR1 by flow cytometry as described previously.31 Briefly, 2 days after transfection, cells were removed from the flasks by incubation with phosphate-buffered saline (PBS) containing 1 mM EDTA, washed, and incubated at 4°C with anti–c-Myc–FITC or anti–CD8-FITC and anti–TfR1-biotin. As a negative control, cells were incubated with MR12. The cells were washed and then incubated with streptavidin-phycoerythrin to reveal TfR1 levels; control MR12-exposed cells were incubated with either streptavidin-phycoerythrin or goat anti–mouse IgG phycoerythrin to give background fluorescence. The cells were washed and fixed and then analyzed using a Becton Dickinson FACSCalibur and Cellquest software (Oxford, United Kingdom). Transfected cells positive for CD8 or c-Myc were gated on and analyzed for their surface TfR1 expression levels.
59Fe accumulation and 59Fe release
For iron accumulation, 293T cells were incubated with CD8, wtFPN, or FPN mutant DNA-transfection complexes for 8 hours, then 40 μg/mL 59Fe-labeled human transferrin, prepared as described,31 was added to the cultures. Twenty hours later cells were harvested, washed, and counted, and iron accumulation per 106 cells was determined as described.31 For iron release experiments, 293T cells were preloaded with 59Fe using 40 μg/mL 59Fe-Tf for 24 hours before being transfected with CD8, wtFPN, or FPN mutants for 15 hours. Cells were then harvested, washed, and counted, and cellular iron per 016 cells was determined before reculturing the cells in serum-free medium (Pro293a-CDM from BioWhittaker [Walkersville, MD], used to reduce the effects of bovine-Tf–mediated reuptake of exported 59Fe during the export incubation period) for up to 36 hours. At various time points, aliquots of 106 cells were removed from the cultures and collected by centrifugation, and 59Fe exported into the supernatant was measured. The percentage of 59Fe export was calculated as 59Fe in the supernatant at each time point divided by cellular 59Fe at time zero multiplied by 100. Cell viability in Pro293a-CDM after 36 hours of culture was more than 95%.
Immunofluorescence
293T cells were grown to 30% confluence on 8-well glass chamber slides (BD Falcon, Heidelberg, Germany) and cotransfected with wtFPN-FLAG and one of the c-Myc–tagged FPN constructs encoding either wtFPN, A77D, V162Δ, or C326Y FPN. Two days later, medium was removed, and cells were fixed and permeabilized using Cytofix/Cytoperm (Becton Dickinson [BD]), then incubated with anti–c-Myc-FITC and anti–FLAG-biotin antibodies diluted in Perm/Wash (BD). Cells were then washed and incubated with strepdavidin-AlexaFluor568 in Perm/Wash, before washing, fixing, and mounting in Vectashield containing the DNA dye DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA). Cells were imaged under Immersol immersion oil using a 100 × objective lens on a Zeiss Axiovert microscope (Zeiss Jena, Germany) equipped with epifluorescence illumination and running a Hal100 camera (Hamamatsu, Hamamatsu City, Japan). Images were captured and processed with OpenLab 3.5 software (Improvision, Lexington, MA).
Western blot
Cells were cotransfected with wtFPN-FLAG and wtFPN- or mutant FPN–c-Myc or CD8 for the immunofluorescence experiments. The cells were lysed and immunoprecipitated with anti–c-Myc, anti-FLAG, anti-FPNct, or antiCD8 at 15 μg/mL. Immunoprecipitates were run on a 12% acrylamide gel and blotted onto nitrocellulose membrane (Amersham, Chalfont St Giles, Buckinghamshire, United Kingdom). The membrane was probed with anti–FLAG-HRP antibody at 5 μg/mL in PBS containing 5% bovine serum albumin (BSA), and proteins were visualized with electrochemiluminescence (ECL) reagent (Amersham)
Results
Effect of FPN and FPN mutants on cell-surface TfR1 and intracellular ferritin
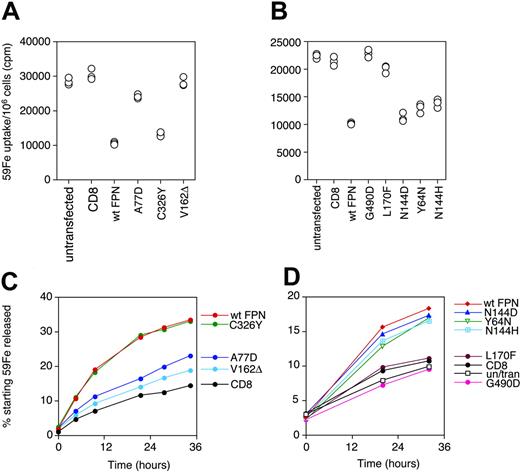
We first assayed the effect of FPN expression on a cell's iron status by looking at levels of expression of IRE-dependent proteins TfR1 and ferritin. 293T cells were transfected with wtFPN and mutant FPN constructs. Two days after transfection, cells were analyzed by 2-color flow cytometry to determine surface TfR1 levels on transfected cells. Cells transfected with CD8 were used as a negative control, as CD8 is a membrane-bound protein with no known role in iron metabolism. Figure 1A top left shows that as expected, untransfected cells have the same amount of TfR1 on the surface as those transfected with CD8. Thus, cells transfected with FPN can be compared with those transfected with CD8. Expression of wtFPN produces an increase in TfR1 on the cell surface, indicating iron deficiency. Disease-associated mutants A77D, V162Δ, and G490D do not cause an up-regulation of TfR1; however, Q248H, C326Y, N144H, N144D, and Y64N up-regulate TfR1 in the same way as wild type. L170F, the mutation found in the weh zebra fish, elicits a partial increase in TfR1 expression, but not to the same extent as wild type. Levels of the intracellular iron storage protein ferritin, whose expression is correlated with and controlled by cellular iron, were measured by ELISA. We performed this assay on cells grown in 1 mg/mL hTf because 293T cells are normally low in ferritin (L.M.S., unpublished data, 2004; 293T cells contain 20-50 ng ferritin per 107 cells depending on cell cycle, while HeLa cells contain 60-100 ng ferritin per 107 cells), and so, in some cases, a clearer result could be obtained by increasing the starting levels. Expression of wtFPN leads to a 4-fold reduction in ferritin (Figure 1B) compared with control cells transfected with CD8. FPN mutants A77D, V162Δ, L170F, and G490D do not cause a significant reduction in ferritin, but C326Y, Q248H, N144H, N144D, and Y64N behave similarly to wtFPN. These ELISA data agree with ferritin levels assayed by permeablizing cells and analyzing for ferritin by flow cytometry (data not shown). The data shown in Figure 1 indicate that as for mouse Fpn,11 human FPN causes iron deficiency when expressed in human cells.
Effect of ferroportin and ferroportin mutants on cell-surface transferrin receptor 1 and intracellular ferritin levels. (A) 293T cells at 30% to 50% confluence were transiently transfected with vectors encoding CD8, wtFPN–c-Myc, and the following c-Myc–tagged FPN mutants: C326Y, A77D, V162Δ, N144H, N144D, G490D, Y64N, Q248H, and L170F. After 2 days cells were double stained for CD8 or c-Myc tag and surface TfR1 and analyzed by 2-color flow cytometry. Transfected cells (> 70% of the population) expressing either CD8 or c-Myc were gated on and compared for their surface TfR1 levels. A comparison (top) is shown of TfR1 levels on cells positive for CD8 (gray filled histogram) versus untransfected cells (black line), demonstrating that expression of CD8 has no effect on TfR1. In all other panels, the gray-shaded histogram is TfR1 on CD8-transfected cells, and TfR1 on cells expressing wtFPN or mutant FPN is shown by a black line. Compared with CD8, TfR1 is increased on cells expressing wtFPN, indicating iron deficiency. C326Y, N144H, N144D, Q248H, and Y64N all increase expression of TfR1 in the same way as wild type, but A77D, V162Δ, and G490D do not increase TfR1 compared with CD8. L170F partially increases TfR1. The filled purple histogram represents the background fluorescence of cells stained with an isotype control antibody. (B) 293T cells were transfected as in panel A, but this time in the presence of 1 mg/mL Tf to increase background ferritin levels. After 2 days, cells were analyzed for total cell ferritin by ELISA. Cells were lysed in NP-40 at 107 cells/mL, doubling dilutions were made, and 20 μL was transferred to ELISA plate in triplicate. Graph shows mean total ferritin in ng/107 cells (± 95% CI [confidence interval]). Cells transfected with wtFPN have around 40 ng ferritin/107 cells, a 4-fold reduction compared with untransfected and control cells transfected with CD8. C326Y, N144H, N144D, Q248H, and Y64N all decrease ferritin levels in the same way as wild type, but A77D, V162Δ, L170F, and G490D do not significantly reduce ferritin. Asterisk indicates significance compared with untransfected or CD8 control (P < .001 by Student t test). Data shown is representative of 2 or more experiments for each mutant.
Effect of ferroportin and ferroportin mutants on cell-surface transferrin receptor 1 and intracellular ferritin levels. (A) 293T cells at 30% to 50% confluence were transiently transfected with vectors encoding CD8, wtFPN–c-Myc, and the following c-Myc–tagged FPN mutants: C326Y, A77D, V162Δ, N144H, N144D, G490D, Y64N, Q248H, and L170F. After 2 days cells were double stained for CD8 or c-Myc tag and surface TfR1 and analyzed by 2-color flow cytometry. Transfected cells (> 70% of the population) expressing either CD8 or c-Myc were gated on and compared for their surface TfR1 levels. A comparison (top) is shown of TfR1 levels on cells positive for CD8 (gray filled histogram) versus untransfected cells (black line), demonstrating that expression of CD8 has no effect on TfR1. In all other panels, the gray-shaded histogram is TfR1 on CD8-transfected cells, and TfR1 on cells expressing wtFPN or mutant FPN is shown by a black line. Compared with CD8, TfR1 is increased on cells expressing wtFPN, indicating iron deficiency. C326Y, N144H, N144D, Q248H, and Y64N all increase expression of TfR1 in the same way as wild type, but A77D, V162Δ, and G490D do not increase TfR1 compared with CD8. L170F partially increases TfR1. The filled purple histogram represents the background fluorescence of cells stained with an isotype control antibody. (B) 293T cells were transfected as in panel A, but this time in the presence of 1 mg/mL Tf to increase background ferritin levels. After 2 days, cells were analyzed for total cell ferritin by ELISA. Cells were lysed in NP-40 at 107 cells/mL, doubling dilutions were made, and 20 μL was transferred to ELISA plate in triplicate. Graph shows mean total ferritin in ng/107 cells (± 95% CI [confidence interval]). Cells transfected with wtFPN have around 40 ng ferritin/107 cells, a 4-fold reduction compared with untransfected and control cells transfected with CD8. C326Y, N144H, N144D, Q248H, and Y64N all decrease ferritin levels in the same way as wild type, but A77D, V162Δ, L170F, and G490D do not significantly reduce ferritin. Asterisk indicates significance compared with untransfected or CD8 control (P < .001 by Student t test). Data shown is representative of 2 or more experiments for each mutant.
Iron-deficient phenotype is due to iron release; some mutants do not release iron as effectively as wtFPN
We measured accumulation of radioactively labeled Fe in cells transfected with wtFPN and mutant FPN, using untransfected cells and cells transfected with CD8 as controls (Figure 2A-B). Over 20 hours CD8-transfected and -untransfected cells accumulated iron, leading to approximately 30 000 cpm/106cells. Cells transfected with FPN displayed a 2- to 3-fold reduction in iron accumulation. The FPN mutants A77D, V162Δ, L170F, and G490D, which were shown in Figure 1 to lose the wtFPN effect of up-regulating TfR1 and down-regulating ferritin, were shown in this assay to have no effect in reducing iron accumulation. However, mutants C326Y, N144H, N144D, and Y64N, which retain the wt effect, were shown to accumulate lower levels of iron, similar to wild type.
We then looked at iron release from the cells over 36 hours (Figure 2C-D). FPN-transfected cells were able to release up to 35% of their initial iron, while release from control CD8-transfected cells was not more than about 14%. In several different experiments, mutants C326Y, N144H, N144D, and Y64N were shown to cause a release of iron in amounts comparable to wild type. However mutants A77D, V162Δ, L170F, and G490D all failed to increase iron release from the cell and released amounts similar to control CD8-transfected cells.
Effect of wtFPN and FPN mutants on iron accumulation and iron release. (A-B) Iron accumulation. 293T cells were transfected as for Figure 1, grown for 8 hours, and then fed 40 μg/mL 59Fe-Tf for 20 hours. Cells were then washed 3 times and counted, and cellular 59Fe in the pellets was measured. Each point represents the uptake into 1 aliquot of 106 cells. Panels A and B show that cells transfected with wtFPN accumulate 35% to 40% as much radioactive iron as untransfected control or CD8 transfectants. Mutants A77D, V162Δ, L170F, and G490D are able to accumulate 59Fe to the same levels as CD8 control cells, while in the case of C326Y, N144D, Y64N, and N144H levels are lower and comparable to wild type. (C-D) Iron release. Cells were grown in 40 μg/mL 59Fe for 24 hours, then washed and transfected as in panels A and B. Fifteen hours later cells were harvested, washed, and resuspended at 106 cells/mL in serum-free medium, and 59Fe export was measured at the intervals shown up to 36 hours after harvesting. Data are presented as a percentage of starting radioactivity when cells were harvested. Panels C and D show that cells transfected with wtFPN release up to 3 times as much 59Fe as untransfected control or CD8 transfectants. Mutants A77D, V162Δ, L170F, and G490D are only able to release 59Fe to levels similar to CD8 control cells, while in the case of C326Y, N144D, Y64N, and N144H iron release is higher and comparable to wtFPN. These results are representative of 3 experiments.
Effect of wtFPN and FPN mutants on iron accumulation and iron release. (A-B) Iron accumulation. 293T cells were transfected as for Figure 1, grown for 8 hours, and then fed 40 μg/mL 59Fe-Tf for 20 hours. Cells were then washed 3 times and counted, and cellular 59Fe in the pellets was measured. Each point represents the uptake into 1 aliquot of 106 cells. Panels A and B show that cells transfected with wtFPN accumulate 35% to 40% as much radioactive iron as untransfected control or CD8 transfectants. Mutants A77D, V162Δ, L170F, and G490D are able to accumulate 59Fe to the same levels as CD8 control cells, while in the case of C326Y, N144D, Y64N, and N144H levels are lower and comparable to wild type. (C-D) Iron release. Cells were grown in 40 μg/mL 59Fe for 24 hours, then washed and transfected as in panels A and B. Fifteen hours later cells were harvested, washed, and resuspended at 106 cells/mL in serum-free medium, and 59Fe export was measured at the intervals shown up to 36 hours after harvesting. Data are presented as a percentage of starting radioactivity when cells were harvested. Panels C and D show that cells transfected with wtFPN release up to 3 times as much 59Fe as untransfected control or CD8 transfectants. Mutants A77D, V162Δ, L170F, and G490D are only able to release 59Fe to levels similar to CD8 control cells, while in the case of C326Y, N144D, Y64N, and N144H iron release is higher and comparable to wtFPN. These results are representative of 3 experiments.
FPN mutations associated with loss of function reduce cell-surface expression
Our next question was how do the mutations at A77D, V162Δ, L170F, and G490D lead to a loss of export function of FPN. We have found that the c-Myc epitope engineered onto the C-terminus of FPN is detectable with antibodies at the cell surface without prior treatment to permeablize the cell membrane. We, therefore, transfected 293T cells with c-Myc–tagged FPN variants and measured the mean fluorescence intensity (MFI) of cells detectable with anti–c-Myc antibody. The fraction of 293Tcells that became positive for c-Myc after transfection was not influenced by any of the mutations (70%-80% positive cells, data not shown). However, the MFI of positive cells (amount of c-Myc on the surface of positive cells) was influenced by some of the mutations (Figure 3A). This quantitative difference is expressed as the MFI of c-Myc–positive cells of FPN mutants divided by the MFI of wtFPN–c-Myc expressed under exactly the same transfection conditions. Figure 3A shows that in C326Y, Q248H, N144H, N144D, and Y64N mutants that retain iron-export function, surface expression levels are comparable to wild type, whereas in A77D, V162Δ, L170F, and G490D mutants that lose iron-export function, surface expression levels are reduced 25% to 40%.
We next investigated whether mutant FPN could influence the surface localization of wild type when coexpressed. To distinguish between the 2 we constructed a version of wtFPN with a FLAG tag instead of a c-Myc tag at the C-terminus. FLAG-tagged wtFPN was transfected into 293T cells along with c-Myc–tagged wtFPN (Figure 3Bi-iii), A77D (Figure 3Biv-vi), V162Δ (Figure 3Bvii-ix), or C326Y (Figure 3Bx-xii) for 2 days. Cells were then stained with anti-FLAG (red color detecting wtFPN) and anti–c-Myc (green color). Staining of FLAG for wild type shows localization of the protein to the cell membrane (arrowheads in Figure 3Bi,vii). When wtFPN–c-Myc is coexpressed, staining for c-Myc also shows localization to the cell surface (arrowhead in Figure 3Bii), and this is confirmed by the merged image, which shows colocalization of wtFPN-FLAG and wtFPN–c-Myc on the cell surface (arrowhead in Figure 3Biii).
In contrast, both A77D and V162Δ have a predominant intracellular localization (arrowheads in Figure 3Bv,vi,viii,ix) with little or no visible surface membrane staining, while the coexpressed wtFPN is still present at the cell membrane (arrowheads in Figure 3Bvii). This shows that neither of these two mutants interferes with the trafficking of the wtFPN to the cell surface. C326Y, which retains export function, is localized to the cell membrane (arrowheads in Figure 3Bxi) in the same way as wtFPN, and the merge shows they colocalize there (arrowhead in Figure 3Bix).
Mutant forms of FPN do not bind to or reduce the function of wild-type protein
The previous immunofluorescence implied that wtFPN and mutant FPN may not physically interact in the cell, but we wanted to confirm this, because a possible dominant mechanism of action of the mutant FPNs in causing disease would be to bind to and inhibit wild-type ferroportin. First, to test whether wtFPN multimerizes we transfected 293T cells with wtFPN–c-Myc and wtFPN-FLAG, and as a control we also transfected cells with CD8 and wtFPN-FLAG (Figure 4A). Cells were lysed and proteins immunoprecipitated with monoclonal antibodies anti-CD8, anti-c-Myc, anti-FLAG, or polyclonal anti-FPNct antibody. The precipitates were run on a gel and blotted, and the membrane was probed with anti–FLAG-HRP conjugate to look for evidence of coprecipitation. Figure 4A shows that the positive control anti-FLAG precipitate was detected as a protein band of around 67 kDa, and this was also present in lysates precipitated with anti-FPN polyclonal serum but was much fainter. (In our experiments this polyclonal antiserum will precipitate FPN but not as efficiently as the anti-FLAG or anti–c-Myc.) The anti–c-Myc or anti-CD8 precipitates did not contain any detectable FLAG-tagged wtFPN, indicating that c-Myc–tagged and FLAG-tagged wtFPN do not coprecipitate.
Cell-surface expression and intracellular localization of wtFPN and FPN mutants. (A) 293T cells were transfected with wtFPN–c-Myc and the mutant FPN–c-Myc variants indicated, and 2 days later stained for surface c-Myc tag and analyzed by flow cytometry for percentage of c-Myc–positive transfected cells and the MFI of the transfected cells (which is proportional to the amount of c-Myc expressed on the cell surface). The percentage of transfected cells did not vary significantly between wtFPN and mutant FPN constructs (70%-80% of cells were routinely c-Myc positive, not shown). However, the degree of fluorescence differed, with Y64N, C326Y, Q248H, N144D, and N144H all showing similar MFI comparable to wild type, while V162Δ, L170F, A77D, and G490D all express at levels less than 50% of wild type. (B) 293T cells were cotransfected with wtFPN-FLAG and either wtFPN–c-Myc or mutant FPN–c-Myc and stained with antibodies against the FLAG tag (red, i,iv,vii,viii) and the c-Myc tag (green, ii,v,viii,xi). The staining in i to iii shows that both FLAG-tagged and c-Myc–tagged wtFPN are expressed at the cell-surface membrane (arrowheads) as well as intracellularly. A77D FPN is located perinuclearly and not visible on the cell surface (arrowheads in v,vi) and does not colocalize with wtFPN, or alter the cell membrane staining of wtFPN (compare red with green in vi). Similarly, V162Δ locates predominantly intracellularly (arrowheads in viii,ix); however, x to xii show that C326Y FPN colocalizes with wtFPN at the cell membrane.
Cell-surface expression and intracellular localization of wtFPN and FPN mutants. (A) 293T cells were transfected with wtFPN–c-Myc and the mutant FPN–c-Myc variants indicated, and 2 days later stained for surface c-Myc tag and analyzed by flow cytometry for percentage of c-Myc–positive transfected cells and the MFI of the transfected cells (which is proportional to the amount of c-Myc expressed on the cell surface). The percentage of transfected cells did not vary significantly between wtFPN and mutant FPN constructs (70%-80% of cells were routinely c-Myc positive, not shown). However, the degree of fluorescence differed, with Y64N, C326Y, Q248H, N144D, and N144H all showing similar MFI comparable to wild type, while V162Δ, L170F, A77D, and G490D all express at levels less than 50% of wild type. (B) 293T cells were cotransfected with wtFPN-FLAG and either wtFPN–c-Myc or mutant FPN–c-Myc and stained with antibodies against the FLAG tag (red, i,iv,vii,viii) and the c-Myc tag (green, ii,v,viii,xi). The staining in i to iii shows that both FLAG-tagged and c-Myc–tagged wtFPN are expressed at the cell-surface membrane (arrowheads) as well as intracellularly. A77D FPN is located perinuclearly and not visible on the cell surface (arrowheads in v,vi) and does not colocalize with wtFPN, or alter the cell membrane staining of wtFPN (compare red with green in vi). Similarly, V162Δ locates predominantly intracellularly (arrowheads in viii,ix); however, x to xii show that C326Y FPN colocalizes with wtFPN at the cell membrane.
A possible mechanism of action of the mutant FPNs in causing disease would be to bind to and inhibit wtFPN. We next cotransfected c-Myc–tagged mutant FPN constructs with wtFPN-FLAG, immunoprecipitated, and probed the immunoprecipitates by Western blot with anti–FLAG-HRP as in Figure 4A. Figure 4B shows that wtFPN was present in anti-FLAG precipitates but not detectable in anti–c-Myc immunoprecipitates of cells cotransfected with wtFPN-FLAG and A77D, V162Δ, or C326Y FPN–c-Myc, indicating these mutants do not physically interact with wtFPN.
To test whether mutant FPNs could functionally interfere with the ability of wtFPN to make cells iron deficient, we performed a ferritin ELISA on cells transfected with combinations of wtFPN and mutant FPN. Figure 4B shows that the presence of mutants A77D, V162Δ, and C326Y did not affect the ability of wtFPN to reduce ferritin. We conclude that the mutant FPNs tested do not interfere physically or functionally with wtFPN.
Discussion
The plethora of papers describing patients with iron overload linked to ferroportin mutations indicates its important role in iron metabolism. We have investigated how these mutations could lead to disease by in vitro expression of the protein. First, we have shown that human wtFPN expressed in human cells is able to export iron, leading to an iron-deficient phenotype characterized by low ferritin and high TfR1 (Figures 1, 2). This extends the initial functional characterization of murine Fpn expressed in Xenopus oocytes and human cells.10-12 In our experiments human FPN exported iron independently of added ceruloplasmin (data not shown); this could be due to ceruloplasmin present in fetal calf serum or oxidases endogenous to the cell. Staining of cells transfected with wtFPN showed the protein to localize to the cell membrane (Figure 3).
Mutant FPNs do not coimmunoprecipitate with or affect the function of wtFPN. (A-B) 293T cells were doubly transfected with wtFPN-FLAG and either CD8, wtFPN–c-Myc, or mutant FPN–c-Myc as shown. Cells were lysed and immunoprecipitated (i.p.) with anti-CD8 (negative control), anti–c-Myc, anti-FLAG, or anti-FPNct. The precipitates were run on a gel, blotted onto membrane, and probed with anti–FLAG-HRP conjugate. Panel A shows cells transfected with CD8 and wtFPN-FLAG and cells transfected with wtFPN–c-Myc and wtFPN-FLAG. In both cases, precipitation with anti-FLAG results in a ferroportin band at around 70 kDa, revealed by probing the membrane with anti–FLAG-HRP. A much fainter band of the same size is precipitated by the anti-FPN polyclonal antisera. The same pattern of bands is seen when wtFPN-FLAG is coexpressed with c-Myc–tagged FPN mutants A77D, V162Δ, and C326Y (B). In each case no anti-FLAG reactive 70-kDa band is precipitated by anti–c-Myc, so that panel A indicates the wtFPN molecule does not form multimers, and panel B indicates that mutant FPNs also do not bind wtFPN. (C) 293T cells transfected with either CD8 alone, wtFPN-FLAG alone, or wtFPN-FLAG and mutant FPN–c-Myc together were lysed and tested for ferritin by ELISA as for Figure 1, except that this time no Tf was present in the media. Graph shows individual (•) and mean (horizontal bars) values for ferritin in ng/107 cells. Error bars depict ± 95% CI. As before, cells expressing wtFPN have reduced ferritin compared with control transfectants expressing CD8. Cells expressing wtFPN along with FPN mutants A77D, V162Δ, and C326Y have a similar reduction in ferritin compared with wtFPN alone, indicating that the mutant FPNs do not interfere with the ability of wtFPN to make cells iron deficient.
Mutant FPNs do not coimmunoprecipitate with or affect the function of wtFPN. (A-B) 293T cells were doubly transfected with wtFPN-FLAG and either CD8, wtFPN–c-Myc, or mutant FPN–c-Myc as shown. Cells were lysed and immunoprecipitated (i.p.) with anti-CD8 (negative control), anti–c-Myc, anti-FLAG, or anti-FPNct. The precipitates were run on a gel, blotted onto membrane, and probed with anti–FLAG-HRP conjugate. Panel A shows cells transfected with CD8 and wtFPN-FLAG and cells transfected with wtFPN–c-Myc and wtFPN-FLAG. In both cases, precipitation with anti-FLAG results in a ferroportin band at around 70 kDa, revealed by probing the membrane with anti–FLAG-HRP. A much fainter band of the same size is precipitated by the anti-FPN polyclonal antisera. The same pattern of bands is seen when wtFPN-FLAG is coexpressed with c-Myc–tagged FPN mutants A77D, V162Δ, and C326Y (B). In each case no anti-FLAG reactive 70-kDa band is precipitated by anti–c-Myc, so that panel A indicates the wtFPN molecule does not form multimers, and panel B indicates that mutant FPNs also do not bind wtFPN. (C) 293T cells transfected with either CD8 alone, wtFPN-FLAG alone, or wtFPN-FLAG and mutant FPN–c-Myc together were lysed and tested for ferritin by ELISA as for Figure 1, except that this time no Tf was present in the media. Graph shows individual (•) and mean (horizontal bars) values for ferritin in ng/107 cells. Error bars depict ± 95% CI. As before, cells expressing wtFPN have reduced ferritin compared with control transfectants expressing CD8. Cells expressing wtFPN along with FPN mutants A77D, V162Δ, and C326Y have a similar reduction in ferritin compared with wtFPN alone, indicating that the mutant FPNs do not interfere with the ability of wtFPN to make cells iron deficient.
Our results show that disease-associated FPN mutations appear to fall into 2 categories, some losing export function, while others behave as wild type; mutations within these 2 categories of mutations do not cluster together in terms of the primary sequence of FPN. FPN mutations A77D, V162Δ, L170F (the human homologue of the L167F mutation found in ferroportin1 from the weh zebra fish), and G490D all lose export function (Figure 2). When cells transfected with these mutants were stained for FPN by immunofluorescence or flow cytometry, FPN was shown to be localized intracellularly, with greatly reduced amounts reaching the cell surface (Figure 3A-B), possibly because of protein misfolding leading to inefficient transport to the plasma membrane. However, Y64N, N144H, N144D, Q248H, and C326Y are expressed on the cell surface as efficiently as wild type and are able to export iron to a similar degree (Figures 1, 2, 3). This leads to the suggestion that iron export through FPN may be dependant on cell-surface expression of the protein. The recent observation that internalization of FPN mediated by hepcidin ablates iron export is consistent with this notion.32
The autosomal dominant nature of disease inheritance led us to investigate whether the mutant form of the protein could influence the function of the wild type when expressed in the same cell. That is, in patients with loss of function mutations, does the mutant allele in some way impede the action of the wild-type allele, or alternatively is disease an effect of haploinsufficiency? Our findings suggest that the FPN molecule functions as a monomeric entity, as forms with different tags show no association by immunoprecipitation (Figure 4A). Wild-type FPN is shown to have no interaction with mutant forms, by immunoprecipitation and localization in the cell (Figures 3B and 4B), and the mutants do not interfere with the function of the wild type when coexpressed (Figure 4C). This leads us to conclude that the loss of function mutants cause disease by haploinsufficiency.
These results with the A77D, V162Δ, and G490D mutations of FPN are, thus, consistent with the scheme proposed by Montosi et al,16 to explain the macrophage iron loading observed in patients with hemochromatosis with these mutations.30 The reduced iron export resulting from half the expressed molecules being nonfunctional leads to iron becoming trapped within macrophages that normally mediate approximately 30 mg/day iron recycling. Enterocytes that take up approximately 1 mg/day iron from the diet may be able to function adequately with one wild-type allele of SLC40A1. Lower serum iron resulting from iron sequestration in macrophages reduces availability to the bone marrow for erythropoiesis. Thus, it might be expected that these FPN mutations would lead to anemia, and, indeed, it has been reported that some patients are mildly anemic in the early stages of disease and respond poorly to phlebotomy26,29 ; in addition the L170F mutation that behaves like A77D, V162Δ, and G490D in our assays, causes anemia in weh zebra fish.12 In humans with A77D, V162Δ, and G490D mutations, eventual iron overload may be a consequence of the erythron signaling to the gut enterocyte to increase iron uptake from the diet to compensate for the anemia. One corollary of this interpretation is that inactivation of one allele of Slc40a1 by gene knock-out should give a similar phenotype to the mutations, through reduced levels of expression of Fpn in the macrophages.
In contrast to A77D, V162Δ, and G490D, in our assays mutations at Y64N, N144H, N144D, Q248H, and C326Y all allow the full export function of FPN (Figure 2). The Q248H mutation has been found in patients with overload but is also relatively common in African and African American populations with or without symptoms of overload.23,28 This suggests that Q248H may be a polymorphism of FPN and infrequently associated with clinical disease. The phenotype of patients with Y64N, N144H, N144D, and C326Y are more severe, with typically almost 100% transferrin saturation and deposition of iron throughout the liver, rather than just the macrophages17,18 (V.V., A.T.M.-C., Y.C., Chanin Lim Wongse, and K.J.H.R., manuscript in preparation: A novel ferroportin mutation (C326Y) causes a dominant form of inherited iron overload in the far east). This suggests a mechanism of disease for these mutations distinct to that for the loss of function mutations.
Njajou et al18 postulated the N144H mutation may lead to a “gain of function” effect for FPN, but we find no evidence for gain of function in our assays. How then could the Y64N, N144H, N144D, and C326Y mutations lead to disease? One possibility is that the mutations confer resistance to homeostatic mechanisms that normally negatively regulate FPN. Recently, it has been found that the antimicrobial peptide hepcidin, which acts as a negative regulator of iron export from enterocytes and macrophages, binds to FPN, causing internalization and degradation of the iron exporter.32 In addition, the hemochromatosis protein HFE has been found to inhibit iron export from the macrophage-like cell line THP-1 and duodenal cell line HT-29 that naturally express FPN31,33,34 (although a direct HFE/FPN interaction has yet to be established). Perhaps the Y64N, N144H, N144D, and C326Y mutations of FPN prevent its interaction with hepcidin or (possibly) HFE. If this were the case, the mutated allele of SLC40A1 would resist negative regulation and be permanently “turned on,” leading to uninhibited iron uptake from the diet and release from macrophages, causing body iron overload and iron deposition throughout the liver.
In summary, we have confirmed that human FPN can mediate iron export from human cells and shown that the FPN mutants associated with type IV hemochromatosis fall into 2 categories, those that traffic poorly to the cell surface and lose iron export function (A77D, V162Δ, and G490D) and those that retain cell-surface expression and export function (Y64N, N144D, N144H, Q248H, and C326Y). The loss-of-function mutations probably cause disease by withholding iron from the bone marrow through sequestration within macrophages, subsequently leading to a signal for increased dietary iron uptake being sent from the erythron to the intestine. The second set of mutations may cause disease by resisting negative feedback mechanisms that normally operate to maintain iron homeostasis. More work is required to confirm these ideas, which have implications for the clinical management of disease.
Prepublished online as Blood First Edition Paper, February 3, 2005; DOI 10.1182/blood-2004-11-4502.
Supported by the Welcome Trust Programme (grant no. 060542/Z/00) (L.M.S., H.D., J.P.E., E.S., J.M.B., D.C., and A.R.M.T.).
L.M.S. and H.D. designed and performed research and wrote the paper; A.T.M.-C., V.V., Y.C., K.J.H.R., J.P.E., E.S., J.M.B., and D.C. performed the research; and A.R.M.T. designed the research and wrote the paper.
L.M.S and H.D. contributed equally to this work.
An Inside Blood analysis of this article appears at the front of the issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Andrew McKie and George Gao for generous gifts of expression vectors.

![Figure 1. Effect of ferroportin and ferroportin mutants on cell-surface transferrin receptor 1 and intracellular ferritin levels. (A) 293T cells at 30% to 50% confluence were transiently transfected with vectors encoding CD8, wtFPN–c-Myc, and the following c-Myc–tagged FPN mutants: C326Y, A77D, V162Δ, N144H, N144D, G490D, Y64N, Q248H, and L170F. After 2 days cells were double stained for CD8 or c-Myc tag and surface TfR1 and analyzed by 2-color flow cytometry. Transfected cells (> 70% of the population) expressing either CD8 or c-Myc were gated on and compared for their surface TfR1 levels. A comparison (top) is shown of TfR1 levels on cells positive for CD8 (gray filled histogram) versus untransfected cells (black line), demonstrating that expression of CD8 has no effect on TfR1. In all other panels, the gray-shaded histogram is TfR1 on CD8-transfected cells, and TfR1 on cells expressing wtFPN or mutant FPN is shown by a black line. Compared with CD8, TfR1 is increased on cells expressing wtFPN, indicating iron deficiency. C326Y, N144H, N144D, Q248H, and Y64N all increase expression of TfR1 in the same way as wild type, but A77D, V162Δ, and G490D do not increase TfR1 compared with CD8. L170F partially increases TfR1. The filled purple histogram represents the background fluorescence of cells stained with an isotype control antibody. (B) 293T cells were transfected as in panel A, but this time in the presence of 1 mg/mL Tf to increase background ferritin levels. After 2 days, cells were analyzed for total cell ferritin by ELISA. Cells were lysed in NP-40 at 107 cells/mL, doubling dilutions were made, and 20 μL was transferred to ELISA plate in triplicate. Graph shows mean total ferritin in ng/107 cells (± 95% CI [confidence interval]). Cells transfected with wtFPN have around 40 ng ferritin/107 cells, a 4-fold reduction compared with untransfected and control cells transfected with CD8. C326Y, N144H, N144D, Q248H, and Y64N all decrease ferritin levels in the same way as wild type, but A77D, V162Δ, L170F, and G490D do not significantly reduce ferritin. Asterisk indicates significance compared with untransfected or CD8 control (P < .001 by Student t test). Data shown is representative of 2 or more experiments for each mutant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/10/10.1182_blood-2004-11-4502/6/m_zh80100578460001.jpeg?Expires=1767696923&Signature=rF2QAAu4Q1NltZ4DUBlItIgw2Z2pSiKSObVy3pezrZjtzWvXTR1MccPqMJiMXNCwVOuG~I7NlGtGAgBntFsBvOh4RA8uqyiE~U3RMCxgmYA48PfUxBiva-9wz62yOTuW~XhYZNeLLXTOhTszpgqeF2bzTcJ4Odsx82RDdvQDR70iv-Qvkc4Hve1KyX45usaafOWLpUDHdcKSg2nN5hmPpM8abWBSMsTnGLsg1d6dGenhK~afNEi2FU43hYO3AV5he8bOkmkhw7FeXMqV0lcoFk-EzGF4dwzZfU14lIXPH1Fd0RISRXCC46vEuXqqpS7mlIgGcRSo1k5wCRoMv-7OEg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal