Abstract
Targeted disruption of the Friend leukemia integration 1 (Fli-1) proto-oncogene results in severe dysmegakaryopoiesis and embryonic lethality. We used morula-stage aggregation as a strategy to further clarify the hematopoietic defects of the Fli-1 gene-targeted mice. Analyses of lineage expression of Fli-1+/- and Fli-1-/- cells in the peripheral blood and bone marrow of chimeric mice consistently demonstrated reduced numbers of neutrophilic granulocytes and monocytes and increased numbers of natural killer (NK) cells. Transplantation studies using sorted Fli-1 mutant cells produced similar findings. Clonal culture studies of bone marrow cells revealed increased numbers of granulocytic and early erythroid progenitors in the Fli-1+/- cells. The sorted Fli-1-/- bone marrow cells revealed specific down-regulation of CCAAT/enhancer binding protein-α (C/EBPα) and C/EBPϵ, and the receptors for granulocyte colony-stimulating factor (G-CSF) and granulocyte-macrophage CSF (GM-CSF), consistent with their critical roles in granulopoiesis. Collectively, these observations suggest previously unknown physiologic roles for Fli-1 in granulocytic, erythroid, and NK cell proliferation and differentiation. Production of chimeras by morula-stage embryo aggregation is an effective way to unravel cell-autonomous hematopoietic defects in gene-targeted mice.
Introduction
The Friend leukemia integration 1 (Fli-1) proto-oncogene is a member of the Ets gene family of transcription factors, which bind to DNA elements containing the consensus sequence GGA(A/T).1-3 Fli-1 is preferentially expressed in cells of hematopoietic lineages and vascular endothelial cells, and has been shown to transactivate such genes as GATA-1,1,2 glycoprotein (gp) IIb,4-6 gpVI,7 gpIX,8 gpIb,8 and c-MPL.9 Fli-1 activation by Friend murine leukemia virus leads to development of erythroleukemia. It has been demonstrated that Fli-1 expression promotes megakaryocytic differentiation in K562 cells.10 Overexpression of Fli-1 in all tissues of transgenic mice results in death from progressive immunologic renal disease associated with an increased number of autoreactive T and B lymphocytes.11 Taken together, these results suggested that Fli-1 plays significant roles in hematopoiesis. To clarify the physiologic role of Fli-1 in hematopoiesis, we12 and others13 generated mice with the targeted disruption of Fli-1. The Fli-1 homozygous mutant (Fli-1-/-) embryos showed hemorrhage from the dorsal aorta into the lumen of the neural tube and the ventricles of the brain beginning on embryonic day 11.0 (E11.0) and were dead on or before E12.0. In addition, severe dysmegakaryopoiesis13,14 and vascular defects13 were found. We also noted that livers of the E11.0 Fli-1-/- embryos were pale and contained primarily polychromatophilic and orthochromatic normoblasts.12 To further define the roles of Fli-1 in hematopoiesis in individuals with longer viability, we generated Fli-1 chimeric mice by morula-stage embryo aggregation between Ly-5.2 Fli-1 heterozygote (Fli-1+/-) intercross embryos and wild-type (WT) Ly-5.1 embryos. Such chimeras were rescued from the embryonic lethality and could be studied for Fli-1-associated cell-autonomous defects. Analysis of the adult chimeric mice demonstrated significant reduction of both mature granulocytes and monocytes in the peripheral blood (PB) cells and striking increases in erythroid burst-forming units (BFU-Es) in the bone marrow (BM) cells that were derived from Fli-1 mutants. We also demonstrated that the expression of genes previously shown to be critical for granulopoiesis is reduced in the BM cells derived from the Fli-1 mutant. Conversely, natural killer (NK) cells were increased in PB and BM of the mutants. These observations suggest that the Fli-1 gene plays important roles in myelomonocytic, erythroid, and NK cell development.
Materials and methods
Monoclonal antibodies
The following monoclonal antibodies were purchased from Pharmingen (San Diego, CA): fluorescein isothiocyanate (FITC)-conjugated or biotin-conjugated 104 (anti-Ly-5.2; mouse immunoglobulin G2a [IgG2a]) and phycoerythrin (PE)-conjugated A20 (anti-Ly-5.1; mouse IgG2a), PE-conjugated RB6-8C5 (anti-Ly-6G [anti-Gr-1]; rat IgG2b), PE-conjugated GK1.5 (anti-CD4; rat IgG2b), PE-conjugated PK136 (anti-Ly-55 [anti-NK1.1]; mouse IgG2a), allophycocyanin (APC)-conjugated M1/70 (anti-Mac-1; rat IgG2b), APC-conjugated 53-6.7 (anti-Ly-2 [anti-CD8a]; rat IgG2a), and APC-conjugated RA3-6B2 (anti-CD45R/B220; rat IgG2a).
Growth factors
Recombinant mouse steel factor (SF) was a gift from Kirin Brewery (Takasaki, Japan). Recombinant mouse interleukin 3 (IL-3) was purchased from R&D Systems (Minneapolis, MN). Recombinant human erythropoietin (EPO) was provided by Amgen (Thousand Oaks, CA). The concentrations of cytokines used were as follows: SF, 100 ng/mL; IL-3, 10 ng/mL; EPO, 2 U/mL.
Mice
C57BL/6J-Ly-5.2 and C57BL/6J-Ly-5.1 mice were purchased from Jackson Laboratories (Bar Harbor, ME). Fli-1 WT mice and Fli-1+/- mice were generated, bred, maintained, and genotyped as previously described.12 Fli-1 chimeric mice were generated by morula-stage embryo aggregation at the Gene Targeting and Knockout Mouse Facility of Hollings Cancer Center (Medical University of South Carolina, Charleston, SC).12 Briefly, E2.5 embryos were isolated from the oviducts of superovulated C57BL/6J-Ly-5.2 Fli-1+/- females that had been mated with C57BL/6J-Ly-5.2 Fli-1+/- males. Similarly, E2.5 embryos were isolated from superovulated C57BL/6J-Ly-5.1 females that had been mated with C57BL/6J-Ly-5.1 males. Zona pellucidas were removed by exposure to acidic Tyrode solution (Sigma, St Louis, MO). In one set of experiments, 1 embryo from the Fli-1+/- intercross mating and 2 embryos from the WT Ly-5.1 mating were aggregated in V-bottomed 96-well culture plate (Corning, Corning, NY) containing M16 medium (Cell & Molecular Technologies, Phillipsburg, NJ), cultured overnight at 37°C in 5% CO2 in a humidified atmosphere, transferred into the uteri of 2.5-day pseudopregnant females and allowed to develop to term. Four chimeric mice containing Fli-1+/- cells and a mouse containing Fli-1-/- cells were derived from this experiment. In another experiment, we used one-to-one aggregation. Three chimeric mice containing Fli-1+/- cells, a mouse containing WT Ly-5.2 cells, and a mouse containing Fli-1-/- cells were derived from this experiment. Chimeric mice were maintained under specific-pathogen-free conditions at the Animal Research Facility of the Veterans Affairs Medical Center (Charleston, SC). All aspects of the animal research have been conducted in accordance with the guidelines set by the Institutional Animal Care and Use Committees of the Department of Veterans Affairs Medical Center and the Medical University of South Carolina.
Cell preparation
PB was obtained through cardiac puncture from individual Fli-1 chimeric mouse. Red blood cells were lysed by 0.15 M NH4Cl and washed twice with Ca2+-, Mg2+-free phosphate-buffered saline (PBS) (Life Technologies, Grand Island, NY) containing 0.1% bovine serum albumin (BSA) (Sigma-Aldrich, St Louis, MO). BM cells were flushed from femurs and tibiae, pooled, and washed twice with PBS containing 0.1% BSA. Samples were made into single-cell suspensions by repeated pipetting and filtering through a 40-μm nylon mesh. Mononuclear cells (MNCs) with densities ranging from 1.063 to 1.077 g/mL were collected by gradient separation with the use of Nycodenz (Accurate Chemical and Scientific, Westbury, NY). Thymus and spleen cell suspensions were initially prepared by mincing organs with scissors and by repeated passages through a syringe and no. 22 gauge needle. The samples were then washed twice with PBS containing 0.1% BSA and passed through a 40-μm nylon mesh. For cell sorting, total nucleated cells or MNCs from BM, PB, and thymi were stained with FITC-conjugated anti-Ly-5.2 and PE-conjugated anti-Ly-5.1. After addition of propidium iodide at a concentration of 1 μg/mL, the cells were washed twice, resuspended in PBS containing 0.1% BSA and 0.1 mg/mL DNase I (Sigma-Aldrich), and kept on ice until cell sorting. Three-color analysis and cell sorting were performed by means of a FACSVantage (Becton Dickinson Bioscience, San Jose, CA) with appropriate isotype-matched controls. Reanalysis revealed that the sorted Ly-5.1 and Ly-5.2 fractions were more than 98% pure. In addition, Fli-1+/+ DNA was absent in Ly-5.2 cells and Fli-1-/- DNA was absent in Ly-5.1 cells isolated from Fli-1-/- chimeric mice.
Genotyping
For genotyping, Ly-5.2 cells sorted from BM or thymi of the chimeric mice were subjected to polymerase chain reaction (PCR) to detect fragments of the WT Fli-1 and the mutant Fli-1 allele. Samples were subjected to 35 cycles of 94°C (1 minute), 68°C (1 minute), and 72°C (1 minute). A 309-bp fragment indicated the presence of WT allele whereas a 406-bp fragment was amplified from the mutated allele. The primers used are as follows: Fli-1 exon IX forward primer, 5′-GACCAACGGGGAGTTCAAAATGACG-3′; Fli-1 exon IX reverse primer, 5′-GGAGGATGGGTGAGACGGGACAAAG-3′; and Pol II reverse primer, 5′-GGAAGTAGCCGTTATTAGTGGAGAGG-3′.
RT-PCR analysis
Total RNA was extracted from the Ly-5.1- and Ly-5.2-sorted BM nuclear cells with Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Total RNA (1 μg) was reverse transcribed with the use of Superscript II (single-strand synthesis reverse transcription [RT]; Invitrogen). Aliquots from this reaction were subjected to PCR by means of the indicated gene-specific primers. Hypoxanthine phosphoribosyl transferase (HPRT) served as the control. The reaction mixture contained 1.5 mM Mg2+, 0.2 mM deoxynucleoside triphosphates, 1 × Taq Gold buffer, 0.88 pmol/μL primers, and 0.02 U/μL Taq Gold. The basic PCR reaction conditions were as follows: 95°C for 10 minutes; then 22, 26, or 32 cycles of 95°C for 30 seconds; Ta (annealing temperature) (specific annealing temperature provided in Table 1) for 45 seconds; 72°C for 1 minute; followed by 72°C for 7 minutes. PCR products were analyzed by electrophoresis on a 1% TBE (tris(hydroxymethyl)aminomethane-borate-ethylenediaminetetraacetic acid) gel containing ethidium bromide and visualized under ultraviolet light.
PCR primer sequences and GenBank accession numbers
Primer name . | Forward primer . | Reverse primer . | Length, bp . | Accession no. . | Ta, °C . |
|---|---|---|---|---|---|
| CD11b | TGTGGACATGGACGCTGATG | GCCTATGATCCGGTGGCTGT | 331 | NM_008401 | 60 |
| CD18 | GCCGAGTGCCTGAAGTTTGAT | TAAGCCACGTTGTGTTTGCAGA | 504 | X14951 | 60 |
| C/EBPα | GCCCCTCAGTCCCTGTCTTTAG | ATGGTCCCCGTGTCCTCCTA | 302 | BC051102 | 60 |
| C/EBPϵ | CAGCTGGACTCAGAAGACAGGC | ACAGCTTCCCTTGTCCAGGTG | 306 | XM_139135 | 60 |
| GM-CSF2Rβ2 | ACCTCCAATCCTCAACCAGACC | CTGTACCCATAAACACGGCCAA | 378 | NM_007781 | 60 |
| GM-CSFRα | ACGGAGGTCACAAGGTCAAGGT | ACACGCCCACTTTGGTGATTG | 454 | M85078 | 60 |
| GM-CSFRβ1 | ATCCAGCTCATGGTGCCACTT | TCACCACCGGAGAGCATTTCT | 520 | NM_007780 | 60 |
| EPOR | ATCTCGTTGTTGCTGACGGTTC | TCCAATACCAAGTAGGTGTCCTGG | 332 | BC046282 | 60 |
| c-FMS | AACCCTGGCTACTCCTCCATCC | GCTTCATCCTACTCGGACCCCT | 301 | BC039758 | 60 |
| G-CSFR | CAAACTGGACCAACAGGCAAA | GGAGCAGTTGTTCTGCCTCTTC | 406 | M58288 | 60 |
| GATA-1 | TAAGGTGGCTGAATCCTCTGCATC | CCGTCTTCAAGGTGTCCAAGAACGT | 445 | NM_008089 | 58 |
| GATA-2 | TTCTTCTGCAGGGGGTAGTGTAG | GGTGACTTCTCTTGCATGCACTT | 693 | NM-008090 | 58 |
| GM-CSF | TGCAGACCCGCCTGAAGATAT | ATACTGCCCTCCAACTGTGGCT | 353 | X03019 | 60 |
| Gr-1 | CCTTCCAGGAACCCTGTACTGA | TGCTGTCCATATCCATGAGCA | 370 | XM_194966 | 60 |
| HPRT | GCTGGTGAAAAGGACCTCTC | ATGGCCACAGGACTAGAACAC | 254 | J00423 | 62 |
| IL-6R | GTTCTACAGAAGCAACGAGTGTCCTC | AGTCTCTGTTGCTGTTGTCATAAGG | 336 | XM_194097 | 56 |
| c-MPL | ATGCCTACCGAGGAGAGAAGCC | GCCATAGCGGAGTTCATGCC | 317 | NM_010823 | 60 |
Primer name . | Forward primer . | Reverse primer . | Length, bp . | Accession no. . | Ta, °C . |
|---|---|---|---|---|---|
| CD11b | TGTGGACATGGACGCTGATG | GCCTATGATCCGGTGGCTGT | 331 | NM_008401 | 60 |
| CD18 | GCCGAGTGCCTGAAGTTTGAT | TAAGCCACGTTGTGTTTGCAGA | 504 | X14951 | 60 |
| C/EBPα | GCCCCTCAGTCCCTGTCTTTAG | ATGGTCCCCGTGTCCTCCTA | 302 | BC051102 | 60 |
| C/EBPϵ | CAGCTGGACTCAGAAGACAGGC | ACAGCTTCCCTTGTCCAGGTG | 306 | XM_139135 | 60 |
| GM-CSF2Rβ2 | ACCTCCAATCCTCAACCAGACC | CTGTACCCATAAACACGGCCAA | 378 | NM_007781 | 60 |
| GM-CSFRα | ACGGAGGTCACAAGGTCAAGGT | ACACGCCCACTTTGGTGATTG | 454 | M85078 | 60 |
| GM-CSFRβ1 | ATCCAGCTCATGGTGCCACTT | TCACCACCGGAGAGCATTTCT | 520 | NM_007780 | 60 |
| EPOR | ATCTCGTTGTTGCTGACGGTTC | TCCAATACCAAGTAGGTGTCCTGG | 332 | BC046282 | 60 |
| c-FMS | AACCCTGGCTACTCCTCCATCC | GCTTCATCCTACTCGGACCCCT | 301 | BC039758 | 60 |
| G-CSFR | CAAACTGGACCAACAGGCAAA | GGAGCAGTTGTTCTGCCTCTTC | 406 | M58288 | 60 |
| GATA-1 | TAAGGTGGCTGAATCCTCTGCATC | CCGTCTTCAAGGTGTCCAAGAACGT | 445 | NM_008089 | 58 |
| GATA-2 | TTCTTCTGCAGGGGGTAGTGTAG | GGTGACTTCTCTTGCATGCACTT | 693 | NM-008090 | 58 |
| GM-CSF | TGCAGACCCGCCTGAAGATAT | ATACTGCCCTCCAACTGTGGCT | 353 | X03019 | 60 |
| Gr-1 | CCTTCCAGGAACCCTGTACTGA | TGCTGTCCATATCCATGAGCA | 370 | XM_194966 | 60 |
| HPRT | GCTGGTGAAAAGGACCTCTC | ATGGCCACAGGACTAGAACAC | 254 | J00423 | 62 |
| IL-6R | GTTCTACAGAAGCAACGAGTGTCCTC | AGTCTCTGTTGCTGTTGTCATAAGG | 336 | XM_194097 | 56 |
| c-MPL | ATGCCTACCGAGGAGAGAAGCC | GCCATAGCGGAGTTCATGCC | 317 | NM_010823 | 60 |
Cytochemical studies
Sorted Ly-5.2 and Ly-5.1 PB and BM cells were centrifuged in a Cytospin 2 (Shandon, Pittsburgh, PA), dried, and stained with May-Grünwald Giemsa solution for 500-cell differential counting.
Transplantation
Ly-5.1 mice were given a single 950 cGy dose of total body irradiation by means of a 4 × 106 V linear accelerator, and within 24 hours, designated numbers of test donor Ly-5.1 cells were injected into the tail vein of these irradiated recipient mice together with 2 × 105 crude BM cells from Ly-5.1 mice as radioprotective cells. Levels of hematopoietic engraftment were analyzed out 2 months after transplantation. PB cells were obtained from the retro-orbital plexus of the recipient mice by means of heparin-coated micropipettes (Drummond Scientific, Broomall, PA) and stained with FITC-conjugated anti-Ly-5.2-PE, APC-conjugated lineage-specific antibodies mentioned under “Monoclonal antibodies” and biotin-conjugated anti-Ly-5.1, followed by streptavidin-conjugated peridinin chlorophyll-alpha protein (PerCP) (Pharmingen). They were analyzed for hematopoietic engraftment on a FACSCalibur (Becton Dickinson).
Methylcellulose culture
Sorted Ly-5.1 and Ly-5.2 populations of BM MNCs from Fli-1+/- chimeric mice were plated in 35-mm Falcon suspension culture dishes (Becton Dickinson Labware, Lincoln Park, NJ). The culture medium consisted of α-medium (ICN Laboratories, Rockville, MD), 1.2% 1500-centipoise methylcellulose (Shinetsu Chemical, Tokyo, Japan), 30% fetal bovine serum (Atlanta Biologicals, Norcross, GA), 1% deionized fraction V BSA, 1 × 10-4 M 2-mercaptoethanol (Sigma-Aldrich), 100 ng/mL SF, 10 ng/mL IL-3, and 2 U/mL EPO. Dishes were incubated at 37°C in a humidified atmosphere flushed with a mixture of 5% CO2, 5% O2, and 90% N2 (vol/vol). On day 12 of culture, colonies consisting of 40 or more cells were scored on an inverted microscope.
Statistical analysis
Student paired t test was used to analyze the statistical significance of the differences between Ly-5.2 and Ly-5.1 populations. The results of clonal culture were analyzed according to the Mann-Whitney U test. P < .05 was considered statistically significant.
Results
Chimeric mice
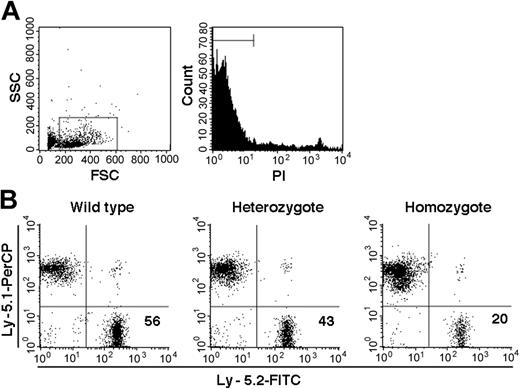
On the basis of PCR analysis of sorted Ly-5.2 cells, we identified a total of 10 chimeric mice, 1 with WT Ly-5.2 cells, 7 with Fli-1+/- Ly-5.2 cells, and 2 with Fli-1-/- Ly-5.2 cells. The levels of Ly-5.2 cells, calculated as percentage of Ly-5.2 cells × 100/(percentage of Ly-5.2 cells + percentage of Ly-5.1 cells), were 56.0% in the chimeric mouse containing WT Ly-5.2 cells (WT chimeric mouse) and 29.5% ± 17.7% (mean ± standard deviation [SD]) in the 7 chimeric mice containing Ly-5.2 Fli-1+/- cells (Fli-1+/- chimeric mice). In the 2 chimeric mice containing Ly-5.2 Fli-1-/- cells (Fli-1-/- chimeric mice), the levels of Ly-5.2 cells were 1.5% and 20.0%, each. Representative analyses of the 3 types of mice are shown in Figure 1.
Examples of chimerism in the PB of chimeric mice. (A) Sorting gates used for viable cells. (B) Analyses of Fli-1 WT, Fli-1+/-, and Fli-1-/- chimeric mice are shown. Numbers indicate percentage of Ly-5.2 cells in each mouse.
Examples of chimerism in the PB of chimeric mice. (A) Sorting gates used for viable cells. (B) Analyses of Fli-1 WT, Fli-1+/-, and Fli-1-/- chimeric mice are shown. Numbers indicate percentage of Ly-5.2 cells in each mouse.
Comparison of lineage expression by Ly-5.2 and Ly-5.1 cells in the chimeric mice
In the next series of experiments, we analyzed lineage expression by Ly-5.2 and Ly-5.1 cells of the chimeric mice. First, we analyzed the PB and BM nucleated cells of the WT chimeric mouse by flow cytometry. There were no significant differences between the Ly-5.2 and Ly-5.1 cell populations in the composition of cells (data not shown).
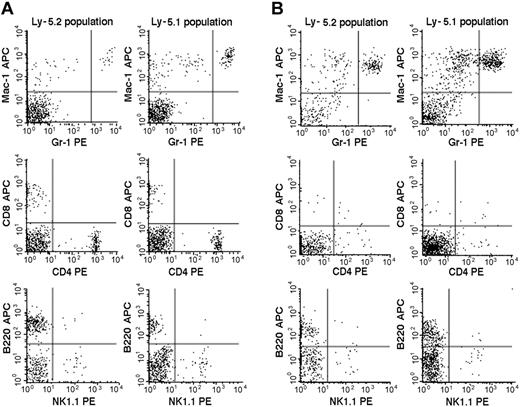
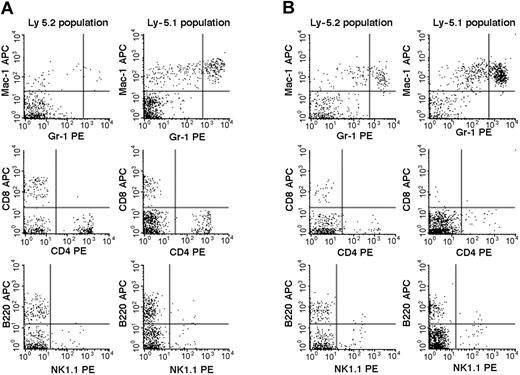
We then studied the 7 Fli-1+/- chimeric mice. An example of the flow cytometric analyses is presented in Figure 2, and the results are summarized in Table 2. The percentages of mature granulocytes (Gr1brightMac1+) and monocytes (Gr1dim/-Mac1+) in PB were significantly different in the Ly-5.2 (Fli-1+/-) and Ly-5.1 populations, and the differences were more pronounced in granulocytes than in monocytes. There were no significant differences in the granulocyte and monocyte counts in normal Ly-5.2 and Ly-5.1 mice (data not shown). Although the percentages of B cells (B220+) and NK cells (NK1.1+) were also higher in the Ly-5.2 (Fli-1+/-) population than in the Ly-5.1 population, studies of Fli-1-/- chimeric mice appear to support changes in NK cells only (Table 2). In contrast to PB, studies of BM revealed no significant differences in the percentages of mature granulocytes and monocytes in the 2 populations. Although BM CD4+ T cells, CD8+ T cells, and NK cells were higher in the Ly-5.2 population than in the Ly-5.1 population, both constituted very minor populations. In the thymus and spleen, percentages of B cells and T cells, including CD4+CD8- T cells, CD4-CD8+ T cells, and CD4+CD8+ T cells, were comparable in the 2 populations (data not shown). Two Fli-1-/- chimeric mice were available for analysis. As discussed in the preceding section, one mouse showed 1.5% and the other 20.0% levels of Ly-5.2 Fli-1-/- cells. The flow cytometric analysis of the mouse showing 20.0% level of Ly-5.2 cells is presented in Figure 3. As shown in Table 2, despite the very different levels of chimerism, PB cells from both mice revealed severe reduction in the number of granulocytes and monocytes in the Ly-5.2 population relative to Ly-5.1 population. Similarly, the levels of NK cells appeared higher in the Ly-5.2 Fli-1-/- cells than in Ly-5.1 cells. Comparison of the numbers of granulocytes in the Fli-1+/- mice and Fli-1-/- mice strongly indicates gene-dose effects of Fli-1 on granulocytes.
Flow cytometric comparison of lineage expression by Ly-5.2 and Ly-5.1 cells of a representative Fli-1+/- chimeric mouse. Analyses of PB (panel A) and BM (panel B) nucleated cells are shown. The results are representative of 7 mice. Gr-1brightMac-1+ cells are neutrophilic granulocytes, and Gr-1dim/-Mac-1+ cells are monocytes.
Flow cytometric comparison of lineage expression by Ly-5.2 and Ly-5.1 cells of a representative Fli-1+/- chimeric mouse. Analyses of PB (panel A) and BM (panel B) nucleated cells are shown. The results are representative of 7 mice. Gr-1brightMac-1+ cells are neutrophilic granulocytes, and Gr-1dim/-Mac-1+ cells are monocytes.
Comparison of lineage expression by Ly-5.2 and Ly-5.1 cell populations of Fli-1+/– and Fli-1-/- chimeric mice
. | Fli-1+/- . | . | Fli-1-/- . | . | ||
|---|---|---|---|---|---|---|
| Tissue and lineage . | Ly-5.2 . | Ly-5.1 . | Ly-5.2 . | Ly-5.1 . | ||
| PB | ||||||
| Gr1brightMac1+, % | 4.1 ± 1.9* | 10.8 ± 3.6 | 1.1, 1.6 (1.4) | 12.2,16.1 (14.2) | ||
| Gr1dim/-Mac1+, % | 10.9 ± 2.8† | 15.8 ± 1.9 | 12.5, 8.3 (10.4) | 14.9,16.1 (15.5) | ||
| B220+, % | 44.9 ± 1.1‡ | 35.2 ± 6.7 | 30.4,26.2 (28.3) | 46.5,40.2 (43.4) | ||
| CD4+, % | 20.1 ± 4.6 | 20.5 ± 3.0 | 24.4,35.6 (30.0) | 14.4,13.6 (14.0) | ||
| CD8+, % | 14.4 ± 3.2 | 14.4 ± 2.3 | 19.9,18.3 (19.1) | 11.2, 9.4 (10.3) | ||
| NK1.1+, % | 4.8 ± 1.1‡ | 3.7 ± 0.6 | 5.4, 4.9 (5.2) | 2.6, 2.2 (2.4) | ||
| BM | ||||||
| Gr1brightMac1+, % | 36.6 ± 2.2 | 43.2 ± 8.5 | 22.1 | 54.7 | ||
| Gr1dim/-Mac1+, % | 22.3 ± 1.8 | 23.0 ± 2.4 | 18.5 | 23.8 | ||
| B220+, % | 30.1 ± 2.5 | 22.3 ± 9.2 | 27.0 | 11.0 | ||
| CD4+, % | 5.1 ± 0.3‡ | 2.3 ± 0.5 | 6.8 | 1.5 | ||
| CD8+, % | 2.8 ± 0.9‡ | 1.7 ± 0.8 | 5.2 | 1.1 | ||
| NK1,1+, % | 3.8 ± 0.2† | 1.8 ± 0.2 | 7.7 | 2.0 | ||
. | Fli-1+/- . | . | Fli-1-/- . | . | ||
|---|---|---|---|---|---|---|
| Tissue and lineage . | Ly-5.2 . | Ly-5.1 . | Ly-5.2 . | Ly-5.1 . | ||
| PB | ||||||
| Gr1brightMac1+, % | 4.1 ± 1.9* | 10.8 ± 3.6 | 1.1, 1.6 (1.4) | 12.2,16.1 (14.2) | ||
| Gr1dim/-Mac1+, % | 10.9 ± 2.8† | 15.8 ± 1.9 | 12.5, 8.3 (10.4) | 14.9,16.1 (15.5) | ||
| B220+, % | 44.9 ± 1.1‡ | 35.2 ± 6.7 | 30.4,26.2 (28.3) | 46.5,40.2 (43.4) | ||
| CD4+, % | 20.1 ± 4.6 | 20.5 ± 3.0 | 24.4,35.6 (30.0) | 14.4,13.6 (14.0) | ||
| CD8+, % | 14.4 ± 3.2 | 14.4 ± 2.3 | 19.9,18.3 (19.1) | 11.2, 9.4 (10.3) | ||
| NK1.1+, % | 4.8 ± 1.1‡ | 3.7 ± 0.6 | 5.4, 4.9 (5.2) | 2.6, 2.2 (2.4) | ||
| BM | ||||||
| Gr1brightMac1+, % | 36.6 ± 2.2 | 43.2 ± 8.5 | 22.1 | 54.7 | ||
| Gr1dim/-Mac1+, % | 22.3 ± 1.8 | 23.0 ± 2.4 | 18.5 | 23.8 | ||
| B220+, % | 30.1 ± 2.5 | 22.3 ± 9.2 | 27.0 | 11.0 | ||
| CD4+, % | 5.1 ± 0.3‡ | 2.3 ± 0.5 | 6.8 | 1.5 | ||
| CD8+, % | 2.8 ± 0.9‡ | 1.7 ± 0.8 | 5.2 | 1.1 | ||
| NK1,1+, % | 3.8 ± 0.2† | 1.8 ± 0.2 | 7.7 | 2.0 | ||
Data for Fli-1+/- chimeric mice represent mean ± SD (n = 7 for PB; n = 4 for BM) of percentage of lineage-positive cells within Ly-5.2 or Ly-5.1 cell population. The mean of the percentage of lineage-positive cells of 2 Fli-1-/- chimeric mice appear in parentheses. BM from only one Fli-1-/- mouse was analyzed.
P values were determined for differences between Ly-5.2 and Ly-5.1 groups.
P < .001.
P < .01.
P < .05.
Flow cytometric comparison of lineage expression by Ly-5.2 and Ly-5.1 cells of a representative Fli-1-/- chimeric mouse. Analyses of PB (panel A) and BM (panel B) nucleated cells are shown.
Flow cytometric comparison of lineage expression by Ly-5.2 and Ly-5.1 cells of a representative Fli-1-/- chimeric mouse. Analyses of PB (panel A) and BM (panel B) nucleated cells are shown.
Differential counts (500 cells) with May-Grünwald Giemsa staining were performed on the sorted PB and BM cells from 2 Fli-1+/- chimeric mice. The mature neutrophils (bands and segmented forms) comprised 8.7% and 4.2% of the Ly-5.2 populations and 22.0% and 27.4% in the Ly-5.1 populations, respectively, confirming the flow cytometric analyses. Differential counts of the BM cells showed no differences between the 2 populations, again confirming the flow cytometric analysis presented in Table 2. May-Grünwald Giemsa staining of the sorted PB cells of the Fli-1-/- chimeric mouse revealed 2.0% and 22.8% mature neutrophils in the Ly-5.2 and Ly-5.1 cells, respectively. This observation was also consistent with the results of the flow cytometric analysis and further supported a correlation between severity of phenotype and Fli-1 gene dosage.
Colony formation in culture from the BM of Fli-1+/- chimeric mice
Although granulocytes and monocytes were reduced in PB of the chimeric mice, differential counts of BM cells were normal. To uncover possible progenitor abnormalities, we analyzed colony formation from sorted BM MNCs from 3 chimeric mice. The results are shown in Table 3. Mature erythrocytes do not express Ly-5, but primitive erythroid progenitors do. Therefore, it is possible to separate mutant BFU-Es from normal BFU-Es on the basis of Ly-5 expression. Strikingly, the number of erythroid bursts derived from Ly-5.2 (Fli-1+/-) BM MNCs was 6 to 11 times higher than that from Ly-5.1 cells. There were, however, no apparent differences in the size and color of the erythroid bursts. The numbers of granulocyte and/or macrophage colonies derived from Ly-5.2 (Fli-1+/-) BM MNCs were also significantly higher than those from Ly-5.1 cells. In a separate experiment, we had confirmed that there are no significant differences in colony formation between Ly-5.1 and Ly-5.2 substrains of B57BL/6J mice. The results presented in Table 3, therefore, strongly indicated the involvement of Fli-1 in not only the granulocyte lineage but also erythroid development.
Colony formation from sorted BM mononuclear cells of Fli-1+/– chimeric mice
. | No. colonies, mean ± SD . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell type plated . | G . | M . | GM . | E . | Mixed . | Total . | |||||
| Experiment 1 | |||||||||||
| Ly-5.2 | 28 ± 6 | 87 ± 6* | 6 ± 1 | 11 ± 4† | 1 ± 1 | 133 ± 8† | |||||
| Ly-5.1 | 15 ± 1 | 48 ± 3 | 10 ± 4 | 1 ± 2 | 0 ± 1 | 74 ± 3 | |||||
| Experiment 2 | |||||||||||
| Ly-5.2 | 24 ± 4* | 73 ± 7* | 5 ± 2 | 11 ± 4† | 1 ± 0 | 113 ± 13† | |||||
| Ly-5.1 | 13 ± 3 | 35 ± 4 | 5 ± 2 | 1 ± 1 | 1 ± 1 | 55 ± 7 | |||||
| Experiment 3 | |||||||||||
| Ly-5.2 | 26 ± 7 | 103 ± 14† | 13 ± 4† | 6 ± 2* | 5 ± 2 | 152 ± 17† | |||||
| Ly-5.1 | 20 ± 4 | 77 ± 6 | 6 ± 3 | 1 ± 2 | 3 ± 1 | 107 ± 12 | |||||
. | No. colonies, mean ± SD . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell type plated . | G . | M . | GM . | E . | Mixed . | Total . | |||||
| Experiment 1 | |||||||||||
| Ly-5.2 | 28 ± 6 | 87 ± 6* | 6 ± 1 | 11 ± 4† | 1 ± 1 | 133 ± 8† | |||||
| Ly-5.1 | 15 ± 1 | 48 ± 3 | 10 ± 4 | 1 ± 2 | 0 ± 1 | 74 ± 3 | |||||
| Experiment 2 | |||||||||||
| Ly-5.2 | 24 ± 4* | 73 ± 7* | 5 ± 2 | 11 ± 4† | 1 ± 0 | 113 ± 13† | |||||
| Ly-5.1 | 13 ± 3 | 35 ± 4 | 5 ± 2 | 1 ± 1 | 1 ± 1 | 55 ± 7 | |||||
| Experiment 3 | |||||||||||
| Ly-5.2 | 26 ± 7 | 103 ± 14† | 13 ± 4† | 6 ± 2* | 5 ± 2 | 152 ± 17† | |||||
| Ly-5.1 | 20 ± 4 | 77 ± 6 | 6 ± 3 | 1 ± 2 | 3 ± 1 | 107 ± 12 | |||||
Numbers represent mean ± SD of colonies from 4000 BM MNCs in quadruplicate culture.
P values were determined for differences between Ly-5.2 and Ly-5.1 groups.
G indicates granulocyte colonies; M, macrophage colonies; GM, granulocyte-macrophage colonies; E, erythroid bursts; mixed, granulocyte-macrophage-erythrocyte colonies.
P < .01.
P < .05.
Transplantation studies
To further confirm that the observed abnormalities are intrinsic to the hematopoietic cells of the mutants, we carried out transplantation studies of the BM cells from the chimeric mice. First, we prepared, by fluorescence-activated cell sorter (FACS) sorting, Ly-5.2 cells from the BM MNCs of one mouse each of WT (72.2% Ly-5.2 cells), Fli-1+/- (26.3% Ly-5.2 cells), and Fli-1-/- (21.5% Ly-5.2 cells) chimeric mice. We then transplanted 1 × 105 sorted Ly-5.2 MNCs into irradiated Ly-5.1 mice together with 2 × 105 BM nucleated cells from a normal Ly-5.1 mouse as radioprotective cells. Two months later, we analyzed lineage expression in the Ly-5.2 and Ly-5.1 cell populations of the PB of the recipient mice. The levels of Ly-5.2 cells in the mice with transplants of WT cells, Fli-1+/- cells, and Fli-1-/- cells were 47.2% ± 12.2%, 28.2% ± 13.3%, and 20.5% ± 15.8%, respectively. A summary of differential counts of the PB nucleated cells of the transplant recipients is presented in Table 4. The findings in general recapitulated those in the donor chimeric mice presented earlier. There was striking reduction in the numbers of granulocytes (Gr-1brightMac-1+ cells) and a significant increase in the numbers of NK cells relative to those in Ly-5.1 populations. There were increases in the CD4+ T cells and CD8+ T cells, but this was in large part due to unusually low levels of Fli-1+/+ Ly-5.2 CD4+ T cells and CD8+ T cells in this experiment. Although the number of monocytes was reduced in the Ly-5.2 population of the mice with transplants of Fli-1+/- cells, studies with Fli-1-/- cells did not show significant differences.
Lineage expression by Ly-5.2 and Ly-5.1 populations of mice with transplants
Donor genotype and population studied . | Lineage+ cells, mean ± SD % . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | GrbrightMac+ . | Grdim/-Mac+ . | B220+ . | CD4+ . | CD8+ . | NK1.1+ . | |||||
| Fli-1+/+ | |||||||||||
| Ly-5.2 | 9.7 ± 2.9 | 15.7 ± 4.7 | 58.1 ± 9.5 | 7.5 ± 1.8 | 3.0 ± 0.7 | 1.8 ± 0.7 | |||||
| Ly-5.1 | 8.5 ± 3.1 | 14.1 ± 2.8 | 36.5 ± 9.1 | 21.0 ± 5.9 | 13.8 ± 4.4 | 1.7 ± 0.3 | |||||
| Fli-1+/- | |||||||||||
| Ly-5.2 | 5.1 ± 3.1* | 8.1 ± 5.4* | 55.4 ± 18.6 | 10.2 ± 2.6 | 12.2 ± 3.0† | 1.2 ± 0.3‡ | |||||
| Ly-5.1 | 8.4 ± 3.6 | 11.7 ± 2.9 | 48.1 ± 10.1 | 11.5 ± 2.3 | 14.6 ± 3.3 | 1.0 ± 0.3 | |||||
| Fli-1-/- | |||||||||||
| Ly-5.2 | 1.5 ± 0.8† | 10.8 ± 3.4 | 37.8 ± 8.1‡ | 25.8 ± 5.6† | 16.6 ± 6.3† | 6.5 ± 2.5‡ | |||||
| Ly-5.1 | 11.8 ± 3.6 | 14.5 ± 2.8 | 41.2 ± 6.4 | 16.4 ± 1.6 | 11.2 ± 0.8 | 1.5 ± 0.5 | |||||
Donor genotype and population studied . | Lineage+ cells, mean ± SD % . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | GrbrightMac+ . | Grdim/-Mac+ . | B220+ . | CD4+ . | CD8+ . | NK1.1+ . | |||||
| Fli-1+/+ | |||||||||||
| Ly-5.2 | 9.7 ± 2.9 | 15.7 ± 4.7 | 58.1 ± 9.5 | 7.5 ± 1.8 | 3.0 ± 0.7 | 1.8 ± 0.7 | |||||
| Ly-5.1 | 8.5 ± 3.1 | 14.1 ± 2.8 | 36.5 ± 9.1 | 21.0 ± 5.9 | 13.8 ± 4.4 | 1.7 ± 0.3 | |||||
| Fli-1+/- | |||||||||||
| Ly-5.2 | 5.1 ± 3.1* | 8.1 ± 5.4* | 55.4 ± 18.6 | 10.2 ± 2.6 | 12.2 ± 3.0† | 1.2 ± 0.3‡ | |||||
| Ly-5.1 | 8.4 ± 3.6 | 11.7 ± 2.9 | 48.1 ± 10.1 | 11.5 ± 2.3 | 14.6 ± 3.3 | 1.0 ± 0.3 | |||||
| Fli-1-/- | |||||||||||
| Ly-5.2 | 1.5 ± 0.8† | 10.8 ± 3.4 | 37.8 ± 8.1‡ | 25.8 ± 5.6† | 16.6 ± 6.3† | 6.5 ± 2.5‡ | |||||
| Ly-5.1 | 11.8 ± 3.6 | 14.5 ± 2.8 | 41.2 ± 6.4 | 16.4 ± 1.6 | 11.2 ± 0.8 | 1.5 ± 0.5 | |||||
Numbers represent mean ± SD (n = 6 for Fli-1+/+, n = 4 for Fli-1+/-, and n = 5 for Fli-1-/-) of percentage of lineage-positive cells within Ly-5.2 or Ly-5.1 cell population. P values were determined for differences compared with Fli-1+/+ groups.
P < .05.
P < .001.
P < .01.
Regulation of hematopoietic genes in the Fli-1-/- chimeric mice
We and others have previously demonstrated that altered Fli-1 expression is associated with changes in erythroid and megakaryocytic differentiation. The phenotypes described in the preceding sections indicated that absence of Fli-1 results in cell-autonomous defects that affect granulocyte production, supporting a previously unrecognized function for Fli-1. On the basis of this novel observation, we initiated analyses of potential targets of Fli-1 that may contribute to the defective granulocyte production seen in Fli-1-/- cells. FACS sorting was used to prepare Ly-5.2 and Ly-5.1 cells from the BM of a Fli-1-/- chimeric mouse. Gene expression was evaluated by semiquantitative RT-PCR of RNA prepared from these cell populations. We first examined whether the expression of 2 genes that are important for megakaryocytic development (c-MPL and GATA-1) are affected by the absence of Fli-1. Consistent with our previous findings,14 c-MPL expression was reduced in RNA prepared from the Ly-5.2 (Fli-1-/-) BM cell population (Figure 4). The level of GATA-1 expression was dramatically reduced in this RNA compared with that prepared from the Ly-5.1 (Fli-1+/+) BM cell population. As is the case for megakaryocytic genes, multiple genes that affect early hematopoiesis as well as granulopoiesis contain Ets binding sites in their promoters and thus represent potential Fli-1 target genes.15 Several transcription factors regulate genes that are involved in myelopoiesis, and targeted inactivation of CCAAT/enhancer binding protein-α (C/EBPα), C/EBPϵ, and PU.1 has demonstrated that these genes are critical for granulocyte development.16-18 While PU.1 expression was not significantly different in the Ly-5.2 (Fli-1-/-) and Ly-5.1 (Fli-1+/+) BM populations, the expression of C/EBPα and C/EBPϵ in the Fli-1-/- cells was significantly reduced (Figure 4).
Altered expression of potential Fli-1 target genes in Ly-5.2 cells of Fli-1-/- chimeric mice demonstrated by semiquantitative RT-PCR. Expression was analyzed for HPRT and the indicated target genes.
Altered expression of potential Fli-1 target genes in Ly-5.2 cells of Fli-1-/- chimeric mice demonstrated by semiquantitative RT-PCR. Expression was analyzed for HPRT and the indicated target genes.
Granulopoiesis is also dependent on specific cytokines and cytokine receptors.19-21 We next evaluated the effect of Fli-1 expression on granulocyte colony-stimulating factor (G-CSF) receptor and granulocyte-macrophage CSF (GM-CSF) receptor. GM-CSF stimulates proliferation and differentiation of granulocyte/macrophage progenitor cells as well as modulating the biologic functions of mature granulocytes. High-affinity GM-CSF binding requires coexpression of the GM-CSF binding subunit (α subunit) and GM-CSF receptor β subunit.22 Consistent with defective granulopoiesis, expression of G-CSF receptor and the α and β subunits of GM-CSF receptor are each significantly reduced in the Fli-1-/- cells. In contrast, expression of the murine β2 subunit (also known as A1C2A) that is more than 90% identical to β1 (A1C2B), but binds IL-3, was not altered in these cell populations. GM-CSF expression was elevated in the Fli-1-/- cells. Mac-1 is composed of αMβ2 integrins, which are encoded by CD11b and CD18. We found that CD18 is reduced in the Fli-1-/- cells, while CD11b expression was elevated. We did not detect a difference in Gr-1 expression in the 2 cell populations.
We also evaluated expression of other genes that are critical for normal hematopoiesis, including Tal-1/SCL, c-FMS (M-CSF receptor), and GATA-2.23-25 While c-FMS expression was not significantly different in the Fli-1-/- and Fli-1+/+ BM cells, GATA-2 was slightly reduced, and we also found a significant decrease in Tal-1/SCL. Expression of IL-6 receptor and EPO receptor was not significantly altered. We did not detect a significant difference between WT Ly-5.1 and WT Ly-5.2 in expression of 3 representative genes (G-CSF-R, CD18, and Tal-1/SCL) that were differentially expressed in the Fli-1+/+ and Fli-1-/- BM cells (data not shown).
Discussion
Target organs/tissues of many Ets-related genes include the lympho-hematopoietic system and the major phenotype of Fli-1-deficient mice is severe thrombocytopenia.13 In our laboratory, we noted a striking reduction in the number of megakayocytes in cultures of aorta-gonad-mesonephros (AGM) cells from Fli-1-/- embryos.14 Fli-1-/- embryos exhibit central nervous system (CNS) hemorrhage beginning on E11.0 and die prior to E12.5. Since the hemorrhage and embryonic lethality occur before BM cells of the fetuses become transplantable to lethally irradiated adult mice,26 it has not been possible to carry out comprehensive studies of the role of Fli-1 in hematopoiesis. To avoid this problem, we generated Fli-1 chimeras by aggregation of Ly-5.2 Fli-1 mutant embryos with Ly-5.1 Fli-1 WT embryos and compared lineage expression by mutant (Fli-1+/+, Fli-1+/-, or Fli-1-/-) cells with that of WT Ly-5.1 cell populations in individual chimeric mouse. We also carried out BM transplantation to further ascertain the cell-intrinsic nature of the abnormalities.
The most striking finding was a reduction in the number of granulocytes seen in both the primary chimeras and transplant recipients. The levels of PB granulocytes in the Fli-1-/- population were almost one tenth of those in the WT Ly-5.1 population. While morphology of the BM Fli-1+/- cells did not show abnormalities, clonal cell culture studies of BM Fli-1+/- cells clearly documented increases in the numbers of granulocyte and granulocyte /macrophage progenitors. These results may indicate either the presence of defective leukocyte maturation or the inability of mature leukocytes to enter the circulation. In either case, the results indicate that Fli-1 plays a physiologic role in production of leukocytes.
Consistent with the cellular phenotype of defective granulopoiesis, expression of C/EBPα, C/EBPϵ, and G-CSF receptor was reduced in the Fli-1-/- cells. Expression of these genes is critical for granulopoiesis. Mice harboring C/EBPα-targeted alleles exhibit selective absence of mature granulocytes.18 These mice showed reduced expression of G-CSF and interleukin-6 receptors while the mRNA levels for GM-CSF receptor was not affected. In contrast, in the Fli-1-/- cells, the expression of GM-CSF receptor was reduced but that of interleukin-6 receptor was not altered. C/EBPϵ-null mice have increased numbers of atypical granulocyte progenitors in the BM, suggesting that C/EBPϵ is required for differentiation and functional maturation of granulocyte progenitor cells.17 Northern blot analysis of BM mRNA from C/EBPϵ-null mice demonstrated elevated expression of GM-CSF receptor, in contrast to results found for Fli-1-/- BM cells. G-CSF receptor-null mice are neutropenic with approximately 12% circulating neutrophils. Interestingly, these mice retain significant levels of BM granulopoiesis,19,21 similar to the Fli-1+/- chimeric mice. The observed altered expression of GM-CSF and its receptor are also significant, as GM-CSF is a known regulator of granulocyte function and hematopoietic progenitor cell migration.20
We also assessed the expression of hematopoietic cell surface markers that define myeloid cells, such as Mac-1 and Gr-1. The myeloid-specific marker integrin αMβ2 (CD18/CD11b, Mac1) regulates granulocyte adherence and migration.27 Consistent with previous studies that have demonstrated that the CD18 promoter contains functional Ets binding sites,28,29 CD18 mRNA was significantly reduced in Fli-1-/- BM cells. In contrast, CD11b expression was elevated and Gr-1 expression was retained in the Fli-1-/- population. While this expression may seem in conflict with the observed dramatic reduction of Gr-1+Mac-1+ cells, expression is not equivalent to cell surface localization. In the context of reduced CD18, CD11b may not be present on the cell surface.
GATA-1 is required for megakaryocytic and erythroid development.30,31 The significantly reduced GATA-1 expression is consistent with our previous demonstration that Fli-1 can activate GATA-1 expression.1,2 Fli-1 and GATA-1 work synergistically to regulate multiple megakaryocytic genes, including gpIIb,4-6 gpVI,7 gpIX,8 gpIbα,8 and c-MPL.9 It is likely that reduced GATA-1, combined with loss of Fli-1, manifested the megakayocytic defects described previously in Fli-1 null mice.13,14
The Tal-1/SCL gene is expressed in hematopoietic stem cells and is essential for the development of both primitive erythropoiesis and definitive hematopoiesis.24,25 The observed reduction of Tal-1/SCL gene expression in Fli-1-/- BM is consistent with the recent demonstration that the SCL enhancer is regulated by Ets factors in vivo32 and may suggest that Fli-1-mediated control of Tal-1/SCL may contribute to multilineage development.
The presence of Ets binding sites and the observed altered expression of specific genes in the Fli-1-/- cells support the model that they may be direct Fli-1 targets. However, it is possible that the block in granulocyte differentiation may result in an insufficient number of cells that express the gene in question; thus, future studies will be required to determine whether these genes are direct targets for Fli-1 in vivo.
In contrast to the chimeric mice, differential counts of PB nucleated cells from nonchimeric heterozygous mice were normal. We believe that the differences between the two are caused by the absence and presence of compensatory mechanisms, respectively. Such compensatory mechanism in the nonchimeric mice may include, for example, elevated levels of cytokines. It has been proposed that targets of Fli-1 include c-MPL,9 the receptor for thrombopoietin. It is possible that the receptor of G-CSF, another single-molecule receptor of hematopoietic cytokine, may also be a target of Fli-1. If so, compensatory increase in G-CSF may account for the normal numbers of granulocytes in the nonchimeric mutant mice. In contrast, in the chimeric mice, the presence of normal Ly-5.1 cells most likely attenuates the feedback compensation mechanisms. If this is the case, use of chimeric mice may prove to be a very sensitive and useful method to study hematopoietic defects of gene-targeted mice that are not obvious in the heterozygous mice.
We could not ascertain the contribution of mature red blood cells by the mutant populations in the chimeric mice because mature red blood cells do not express Ly-5. Clonal cell culture studies of the BM MNCs, however, revealed a striking increase in the number of primitive erythroid progenitors, BFU-Es, in the Fli-1+/- cell population. Earlier, we noted paucity in the early erythroid precursors such as pronormoblasts and basophilic normoblasts in the livers of Fli-1-/- embryos.12 In addition, precursors for these cells were also significantly diminished in cultures of AGM cells from Fli-1-/- mice.14 These observations appear to complement the earlier observation that Fli-1 activation by Friend murine leukemia virus induces erythroleukemia33 and supports the notion that Fli-1 promotes proliferation at the expense of differentiation of erythroid lineage.34 These observations, together with the observed leukopenia and concomitant increase in their progenitors in BM of the chimeric mice, may suggest common regulatory mechanisms for the myeloid and erythroid lineages.
Prepublished online as Blood First Edition Paper, September 14, 2004; DOI 10.1182/blood-2003-12-4345.
Supported by Office of Research and Development, Medical Research Services, Department of Veterans Affairs, and National Institutes of Health grants PO1-CA78582 and RO1-DK54197.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Dr Yong Gong for technical assistance in the generation of chimeric mice, Mrs Anne G. Livingston and Dr Amanda C. LaRue for assistance in preparation of this manuscript, and the staff of the Radiation Oncology Department of the Medical University of South Carolina for assistance in the irradiation of mice.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal