Abstract
Most patients with chronic granulomatous disease (CGD) have mutations in the X-linked CYBB gene that encodes gp91phox, a component of the phagocyte NADPH oxidase. The resulting X-linked form of CGD is usually manifested in boys. Rarely, X-CGD is encountered in female carriers with extreme expression of the mutated gene. Here, we report on a woman with a novel mutation in CYBB (CCG[90-92] → GGT), predicting Tyr30Arg31 → stop, Val in gp91phox, who presented with clinical symptoms at the age of 66. The mutation was present in heterozygous form in genomic DNA from her leukocytes but was fully expressed in mRNA from these cells, indicating that in her leukocytes the X chromosome carrying the nonmutated CYBB allele had been inactivated. Indeed, only 0.4% to 2% of her neutrophils showed NADPH oxidase activity. This extreme skewing of her X-chromosome inactivation was not found in her cheek mucosal cells and is thus not due to a general defect in gene methylation on one X chromosome. Moreover, the CYBB mutation was not present in the DNA from her cheek cells and was barely detectable in the DNA from her memory T lymphocytes. Thus, this patient shows a somatic mosaic for the CYBB mutation, which probably originated during her lifetime in her bone marrow.
Introduction
Chronic granulomatous disease (CGD) is a rare, heterogeneous, inherited disorder that affects about 1 in 250 000 individuals.1,2 The main defect in CGD is a failure of neutrophils, monocytes, macrophages, and eosinophils to mount a respiratory burst and, therefore, to generate superoxide anions and other reactive oxygen species derived from superoxide, such as hydrogen peroxide. This renders the patients susceptible to severe, recurrent bacterial and fungal infections.3-5 The enzyme that generates superoxide is a phagocyte-specific nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, consisting of 2 membrane-bound subunits and 4 cytosolic proteins involved in activity regulation of the enzyme.1,2 The central, catalytic subunit is gp91phox, a flavocytochrome that forms a heterodimer with p22phox in the plasma membrane of phagocytic leukocytes. Defects in gp91phox are inherited in an X-linked fashion because its gene, CYBB, is located on the X chromosome. About 70% to 75% of the CGD patients are X-CGD patients.1,2,6 Autosomal forms of CGD are due to mutations in CYBA, the gene that encodes p22phox in approximately 3% of the CGD patients6,7 or to mutations in the genes encoding the cytosolic proteins p47phox (∼ 25%) or p67phox (∼ 3%).1,2,6
For many years the onset of CGD was regarded to occur early in infancy, with fatal outcome in adolescence, due to the recurrent severe infections and the secondary granulomatous and fibrotic tissue formation that develops in many organs. Changes frequently occur in the skin, lymph nodes, and lungs, but also in bones, joints, liver, and kidneys, leading eventually to pulmonary, gastrointestinal, musculoskeletal, and renal insufficiency.1,2,5,8,9
In the last decades, with the rapid development of sophisticated diagnostic tools and techniques, a better understanding has been gained of the pathophysiologic mechanisms of the disease, and the genetic basis of CGD has been elucidated.1,2,6 Moreover, a more accurate determination of the infectious etiological agents has led to a more appropriate, specific therapy.8-11 Together with improved techniques of bone marrow transplantation, this has brought about the prolongation of life and/or the correction of the basic disorder in CGD.12-16
CGD is a very heterogeneous disease, with different genetic causes and with a variable and heterogeneous clinical spectrum, from a mild disorder to a very severe form. The patient here reported is unique because the disease manifested very late in life and the genetic cause of her disease is unusual. She appeared to be a carrier of the X-linked form of CGD, who usually have no, or only mild clinical, complaints because these women have a mixture of normal and abnormal phagocytes in their circulation. However, this woman had very few normal phagocytes, due to a skewed X-chromosome inactivation pattern. Moreover, the mutation in CYBB was found in her short-living blood cells, but not in her memory T lymphocytes or in her cheek mucosal cells. This indicates that the skewed X-chromosome inactivation pattern in her leukocytes may be due to a clonal expansion of a mutated bone marrow stem cell.
Patient, materials, and methods
We report an 80-year-old woman of Iraqi origin who was born to related parents. As shown in the family pedigree (Figure 1), she is one of 2 pairs of twin siblings; 1 of each pair died soon after birth from an undetermined etiology. Of her offspring, 9 were healthy and 1 of the twins died at the age of 3 months from a severe bacterial infection. A great-grandchild died at 10 days of age from a fulminant infection.
Pedigree of the patient and her family. The patient is indicated in black. Deceased family members are indicated with a diagonal in their symbol. Age at death of no. 3, 54 years; no. 6 and no. 8, at birth; no. 11, 3 months; no. 22, 10 days. Healthy grandchildren and great-grandchildren of the patient are not indicated.
Pedigree of the patient and her family. The patient is indicated in black. Deceased family members are indicated with a diagonal in their symbol. Age at death of no. 3, 54 years; no. 6 and no. 8, at birth; no. 11, 3 months; no. 22, 10 days. Healthy grandchildren and great-grandchildren of the patient are not indicated.
The patient had a normal and healthy life until age 66; since then she underwent about 30 hospitalizations within 8 years, for Serratia marcescens sepsis, recurrent pneumonia (5 times) and sinusitis (two times), Staphylococcal aureus pretibial abscess, Acinetobacter skin abscess, Escherichia coli and Candida albicans urinary tract infection, Providentia osteomyelitis and septic arthritis, suppurative adenitis, liver cysts and calcified lesions (detected by computerized tomography), pan-uveitis, anthralgia, vaginal ulcers, aphthous stomatitis, pyoderma gangrenosum, and vasculitis-like skin rash on face and limbs. Elevated red blood cell sedimentation rate, leukocytosis, neutrophilia, and remarkable dimorphic anemia manifested by anisocytosis as sometimes seen in chronic disorders (microcytichypochromic anemia associated with macrocytic red blood cells) as well as hypergammaglobulinemia became apparent with time. Many tentative diagnoses were raised, such as Behçet disease, vasculitis, systemic lupus erythematosus, and Wegener granulomatosis. Insufficient clinical and laboratory manifestations supported these tentative diagnoses. She was treated with antibiotics, surgical drainage, nonsteroidal anti-inflammatory drugs, steroids, and colchicine.
Following comprehensive studies of leukocyte functions, Western blot analysis of neutrophil extracts, and molecular DNA analysis, the diagnosis of chronic granulomatous disease was established at the age of 74 years. Since then, she was successfully treated with trimethoprim-sulphamethoxazole on a prophylactic daily basis, and no more admissions or relevant infections were recorded. At present in her eighties, she is leading a completely normal life.
Purification of blood cells
Neutrophils were purified from heparinized venous blood of the patient and her relatives. Simultaneously, a healthy volunteer served as control. Informed consent was obtained from all participants. Neutrophils were isolated from 10 milliliters of heparinized blood by dextran sedimentation, followed by erythrocyte lysis, as described by Böyum.17 The neutrophils were washed and resuspended in phosphate-buffered saline with 1% (vol/vol) albumin and kept on ice until tested. The purity of these cells was 99% and the viability more than 95%.
Monocytes and lymphocytes were purified as described by Roos and de Boer.18 CD45RO+ and CD45RO– T lymphocytes were purified from mononuclear leukocytes by incubation with CD45RO magnetic microbeads (Miltenyi, Gladbach, Germany) and isolation over a VarioMacs column. The fraction with CD45RO+ cells was further purified by incubation with CD45RO-phycoerythrin and CD3-fluorescein isothiocyanate (Becton Dickinson, San Jose, CA) and fluorescence-activated cell-sorter scanner (FACS) sorting of the double-positive cells in a MoFlow high-speed cell sorter (Dako Cytomation, Carpinteria, CA).
Neutrophil function tests
Superoxide production by neutrophils was measured as superoxide dismutase–inhibitable reduction of ferricytochrome c by the method of Weisbart et al.19 Neutrophils (106) were suspended in Hanks balanced salt solution with 60 μM ferricytochrome c, with (control) or without 214 U of superoxide dismutase. The rate of superoxide anion release was measured after addition of 100 nM N-formyl-methionyl-leucyl-phenylalanine (fMLP) or 10 ng/mL phorbol myristate acetate (PMA) for 10 minutes at 37°C in a UV-260 Shimadzu spectrophotometer at 550 nm. The superoxide anion release was calculated with the Massey extinction coefficient for ferrocytochrome c of 2.1 × 104 M–1 cm–1.
Superoxide production by neutrophils was also measured as nitroblue tetrazolium (NBT) reduction by the NBT slide test reported by Baehner et al20 with slight modifications.
Hydrogen peroxide production by neutrophils was measured with the dihydrorhodamine-1,2,3 (DHR) assay as described by Vowells et al.21 In short, neutrophils were incubated with DHR and catalase and were activated with 100 ng/mL PMA. The reaction was stopped at various times, and the amount of DHR oxidation product rhodamine was measured by FACS analysis.
The bactericidal activity was assessed as previously reported22 and expressed as the decrease in the number of viable bacteria after incubation of bacteria with neutrophils in the presence of autologous and homologous serum.
The chemotactic response was assessed by 48-well chemotaxic microchamber (Neuro Probe, Bethesda, MD) to determine random migration and chemotaxis as previously reported.23
Western blot was performed for protein analysis of the NADPH-oxidase system.24 Neutrophil lysate (20 μg) was electrophoresed on 10% sodium dodecyl sulfate–polyacrylamide gel and transferred to a nylon membrane. The membrane was blocked with 5% skimmed milk, incubated with a monoclonal antibody against gp91phox (mAb 48) or p22phox (mAb 449), washed, incubated with alkaline phosphatase–conjugated rabbit–anti–mouse Ig, and developed with Alkaline Phosphatase Stain.
DNA and RNA analysis
DNA was purified from leukocytes, cultured fibroblasts, or cheek mucosal cells by standard methods.25 Polymerase chain reaction (PCR) amplification and sequence analysis of all CYBA and CYBB exons with their intron/exon boundaries and of approximately 400 basepairs of the CYBB promoter region was performed as described previously.7,26 RNA was isolated from mononuclear leukocytes and converted to cDNA by standard methods.7,27 A 627-basepair fragment containing the nucleotide sequence of the first five and a half exons of CYBB was amplified and sequenced as described previously.26
Detection of X-chromosome inactivation at the HUMARA locus
The HUMARA locus (human androgen-receptor gene) on the X chromosome contains 2 methylation-sensitive HpaII restriction sites and is 90% polymorphic in Caucasian females for varying allele sizes.28 Genomic DNA was incubated with HpaII overnight to digest all unmethylated, active DNA at this locus. The mixture of digested and undigested DNA was PCR amplified with FAM-labeled fluorescent primers flanking a region comprising both the polymorphic and the restriction sites, according to Allen et al.28 Capillary PCR conditions were as follows: 5 seconds at 95°C, 30 seconds at 60°C, and 15 seconds at 72°C, for 50 cycles. The PCR products were diluted 1:5 with loading buffer. The samples were then heat-inactivated at 98°C for 3 minutes and cooled on ice. Two microliters of each sample were run on an ABI Prism 377 sequencer (Applied Biosystems, Foster City, CA) for separation of the PCR products. The intensities of the bands corresponding to the 2 alleles before and after HpaII digestion were compared to determine the X-chromosome inactivation ratio between the 2 alleles in each female by means of the program Genescan 3.1 (Applied Biosystems).
Polymorphic marker analysis
Isolated DNA was amplified with the commercially available fluorescent short tandem repeat multiplex systems SGM+ (Applied Biosystems) and PowerPlex 16 (Promega, Madison, WI). The set-up of the SGM+ kit was modified by reducing the amplification mixture per sample to 10 μL and a template volume of 1 μL.29 The PowerPlex-16 set-up was modified by reducing the mixture per sample to 10 μL, with 1:2 diluted 10 primer-pair mix.30 PCR products were separated on the ABI 310 analyzer (Applied Biosystems).
Results
The patient's neutrophils displayed a significantly reduced bactericidal activity both in the presence of autologous and homologous serum (Table 1). The NBT slide test with PMA stimulation showed only 2% positive cells, and the DHR test with PMA only 0.4% positive cells. The fMLP- or PMA-stimulated superoxide generation by the patient's neutrophils was undetectable (Table 1). As shown in Table 1, all tests performed with the neutrophils from 4 of her 5 daughters and 3 of her 4 living sons were within the normal range (son no. 12 and daughter no. 14 were not available for analysis). The chemotactic activity and random migration of the patient's neutrophils were found to be normal (data not shown). Western blot with mAb 48 anti-gp91phox or mAb 449 anti-p22phox showed negative results with the patient's neutrophils. This test was normal in her daughters (the sons were not investigated) (Figure 2).
Results of family studies
. | . | \(\mathrm{O}_{2}^{-}\) generation*. | . | Bactericidal activity† . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|
| Subject . | DHR (% positive) . | fMLP . | PMA . | Autologous serum . | Homologous serum . | Mutation present . | Skewing‡(%) . | ||
| Control (♀) | 99 | 3.35 | 4.24 | 1.35 | 1.17 | No | 72 | ||
| Patient no. 9 | 0.4 | 0 | 0 | 0.15 | 0.35 | Yes | 99 | ||
| Son no. 13 | NT | 3.88 | 6.07 | 1.25 | 1.55 | No | NA | ||
| Daughter no. 15 | 95 | 5.15 | 3.31 | 1.07 | 0.94 | No | 75 | ||
| Daughter no. 16 | NT | 4.99 | 4.98 | 0.92 | 0.84 | No | 54 | ||
| Daughter no. 17 | 100 | 4.27 | 2.92 | 0.98 | 1.00 | No | 71 | ||
| Son no. 18 | NT | 4.48 | 2.94 | 1.31 | 1.45 | No | NA | ||
| Son no. 19 | NT | 2.59 | 3.62 | 1.14 | 1.34 | No | NA | ||
| Daughter no. 20 | 100 | 5.94 | 8.39 | 1.06 | 0.94 | No | 50 | ||
. | . | \(\mathrm{O}_{2}^{-}\) generation*. | . | Bactericidal activity† . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|
| Subject . | DHR (% positive) . | fMLP . | PMA . | Autologous serum . | Homologous serum . | Mutation present . | Skewing‡(%) . | ||
| Control (♀) | 99 | 3.35 | 4.24 | 1.35 | 1.17 | No | 72 | ||
| Patient no. 9 | 0.4 | 0 | 0 | 0.15 | 0.35 | Yes | 99 | ||
| Son no. 13 | NT | 3.88 | 6.07 | 1.25 | 1.55 | No | NA | ||
| Daughter no. 15 | 95 | 5.15 | 3.31 | 1.07 | 0.94 | No | 75 | ||
| Daughter no. 16 | NT | 4.99 | 4.98 | 0.92 | 0.84 | No | 54 | ||
| Daughter no. 17 | 100 | 4.27 | 2.92 | 0.98 | 1.00 | No | 71 | ||
| Son no. 18 | NT | 4.48 | 2.94 | 1.31 | 1.45 | No | NA | ||
| Son no. 19 | NT | 2.59 | 3.62 | 1.14 | 1.34 | No | NA | ||
| Daughter no. 20 | 100 | 5.94 | 8.39 | 1.06 | 0.94 | No | 50 | ||
NT indicates not tested; NA, not applicable.
In nmoles per 106 PMN per minute.
Toward S aureus, in log decrease of colony-forming units.
Skewing was defined as percentage change in HUMARA PCR signal caused by HpaII; 50% means equal distribution of methylated and unmethylated gene, that is, no skewing; whereas 99% means that HUMARA on one X chromosome is preferentially methylated, that is, very strong skewing.
Western blot analysis of neutrophil membranes from the patient and 4 of her daughters. Neutrophil membranes were solubilized and subjected to electrophoresis, blotted, and probed with mAbs against gp91phox (A) or against p22phox (B) as indicated in “Methods.” Lane 1, control neutrophil from a patient with X-CGD; lane 2, control neutrophils from a healthy individual; lane 3, neutrophils from the patient (no. 9); lane 4, neutrophils from daughter no. 20; lane 5, neutrophils from daughter no. 16; lane 6, neutrophils from daughter no. 17; and lane 7, neutrophils from daughter no. 15.
Western blot analysis of neutrophil membranes from the patient and 4 of her daughters. Neutrophil membranes were solubilized and subjected to electrophoresis, blotted, and probed with mAbs against gp91phox (A) or against p22phox (B) as indicated in “Methods.” Lane 1, control neutrophil from a patient with X-CGD; lane 2, control neutrophils from a healthy individual; lane 3, neutrophils from the patient (no. 9); lane 4, neutrophils from daughter no. 20; lane 5, neutrophils from daughter no. 16; lane 6, neutrophils from daughter no. 17; and lane 7, neutrophils from daughter no. 15.
Thus, the patient suffers from CGD caused by a defect either in gp91phox or p22phox, because these 2 proteins stabilize each other's expression in the neutrophil membrane—absence of one of these leads to simultaneous absence of the other.24
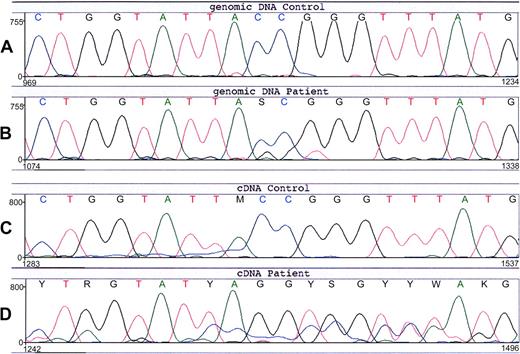
Sequencing of the patient's CYBA (encoding p22phox) did not reveal any mutations. However, in the patient's CYBB (encoding gp91phox) we found an unusual substitution of 3 nucleotides (Figure 3), that is, the sequence TACCGG at position 88-93 (A of the start codon ATG is number 1), which encodes Tyr30 and Arg31, was changed into TAGGTG, coding for Stop and Val. This predicts a premature termination of protein synthesis at amino-acid position 30. The patient was found to be heterozygous for this mutation. None of her relatives, including 5 grandsons and 3 granddaughters, had this mutation in their DNA (not shown). In contrast to the findings with the genomic DNA of the patient, the cDNA from the patient showed an apparently homozygous expression of the mutated sequence (Figure 3), which explains the total lack of NADPH oxidase activity in her neutrophils.
Nucleotide sequence of CYBB around predisposition 88-93 from the patient and a healthy control. (A) Genomic DNA from the leukocytes of a healthy control. (B) Genomic DNA from the leukocytes of the patient (no. 9). The normal CYBB sequence TACCGG is changed in the patient into a mixture with TAGGTG, indicating that she is a heterozygote for this 3-nt mutation. (C) Leukocyte cDNA from a healthy control. (D) Leukocyte cDNA from the patient (no. 9) shows an apparent homozygous mutant sequence TAGGTG. The underlying signal is from another exon, apparently due to some incorrect mRNA splicing.
Nucleotide sequence of CYBB around predisposition 88-93 from the patient and a healthy control. (A) Genomic DNA from the leukocytes of a healthy control. (B) Genomic DNA from the leukocytes of the patient (no. 9). The normal CYBB sequence TACCGG is changed in the patient into a mixture with TAGGTG, indicating that she is a heterozygote for this 3-nt mutation. (C) Leukocyte cDNA from a healthy control. (D) Leukocyte cDNA from the patient (no. 9) shows an apparent homozygous mutant sequence TAGGTG. The underlying signal is from another exon, apparently due to some incorrect mRNA splicing.
We then investigated whether this strongly skewed expression of one of her 2 CYBB loci was due to a generalized defect in X-chromosome inactivation in all somatic cells or confined to her blood cells. For this purpose, we analyzed a highly polymorphic microsatellite region near a cytosine methylation-sensitive restriction site for HpaII at the HUMARA locus on the X chromosome (Figure 4). Predigestion of DNA with HpaII cuts those alleles that are not inactivated by X-chromosome inactivation–related methylation and are therefore not amplified. In DNA isolated from the patient's leukocytes, the ratio between her paternal and maternal microsatellite signal was strongly affected by the HpaII treatment—one of these signals was almost completely suppressed, whereas the other was unaffected. This confirms our cDNA findings that in her leukocytes only one of both CYBB alleles on her X chromosomes is active and transcribed into mRNA. In contrast, in DNA isolated from a buccal smear, HpaII treatment did not affect the ratio between the 2 microsatellite signals, indicating that in her cheek mucosal cells no skewing in X-chromosome inactivation has taken place. Thus, the skewed X-chromosome inactivation in the patient is not a phenomenon found in all of her somatic cells. In the leukocyte DNA from the 4 daughters (Table 1) and 3 granddaughters (not shown) that we investigated, the ratio between maternal and paternal region changed much less dramatically by the HpaII treatment, indicating that they had not inherited this trait from their mother or grandmother. In contrast, in DNA isolated from the leukocytes and from cultured fibroblasts of a patient with a proven generalized defect in X-chromosome inactivation,31 we found that the HpaII treatment eliminated the formation of one of the parental microsatellite PCR products in both DNA preparations (Figure 4).
X-chromosome inactivation pattern of the patient and a control with an X-chromosome inactivation defect. DNA was isolated from the leukocytes and from a buccal smear of the patient (no. 9) as well as from the leukocytes and from cultured fibroblasts of a control with an X-chromosome inactivation defect.31 The DNA samples were incubated with or without the restriction enzyme HpaII and subjected to PCR amplification of the HUMARA locus (see “Methods”). The labeled products were separated by size chromatography and scanned for comparison. Panels A and B, DNA from patient (no. 9) buccal smear; panels C and D, DNA from patient (no. 9) leukocytes; panels E and F, DNA from control fibroblasts; panels G and H, DNA from control leukocytes. The patient shows equal inactivation of X chromosomes in the buccal smear but strongly skewed X-chromosome inactivation in the leukocytes. The control shows strongly skewed X-chromosome inactivation in both fibroblasts and leukocytes. Peaks indicated in gray are the main PCR product peaks; those in white are shorter products due to incorrect polynucleotide synthesis. The calculation of skewing percentage (Table 1) is based on the peak heights of the main PCR products.
X-chromosome inactivation pattern of the patient and a control with an X-chromosome inactivation defect. DNA was isolated from the leukocytes and from a buccal smear of the patient (no. 9) as well as from the leukocytes and from cultured fibroblasts of a control with an X-chromosome inactivation defect.31 The DNA samples were incubated with or without the restriction enzyme HpaII and subjected to PCR amplification of the HUMARA locus (see “Methods”). The labeled products were separated by size chromatography and scanned for comparison. Panels A and B, DNA from patient (no. 9) buccal smear; panels C and D, DNA from patient (no. 9) leukocytes; panels E and F, DNA from control fibroblasts; panels G and H, DNA from control leukocytes. The patient shows equal inactivation of X chromosomes in the buccal smear but strongly skewed X-chromosome inactivation in the leukocytes. The control shows strongly skewed X-chromosome inactivation in both fibroblasts and leukocytes. Peaks indicated in gray are the main PCR product peaks; those in white are shorter products due to incorrect polynucleotide synthesis. The calculation of skewing percentage (Table 1) is based on the peak heights of the main PCR products.
When we investigated the cheek mucosal DNA from the patient, we found that this DNA did not contain the TAGGTG sequence in CYBB, although polymorphic marker analysis proved this mucosal DNA to be otherwise identical to her leukocytic DNA (not shown). This indicates that the patient has a somatic mosaicism for the CYBB mutation and prompted us to search for this mutation in DNA isolated from various purified blood cells of the patient. In DNA from purified neutrophils and monocytes, we found the signals of the 3 aberrant nucleotides (GGT) to be about equal in intensity to the signals of the wild-type CCG, indicating that all neutrophils and monocytes contained a mutated X chromosome and an X chromosome with a normal CYBB gene. In DNA from CD45RO-positive memory T lymphocytes (still containing 10% monocytes), the mutation signals were only about 20% of the wild-type signals. In DNA from CD45RO-negative, naive lymphocytes, the mutation signals were about half of the wild-type signals. These results indicate that the CYBB mutation is present in heterozygous form in the short-living myeloid cells, but not in the long-living memory T lymphocytes and only to a limited extent in the shorter-living naive lymphocytes. Similarly, the skewing of the X-chromosome inactivation was found to be extreme in the myeloid cells and in the naive T lymphocytes but not in the memory T cells. Examination of bone marrow taken in 1992 revealed normocellular tissue with diffuse fibrosis replacing normal trabecular bone and architecture, as well as an increased (left-shifted) myeloid/erythroid ratio without increased blast counts.
Discussion
The impaired respiratory burst of CGD phagocytes is caused by a defect in one of the 4 components of the phagocyte NADPH oxidase, either in one of the 2 subunits that reside in the cell membrane (gp91phox, p22phox) or in 1 of 2 cytosolic subunits (p47phox, p67phox). Our patient has a gp91phox deficiency, type X910 (no gp91phox protein expression). She occasionally suffered from banal infections until the age of 66 years. Since then, she was hospitalized about 30 times for severe, recurrent bacterial or fungal infections. No clinical or laboratory evidence of an autoimmune disorder or other disease was established. The family pedigree reported here showed 2 twin siblings who died at birth for reasons not well established, and 2 additional members, 1 son and 1 great-grandson who died at 3 and 10 months of age, respectively, from a severe generalized infection of nondetermined etiology. Although the primary disorder causing the fatal infections could not be established, knowing the family history, we could not discard the possibility of a primary immune deficiency. Therefore, we performed a full screen for phagocytic disorders and indeed, the functional, biochemical, and molecular diagnosis of CGD was established in the indicator patient. She was put on trimethoprim-sulphamethoxazole prophylaxis and her clinical condition remarkably improved.
The most common form of CGD is due to gp91phox absence, as we found in the patient reported here. The mutation in this patient is unusual, because in general the mutations in CYBB are either point mutations (substitutions, deletions, or insertions) or larger insertions or deletions.32 However, occasionally, more complicated mutations are found. Since CYBB is on the X chromosome, CGD patients with mutations in this gene are usually male, but a few female patients are known.1,6,32 Until now, these all are heterozygotes for CYBB mutations with a low number of functionally active neutrophils (< 15%). This is caused by skewed X-chromosome inactivation, which is a random process early in the fetal development of female individuals. The patient described by us is unusual in her extreme low percentage of neutrophils with an active NADPH oxidase (0.4%-2%), in agreement with a lack of wild-type gp91phox mRNA. These findings can be due either to a random unfavorable X-chromosome inactivation or to a concurrent deficiency of X-chromosome inactivation caused by a mutation in a gene involved in X-chromosome inactivation. X-chromosome inactivation is a process in which one or more genes on the X-chromosome itself are involved.33,34 Products from these genes inactivate the genes located on the same X-chromosome in female cells. If in our patient an additional mutation is located on the X-chromosome that carries the CYBB mutation, this could lead to a lack of inactivation of this CYBB-mutated chromosome, and thus to unique expression of proteins encoded by this CYBB-mutated chromosome. Similar cases of female patients with Wiscott-Aldrich syndrome, Duchenne muscular dystrophy, or glucose-6-phosphate dehydrogenase deficiency in combination with inherited skewed X-chromosome inactivation have been described.31,35,36 However, in our patient this is unlikely, because in that case her clinical symptoms might be expected to have become apparent much earlier in life.
To rule out an inherited X-chromosome inactivation defect, we investigated whether the skewed X-chromosome inactivation pattern was present not only in her leukocytes but also in other somatic cells. With the HUMARA assay we found that this was not the case, in contrast to the situation in the previously mentioned female patient with G6PD deficiency and inherited X-chromosome inactivation defect.31 It is therefore more probable that our patient acquired the skewed X-chromosome inactivation pattern in her leukocytes in the course of her life. Indeed, it is known that this pattern can shift in the hematopoietic system over a woman's lifetime.37-40 This may be due to stem cell depletion, true clonal hematopoiesis, or a growth advantage conferred by parental-specific X chromosomes.41-43 Two recent reports describe this phenomenon as the cause of X-CGD in adult females, at age 45 and 43, respectively.44,45 Apparently, in our patient, it took even longer before the X-chromosome inactivation pattern was skewed to a degree that caused the clinical problems.
The fact that the mutation in CYBB was not transmitted from the indicator patient to any of her 7 children who we investigated is in accordance with our hypothesis that the disorder was acquired late in life. Also, the 2 living children that were unavailable for study have no medical records indicative of CGD. Our finding that the patient has a somatic mosaicism for the CYBB mutation (present in her leukocytes but not in her cheek mucosal cells) provides a clue to explain this phenomenon, because we suppose that the mutation is also absent from her germ-line cells and thus cannot be transmitted to her offspring.
The most plausible explanation for the origin of the clinical symptoms observed in this patient is, in our opinion, as follows. During her lifetime, the mutation in CYBB arose in the stem cells in her bone marrow. This may have happened later in her lifetime or during her embryonic development, but in the latter case these stem cells did not take part in hematopoiesis until later, because we found long-living memory T lymphocytes that did not contain the mutation. Exchange of hematopoietic stem cells with her twin sister during embryonic development46 was rendered unlikely by polymorphic marker comparison between the patients cheek mucosal DNA and her leukocytic DNA. Probably, the mutated stem cells took over hematopoiesis either by chance or by a certain clonal expansion. The latter possibility is the most likely, given the highly skewed pattern of X-chromosome inactivation in the short-living leukocytes. This clonal expansion may perhaps be due to a concomitant growth advantage over the wild-type cells. Since there is no known growth advantage of hematopoietic cells associated with X-CGD mutations, this suggests the influence of a parental gene expressed on the active X chromosome that bears the CGD mutation.41-43 The observed increase in myeloid/erythroid ratio and the diffuse fibrosis in the bone marrow at the time of diagnosis give indications of dysplastic hematopoiesis leading to clonal expansion but may also be due to coexisting and recurrent infections.
Prepublished online as Blood First Edition Paper, August 12, 2004; DOI 10.1182/blood-2004-02-0675.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Carlijn Voermans, Dr Paula van Hennik, and Mr Erik Mul for purification of CD45RO cells, and Dr Ellen van der Schoot and Dr Jaap Jan Zwaginga for helpful discussions.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal