Abstract
Follicular lymphoma (FL) is a disease characterized by a long clinical course marked by frequent relapses that vary in clinical aggressiveness over time. Therefore, the main dilemma at each relapse is the choice for the most effective treatment for optimal disease control and failure-free survival while at the same time avoiding overtreatment and harmful side effects. The selection for more aggressive treatment is currently based on histologic grading and clinical criteria; however, in up to 30% of all cases these methods prove to be insufficient. Using supervised classification on a training set of paired samples from patients who experienced either an indolent or aggressive disease course, a gene expression profile of 81 genes was established that could, with an accuracy of 100%, distinguish low-grade from high-grade disease. This profile accurately classified 93% of the FL samples in an independent validation set. Most important, in a third series of FL cases where histologic grading was ambiguous, precluding meaningful morphologic guidance, the 81-gene profile shows a classification accuracy of 94%. The FL stratification profile is a more reliable marker of clinical behavior than the currently used histologic grading and clinical criteria and may provide an important alternative to guide the choice of therapy in patients with FL both at presentation and at relapse. (Blood. 2005;105:301-307)
Introduction
Follicular lymphoma (FL) is a disease characterized by a long clinical course for most patients and marked by frequent relapses that vary in clinical aggressiveness over time. Transformation to diffuse large B-cell lymphoma (DLBCL) is a common event and occurs in approximately 30% to 85% of the patients.1-3 Primary treatment may vary from watchful waiting to high-dose chemotherapy with stem cell transplantation with the aim to reach long-lasting disease-free survival and in a small subset of patients possibly cure. At subsequent relapse, treatment is again stratified for clinical aggressiveness. Transformation and development of unresponsiveness to chemotherapy in the course of the disease are the main causes of death in patients with FL and in these situations more aggressive and possibly experimental therapy is justified. In individual patients and at each relapse the diagnosis of the phase of the disease (indolent or aggressive) is important for the choice of optimal therapy. Timing of the most optimal treatment is of particular importance for the benefit of patients because both overtreatment and undertreatment in relation to the clinical aggressiveness of the disease affect event-free survival and overall survival. Unnecessary or premature use of anthracycline is harmful with respect to quality of life and morbidity, and the use is limited by an absolute maximum tolerable dose. Conversely, withholding aggressive chemotherapy (in clinically aggressive disease) will result in insufficient therapy response with consequences for disease-free and overall survival, especially in the first line.
Thus far, morphologic subclassification has been used as a major guide in the choice of therapy in FL. In the World Health Organization (WHO) classification, FL is graded as 1, 2, 3a, and 3b according to the number of large transformed cells (centroblasts) per high-power field.4 Even though these criteria appear objective, grading has a notoriously poor reproducibility among community pathologists as well as experienced hematopathologists (agreement 61%-73%).5 This is due to the subjective nature of morphologic grading and the inherent inadequacy of the criteria set for “transformed cells.” The cellular morphology of transformed cells may be highly variable and the component of transformed cells may be heterogeneous within the biopsy specimen. Therefore, classical morphologic grading results in too many inconclusive cases (10%-30% of all cases) and is not optimally suitable as a guideline in the choice of therapy in daily practice. A major effort should be put into finding an alternative method with objective and reproducible parameters to judge clinical aggressiveness at any decision point for treatment in patients with FL. Clinical prognostic indices, such as the International Prognostic Index (IPI)6,7 and variations that are more tailored to FL (Follicular Lymphoma International Prognostic Index [FLIPI]),8,9 are in principle suitable markers of clinical course. However, the discriminative value for most patients is limited because most patients will fall in the low and low-intermediate risk groups10,11 due to the limitations and resolution of the system. Moreover, these indices are only formally validated for predictive value at diagnosis. Hence, other means of stratification would add significantly to guide the choice of therapy.
Here, we present a FL stratification gene expression profile of 81 genes that, with an accuracy of 93%, can distinguish between FLs that behave clinically indolent and those that behave aggressively at the time of biopsy, both at presentation and at relapse. Most important, this profile is also highly discriminative for biopsies with inconclusive morphologic features in which the pathologist cannot provide meaningful information (accuracy 94%). Therefore, it provides an important improvement in comparison to the currently available morphologic and clinical markers.
Patients, materials, and methods
Design of the study
The aim of this study was to search for a gene expression profile that can assess indolent versus aggressive clinical behavior both at diagnosis and at relapse for the disease episode at the moment of biopsy to replace current inadequate morphologic and clinical methods. Because adequate upfront criteria and gold standards for clinical behavior are not available, the complete course of the disease was evaluated retrospectively for all patients and each disease episode was defined as either indolent (nonaggressive) or aggressive. To qualify as a clinically nonaggressive or indolent disease episode, the following criteria had to be met: (1) absence of B symptoms, elevated level of lactate dehydrogenase (LDH), rapid generalized disease progression in the preceding 3 months, and (2) if treated, a good tumor response (partial or complete remission) after at least one of 2 non-anthracycline-containing, non-high-dose chemotherapy regimens (eg, chlorambucil, CVP [cyclophosphamide, vincristine, prednisone], or fludarabine).12 An aggressive disease episode was defined as development of B symptoms and LDH and/or rapid generalized disease progression within the preceding 3 months, and/or progressive disease during successive treatment with 2 or more non-anthracycline-containing, non-high-dose chemotherapy regimens (Table 1). In case of ambiguous clinical situations, the patient was not included in this study. Because FL is a genetically very homogeneous disease and the variation in expression patterns is within small limits, a supervised approach was chosen. A gene expression profile was developed in a set of paired samples and validated in a separate series of patients. Most important, the profile was tested in a third series of cases with ambiguous morphologic features.
Criteria for retrospective assessment of the actual clinical behavior of at any stage of the disease in patients with FL
Indolent clinical behavior . | Aggressive clinical behavior . |
|---|---|
| Slowly progressive or waxing and waning disease during watchful waiting over at least 3 mo | Rapid progression/enlargement of a tumor mass at one or more localizations within 3 mo |
| OR | AND/OR |
| On treatment: good tumor response (CR/PR)36 after treatment with CVP or chlorambucil or chlorambucil/prednisone or fludarabine in first or second line | Development of B symptoms and elevation of LDH level after previously normal levels in the preceding 3 mo |
| AND | AND/OR |
| Normal levels of LDH and no B symptoms* | Progressive disease during successive treatment with 2 or more of the following treatment protocols: CVP, chlorambucil, chlorambucil/prednisone, and fludarabine. |
Indolent clinical behavior . | Aggressive clinical behavior . |
|---|---|
| Slowly progressive or waxing and waning disease during watchful waiting over at least 3 mo | Rapid progression/enlargement of a tumor mass at one or more localizations within 3 mo |
| OR | AND/OR |
| On treatment: good tumor response (CR/PR)36 after treatment with CVP or chlorambucil or chlorambucil/prednisone or fludarabine in first or second line | Development of B symptoms and elevation of LDH level after previously normal levels in the preceding 3 mo |
| AND | AND/OR |
| Normal levels of LDH and no B symptoms* | Progressive disease during successive treatment with 2 or more of the following treatment protocols: CVP, chlorambucil, chlorambucil/prednisone, and fludarabine. |
CR indicates complete remission; PR, partial remission.
B symptoms included more than 10% weight loss within 6 months, night sweats, and fever more than 38†C.
Patient selection
Tumor samples from patients with primary nodal FL treated between 1984 and 2002 were selected from the fresh-frozen tissue banks of the Netherlands Cancer Institute, Central Laboratories Friesland, University Medical Center Nijmegen, Leiden University Medical Center, and Groningen University Medical Center according to the following criteria: availability of a representative frozen sample and paraffin-embedded tissue, proven diagnosis of FL, and availability of complete clinical data at presentation and during follow-up. The study was approved by the medical ethics committee of the Netherlands Cancer Institute. For patients whose diagnosis was made after 2002, informed consent to use human material for research purposes after prior use for diagnostic purposes was given for all patients in all participating hospitals.
A total of 106 samples from 80 patients were included in this study. Of these patients, the full medical history from diagnosis to last follow-up or death was evaluated retrospectively including data on IPI parameters,6 treatment, and treatment results. All relevant biopsy material was reviewed, including full immunohistochemical workup (CD20, CD3, CD5, bcl-2, bcl-6, CD10, CD21) by 3 hematopathologists (D.dJ., R.K., J.vK.), classified, and graded according to the WHO classification. A selection of clinical and histologic data is summarized in Table S1 (available on the Blood website; see the Supplemental Table link at the top of the online article). IPI parameters are grouped in a low/low-intermediate risk group for IPI score 0, 1, or 2, and a high-intermediate/high-risk group for IPI score 3, 4, or 5. For each patient, indolent and aggressive disease episodes were assigned in retrospect according to the criteria described (see “Design of the study”). It should be noted that for the retrospective clinical classification histologic grading was not included. We excluded all patients for whom due to lack of data the clinical episode of the available biopsy sample could not be reliably assigned to the indolent or aggressive phase of the disease.
Treatment varied and was administered according to local protocols at the time of diagnosis, including cyclophosphamide/vincristine/prednisone (CVP), fludarabine, chlorambucil, chlorambucil/prednisone, and cyclophosphamide/doxorubicin/vincristine/prednisone (CHOP-like) with or without radiotherapy. Different “indolent-type” treatment protocols have been shown to be equally effective in FL and do not result in different disease-free and overall survival.13,14 Because the aim of the study was to find a profile related to clinical behavior at the time of biopsy and not a survival-related or treatment response-related profile, variability in treatment will not influence our study results.
The patient samples were divided in 3 different groups. (1) Training series: Patients for whom paired samples corresponding to the concordant clinical/morphologic indolent and to concordant clinical/morphologic aggressive episodes were available; that is, combined indolent disease course and FL grade 1 or 24 versus aggressive episode and FL grade 3b or DLBCL.4 Twenty-four paired samples of both phases in 12 patients were used to build the classifier profile and to minimize patient-specific variation. Age at diagnosis of these selected patients ranged from 24 to 74 years (median, 49.5 years; median follow-up, 80 months; range, 29-304 months). Transformation to aggressive disease occurred with a median interval of 64 months (range, 22-288 months). (2) Validation series: 58 independent samples (54 patients) of clinical/morphologic indolent phases or clinical/morphologic aggressive phases. Age at diagnosis ranged from 27 to 78 years (median, 53 years). The median duration of follow-up was 71 months (range, 4-206 months). The aggressive-phase samples consisted of 18 patient samples, with a median interval to transformation of 36.5 months (range, 3-139 months). The samples of the indolent group consisted of 18 patient samples (median follow-up, 91 months; range, 24-68 months) as well as 22 samples from the indolent phases previous to transformation (median interval to transformation, 47 months; range, 21-168 months). (3) Validation (series) for difficult cases: Eighteen patient samples were selected that showed ambiguous morphologic features according to the review panel (D.d. J., R.K., H.v. K.) and were scored as inconclusive in the past; borderline between FL grade 2 and grade 3a or histology characterized by the dominant presence of small centroblast-like cells that fell outside the consensus criteria for classical “large transformed cells.”4
In addition, 6 patients were selected who presented with a grade 1 FL and clinically indolent behavior in the first biopsy, but showed histologic transformation to DLBCL and progression to clinically high-grade and aggressive disease within 10 months (median time to progression, 6.5 months; range, 3-10 months).
RNA isolation, amplification, labeling, and hybridization
Detailed protocols for RNA isolation, amplification, labeling, and hybridization can be found at http://www.nki.nl/nkidep/pa/microarray/protocols.htm. All samples were cohybridized with a standard reference of pooled and amplified RNA isolated from tonsillectomy specimens of 5 patients who underwent routine tonsillectomy for chronic ear/nose/throat infections. This tissue reference was chosen to provide a lymphoid reference containing all genes that are potentially expressed in the tumor tissues at a significant level and biologically closely related to the tumor samples to be able to identify small changes in expression levels between the tumor groups.
Microarray slides
Microarray slides were prepared at the central microarray facility (CMF) at the Netherlands Cancer Institute. Sequence-verified cDNA clones (Invitrogen, Huntsville, AL) were spotted onto poly-l-lysine-coated glass slides using the Microgrid II arrayer (Apogent, Cambridge, United Kingdom) with a complexity of 19 200 spots/slide. A complete list of genes and controls included on the slides is available on the CMF Web site (http://microarrays.nki.nl/download/geneid.html, http://microarrays.nki.nl/download/protocols.html), as well as details on the process of preparing the DNA for spotting and preparation of the slides.
Normalization
Unsupervised clustering
Gene clustering and tumor clustering were performed independently using agglomerative hierarchical clustering in the software program genesis.17 For gene clustering, complete linkage similarity metrics among genes were calculated on the basis of expression ratio measurement across all tumors. Similarly, for tumor clustering complete linkage clustering was calculated based on expression ratio measurements across all significant genes.
Supervised classification
To reliably discriminate between the indolent and aggressive tumors, the following supervised classification method was used to build the FL stratification classifier: (1) From the 19 200 genes on the microarray, genes that were significantly different from the reference in at least 2 tumors were selected (significance was based on P < .01 computed with the Rosetta error model16 ). (2) Calculation of the paired signal-to-noise ratio (SNR) for each of the selected genes, and ranking of the genes (top-ranked genes being genes that are best suited to separate indolent from aggressive samples). (3) Determination of top-ranked genes that separate the 2 classes best when used in a nearest prototype classifier.18 Steps 2 and 3 are performed in a cross-validation procedure, where at each cross-validation iteration a matched pair is left out and used to test the performance of the classifier trained on the remaining pairs.
The optimal number of genes that could separate molecular low-grade from molecular high-grade disease was determined in a leave-pair-out-cross-validation method. More specifically, in 12 repetitive steps, each paired sample was left out once, and for the remaining 22 samples, genes were ranked on the SNR. Then starting with the 4 most informative genes the classifier was trained on the 22 samples and used to predict the outcome of the left-out pair. The classification of the left-out pair was predicted on the basis of the largest Pearson correlation of the expression profile of each of the left-out samples with the mean expression levels of the remaining samples from the indolent or the aggressive patient samples for the selected reporter genes. Subsequently, the whole procedure was repeated by adding 1 gene at a time. Note that to avoid selection bias,19,20 that is, underestimation of the error rate, samples that were left out were not involved in any of the reporter selection steps.
The performance was measured as the average of the false-positive and false-negative rates of the left-out samples. No further increase in performance was observed when the number of included reporter genes exceeded 81. Because the ranking in every cross-validation iteration produces a slightly different set of marker genes, the final set of 81 classifier genes in the FL stratification classifier was determined by taking the 81 genes that occurred most frequently in each of the 12 steps.
The description of this study followed the Minimum Information About a Microarray Experiment (MIAME) guidelines issued by the Microarray Gene Expression Data group.21
Results
To provide an accurate and clinically relevant classification of FL, 106 specimens from 80 patients (Table S1) were analyzed for gene expression profiles using 19 200 cDNA microarrays.
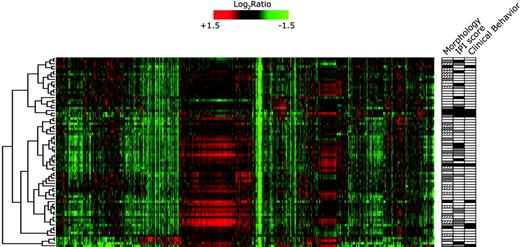
As a first step, unsupervised 2-dimensional hierarchical clustering was performed on 72 FL grade 1, 2, 3a, and 3b samples to group genes on the basis of similarity across all tumors and to group the tumor samples according to similarities in gene expression (Figure 1). This shows a relative homogeneity of FL as a single disease entity and shows a division in 3 main groups with no enrichment for morphologic grade or clinical behavior. It should be noted that multiple samples of single patients do not dominantly cluster together and both IPI score and treatment are not components in the clustering. Unsupervised clustering is by far insufficient for clinical use; therefore, a supervised classification approach was chosen.
Unsupervised clustering of 72 FL samples, grades 1, 2, and 3, shows a relative biologic homogeneity of FL. There is a separation in 3 main groups with no enrichment for morphologic grades or clinical behavior in either of the groups. Each row represents a tumor and each column a single gene. Gene expression is depicted according to the color bar. Red indicates a high level of mRNA expression relative to the reference and green indicates a low level of expression. Selected morphologic and clinical data are depicted in the right panel. For clinical features, □ indicates indolent and ▪ indicates aggressive disease behavior (Table 1 lists criteria). For morphologic grading, □ indicates grade 1; ▨, grade 2; and ▪, grade 3. For IPI, □ denotes scores 0, 1, or 2, and ▪, 3, 4, or 5. ▦ denotes insufficient data.
Unsupervised clustering of 72 FL samples, grades 1, 2, and 3, shows a relative biologic homogeneity of FL. There is a separation in 3 main groups with no enrichment for morphologic grades or clinical behavior in either of the groups. Each row represents a tumor and each column a single gene. Gene expression is depicted according to the color bar. Red indicates a high level of mRNA expression relative to the reference and green indicates a low level of expression. Selected morphologic and clinical data are depicted in the right panel. For clinical features, □ indicates indolent and ▪ indicates aggressive disease behavior (Table 1 lists criteria). For morphologic grading, □ indicates grade 1; ▨, grade 2; and ▪, grade 3. For IPI, □ denotes scores 0, 1, or 2, and ▪, 3, 4, or 5. ▦ denotes insufficient data.
Development of a molecular profile for low-grade and high-grade disease in FL on the basis of paired samples
To build a classifier a group of 24 paired samples from 12 patients was used for whom samples of both the clinically and morphologically indolent phase as well as the clinically and morphologically aggressive phase of the disease were available. Using the Rosetta error model,16 a total of 4760 mRNAs were found to be differentially expressed compared to the reference RNA (P < .01 in 2 or more samples). From these, a set of 81 genes emerged from the cross-validation procedure with the optimal classification performance of 100%.
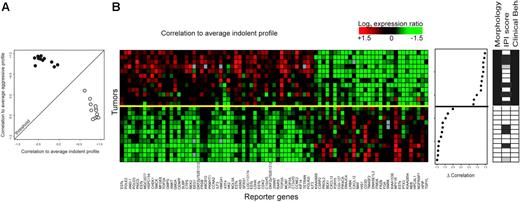
The correlation coefficients for each of the tumor samples with the average expression of these 81 genes in either low-grade or high grade samples in the cross-validation were calculated and are shown in Figure 2A. Tumors positioned above the threshold were classified as aggressive; tumors below the threshold were classified as low-grade. The line of equal correlation was chosen as threshold because misclassification in both directions (false positives and false negatives) would result in equally adverse treatment consequences for the patient.
Supervised classification of 12 paired lymphoma samples (training series). (A) Correlation plot of the FL stratification profile. Correlation of the expression profile of each tumor sample with the average expression profile of all indolent samples is depicted on the horizontal axis, and the correlation of the expression profile of each tumor with the average expression profile of all aggressive tumor samples is shown on the vertical axis. Tumors classified above the threshold are classified as molecular high-grade; tumors below the threshold are classified as molecular low-grade. (B) Expression data matrix of 81 marker genes from tumors of the indolent as well as the aggressive phase of 12 patients with FL. Each row represents a tumor and each column a gene. Genes are ordered on the basis of their SNR. Tumors are rank ordered according to the difference in correlation with the average high-grade profile and the correlation with the low-grade profile (middle panel). The solid yellow line is the classifier with optimal accuracy; patients above the yellow line have an aggressive disease course; below the yellow line, they have an indolent disease course at the disease episode at the time of biopsy. Selected clinical data are shown in the right panel. For morphologic data, □ indicates FL grade 1 or 2; ▪ indicates FL grade 3b or DLBCL. IPI scores and clinical behavior at time of biopsy are as described in Figure 1.
Supervised classification of 12 paired lymphoma samples (training series). (A) Correlation plot of the FL stratification profile. Correlation of the expression profile of each tumor sample with the average expression profile of all indolent samples is depicted on the horizontal axis, and the correlation of the expression profile of each tumor with the average expression profile of all aggressive tumor samples is shown on the vertical axis. Tumors classified above the threshold are classified as molecular high-grade; tumors below the threshold are classified as molecular low-grade. (B) Expression data matrix of 81 marker genes from tumors of the indolent as well as the aggressive phase of 12 patients with FL. Each row represents a tumor and each column a gene. Genes are ordered on the basis of their SNR. Tumors are rank ordered according to the difference in correlation with the average high-grade profile and the correlation with the low-grade profile (middle panel). The solid yellow line is the classifier with optimal accuracy; patients above the yellow line have an aggressive disease course; below the yellow line, they have an indolent disease course at the disease episode at the time of biopsy. Selected clinical data are shown in the right panel. For morphologic data, □ indicates FL grade 1 or 2; ▪ indicates FL grade 3b or DLBCL. IPI scores and clinical behavior at time of biopsy are as described in Figure 1.
The expression pattern of the 81 genes in the 24 learning samples is shown in the color plot of Figure 2B, where the tumors are ranked according to the difference in correlation with the average high-grade profile and the correlation with the low-grade profile (middle panel).
Expression data were associated with data on clinical and histologic parameters (histologic grading, IPI score, and clinical classification; Figure 2B right panel) and correlated perfectly with histologic grading and clinical behavior. This is not unexpected because the tumors were selected for unambiguous morphologic features concordant with clinical behavior. Importantly, IPI score was not found to be a very strong discriminative factor for clinical behavior.
The FL stratification profile (Figure 2B) contains genes significantly up-regulated in the aggressive phase of the disease that are involved in cell cycle control (eg, CCNE2, CCNA2, CDK2, CHEK1, MCM7) and DNA synthesis (eg, TOP2A, POLD3A, HMGA1, POLE2, GMPS, CTPS) as well as genes reflecting increased metabolism (FRSB, RARS, HK2, LDHA). Genes involved in signal transduction reflecting activation of several signaling pathways are differentially expressed (eg, FRZB, HCFCR1, PIK4CA, MAPK1). Genes derived from the reactive infiltrate of T cells and macrophages (CD3D, CXCL12, TM4SF2) were up-regulated in the indolent phase of the disease as expected from immunohistochemical data from corresponding paraffin-embedded material of all samples, showing 40% to 70% T-cell infiltration in FL and 20% to 50% T-cell infiltration in DLBCL.
Validation of the FL stratification profile in an independent series of transformed FL
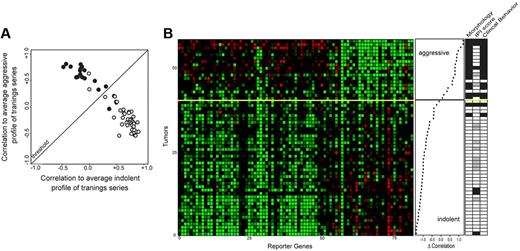
To validate the classifier, an additional independent set of 58 FL samples was investigated: 40 morphologic/clinical indolent samples and 18 samples from the morphologically/clinically aggressive phase of the disease. Like the training series, cases were selected for concordant histologic grade and clinical behavior. The gene expression ratios of the 81 genes for these tumors and their correlation coefficients of the tumor samples with the average expression of these 81 genes with either indolent or aggressive samples of the training series are shown in Figure 3. The 18 samples with values above the threshold were assigned to the high-grade category; the 40 samples below the threshold were assigned to the low-grade category. When compared to morphologic and clinical data (Figure 3B), this resulted in 4 incorrect of 58 classifications: 2 false negatives, that is, morphologically/clinically aggressive samples classified as molecular low-grade and 2 false positives, that is, morphologically/clinically indolent classified as molecular high-grade (93% accuracy).
Correlation and pattern of expression of genes used to determine the clinical characteristics of 58 lymphoma samples (independent validation series). (A) Correlation plot of the FL stratification profile. Correlation of the expression profile of each tumor sample with the average expression profile of all low-grade samples of the training series is depicted on the horizontal axis, and the correlation of the expression profile of each tumor with the average expression profile of all high-grade tumor samples of the training series is shown on the vertical axis. Tumors classified above the threshold are classified as molecular high-grade; tumors below the threshold are classified as molecular low-grade. (B) Expression data matrix of 81 marker genes from tumors of the indolent as well as the aggressive phases of patients with FL. Each row represents the FL stratification profile for one tumor. Each column represents the relative expression of one gene. The genes in the horizontal direction are arrayed in the same order as in Figure 2B. Tumors are rank-ordered according to the difference in correlation with the average aggressive profile and the correlation with the low-grade profile of the training set (middle panel). The yellow line is the threshold as determined in Figure 2 (see Figure 2 for color scheme). In the panel on the right morphologic and clinical features are shown of the samples similar to features shown in Figure 2B. Of the 58 samples, 4 were misclassified (93% accuracy).
Correlation and pattern of expression of genes used to determine the clinical characteristics of 58 lymphoma samples (independent validation series). (A) Correlation plot of the FL stratification profile. Correlation of the expression profile of each tumor sample with the average expression profile of all low-grade samples of the training series is depicted on the horizontal axis, and the correlation of the expression profile of each tumor with the average expression profile of all high-grade tumor samples of the training series is shown on the vertical axis. Tumors classified above the threshold are classified as molecular high-grade; tumors below the threshold are classified as molecular low-grade. (B) Expression data matrix of 81 marker genes from tumors of the indolent as well as the aggressive phases of patients with FL. Each row represents the FL stratification profile for one tumor. Each column represents the relative expression of one gene. The genes in the horizontal direction are arrayed in the same order as in Figure 2B. Tumors are rank-ordered according to the difference in correlation with the average aggressive profile and the correlation with the low-grade profile of the training set (middle panel). The yellow line is the threshold as determined in Figure 2 (see Figure 2 for color scheme). In the panel on the right morphologic and clinical features are shown of the samples similar to features shown in Figure 2B. Of the 58 samples, 4 were misclassified (93% accuracy).
Performance of the FL stratification profile in histologically difficult cases
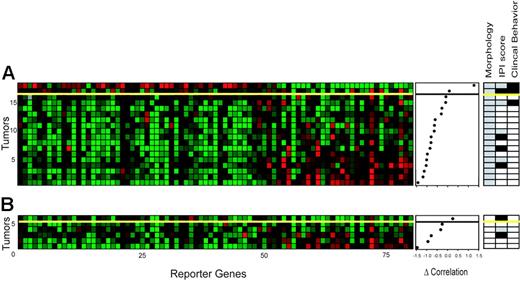
To further confirm the performance of the FL stratification classifier, 18 diagnostically difficult cases with ambiguous morphologic features were selected that precluded a meaningful decision (based on grade) by the pathologist; clinical behavior was determined in retrospect as in the training and validation series. The actual clinical course as predicted by the FL stratification classifier revealed 2 samples classified as high-grade and 16 samples as low-grade. One of these samples was incorrectly classified; this patient was clinically assigned as having aggressive disease, whereas the FL stratification classifier predicted low-grade disease (Figure 4A). Of these 18 patients, IPI scores at the time of biopsy indicated low/low-intermediate risk in 10 patients (Figure 4A, right panel; □) and high/high-intermediate risk in 4 patients (▪); in 4 patients no scores could be assigned due to lack of data (▦).
Pattern of gene expression of genes used to determine the clinical characteristics of 24 FL samples (test series). (A) The expression data matrix of 18 tumor samples with an unclear morphology across the 81 gene profile. The genes in the horizontal direction are arrayed in the same order as in Figure 2B. Tumors are rank ordered according to the difference in correlation to the molecular high-grade and low-grade profile as in Figure 2 (middle panel). The yellow line is the threshold as determined in Figure 2B (see Figure 2 for color scheme). Morphologic data, IPI scores, and clinical behavior are shown on the right. □,  , and ▪ are as indicated in Figure 1B. Of the 18 samples, the classifier predicted the actual clinical behavior correctly in 17 patients (accuracy 94%). (B) Expression data of 6 tumor samples from patients with unequivocal morphologic and clinical indolent disease, but who presented with full-blown aggressive disease within 10 months. One sample was classified as molecular high-grade.
, and ▪ are as indicated in Figure 1B. Of the 18 samples, the classifier predicted the actual clinical behavior correctly in 17 patients (accuracy 94%). (B) Expression data of 6 tumor samples from patients with unequivocal morphologic and clinical indolent disease, but who presented with full-blown aggressive disease within 10 months. One sample was classified as molecular high-grade.
Pattern of gene expression of genes used to determine the clinical characteristics of 24 FL samples (test series). (A) The expression data matrix of 18 tumor samples with an unclear morphology across the 81 gene profile. The genes in the horizontal direction are arrayed in the same order as in Figure 2B. Tumors are rank ordered according to the difference in correlation to the molecular high-grade and low-grade profile as in Figure 2 (middle panel). The yellow line is the threshold as determined in Figure 2B (see Figure 2 for color scheme). Morphologic data, IPI scores, and clinical behavior are shown on the right. □,  , and ▪ are as indicated in Figure 1B. Of the 18 samples, the classifier predicted the actual clinical behavior correctly in 17 patients (accuracy 94%). (B) Expression data of 6 tumor samples from patients with unequivocal morphologic and clinical indolent disease, but who presented with full-blown aggressive disease within 10 months. One sample was classified as molecular high-grade.
, and ▪ are as indicated in Figure 1B. Of the 18 samples, the classifier predicted the actual clinical behavior correctly in 17 patients (accuracy 94%). (B) Expression data of 6 tumor samples from patients with unequivocal morphologic and clinical indolent disease, but who presented with full-blown aggressive disease within 10 months. One sample was classified as molecular high-grade.
To evaluate the notion that the 81-gene profile reflects the actual clinical behavior at the time of biopsy rather than predicts future transformation, an additional set of 6 patients was selected who had morphologically evident indolent, grade 1 disease in their biopsy samples and had indolent disease course at the time of the biopsy, but who presented with full-blown morphologically and clinically aggressive disease within a period of 10 months. The classifier marked 5 of 6 cases as low-grade disease in support of this hypothesis.
Discussion
We have developed an FL stratification gene expression profile to assess the actual clinical behavior in FL to replace current insufficient morphologic and clinical methods. The classifier of 81 genes was developed in a paired-samples series of 12 patients with FL whose disease later transformed to aggressive disease to discriminate at 100% accuracy. In an independent validation series of 40 patients in a morphologic/clinical indolent disease phase and 18 patients with a morphologic/clinical aggressive disease phase, the molecular profile showed an accuracy of 93%. Only samples with unequivocal morphologic features of indolent FL (FL grade 1 or 2) and aggressive disease (FL grade 3b or DLBCL morphology) were included, to select for an expression profile that is optimized for genes that are involved in clinically relevant transformation to the aggressive phase of the disease. The profile would therefore be most relevant in the stratification of FL. This profile applies to FL and transformed FL including DLBCL, but excludes de novo DLBCL. It should be noted that de novo DLBCL represents a different disease. Prognostic profiles for this disease have been developed by Staudt and coworkers22,23 and Shipp and coworkers.24
The selection of the morphologic spectrum of samples used in the training and the validation set (FL grade 1 and 2, and FL grade 3b and DLBCL) has a good reproducibility among hematopathologists and accounts for the significant correlation of morphologic stratification and clinical course in large series.25 However, in practice, the largest problem is in borderline cases that show heterogeneity with respect to the number of large transformed cells or that contain blastlike cells with a variant morphology. In these patients, whose tissues represent approximately 10% to 30% of all FL biopsies, the pathologist cannot provide meaningful information on which to base treatment strategies. In view of the frequent relapsing nature of the disease, histologic grading problems are encountered in the far majority of the patients. We assumed that the genes selected in a profile that was based on the extremes of the spectrum of FL would also be of predictive relevance in the morphologically “gray zone.” Our molecular profile proves its value mostly in this group of FL, as was supported by the findings in a test series containing this type of ambiguous case, in which our FL stratification profile could adequately assess the clinical behavior at the disease episode at the moment of biopsy in 17 of 18 patients. Notably, of the 16 patients classified as having molecular low-grade disease, 5 were overtreated with aggressive therapy if currently used methods to guide therapy are compared to the stratification profile (data not shown). IPI scores in these patients showed mainly low and low-intermediate risk scores that would not add significantly as a guide in the choice of therapy. Our gene expression-based profile outperforms IPI classification in all cases. Especially in difficult cases, the profile outperforms histologic grading and significantly adds to the meaningful stratification of patients with FL and prevents patients from unnecessary aggressive treatment.
A control group of 6 patients with clinically and morphologically indolent disease at the time of biopsy presented with clinically aggressive disease and overt transformation to DLBCL at another localization within 10 months. Five of 6 tumors showed a low-grade gene expression profile. This is not unexpected because the FL stratification profile was not developed to predict the risk for the development of aggressive behavior/transformation nor to predict survival, but only to assess the clinical behavior at the time of biopsy. This is an important, but quite different, question for which it remains to be shown if the molecular reflection of the risk for transformation is an inherent property of the lymphoma ab initio, as has been shown for metastatic behavior in breast cancer,18,26 or is a feature that is only present once actual transformation has occurred. In the latter case, sampling error will remain a confounding factor also in molecular analysis. This may be an explanation for the misclassified cases in our validation series.
Functional annotation for the 81 genes in the FL stratification classifier and in the full set of genes that are differentially expressed in indolent and aggressive disease may provide more insight in the biologic mechanisms underlying transformation in FL. Genes involved in cell cycle control and DNA synthesis and metabolism were significantly up-regulated in the aggressive phase of the disease as expected. Three genes of the molecular profile, CXCL12, which is involved in signaling transduction and NEK2, which is involved in mitotic regulation, and MAPK1 have also been described by others to be differentially expressed in transformed FL.27-29 Other genes that have previously been described to be involved in transformation of indolent lymphomas (CD69, DNA polymerases, WEE1, HMGA1, RAS pathway genes23,24,27,28,30 ) and as single prognostic markers in aggressive lymphomas (eg, survivin/BIRC5,31 LDHA,7 and c-MYC32 ) are only present in the top 1000 ranked genes that are differentially expressed in the indolent and the aggressive phase of the disease, but as such are not a part of the 81-gene classifier. MYC, as a known oncogene, was found up-regulated on transformation, as also reported by others and may be implicated as a direct transforming factor. This may be suggested by concomitant up-regulation of known MYC-target genes as SFRS7, LDHA, MTHFD1, NME1, MSH2, and CKS2 and down-regulation of CDKN1B.30,33 The higher density of the T-cell infiltrate in low-grade FL as compared to high-grade disease is reflected by several T cell-related genes (CD3, CD2, CD69). However, genes related to T-cell and macrophage activation including several chemokine receptors (CCR1, CCL3, CCL5, CCL8, AKAP12, ILF3, GEM) are significantly up-regulated on transformation, suggesting an important biologic role. Notably, specific antagonists to several of the above-mentioned chemokine receptors are available and offer an attractive possibility for therapeutic interventions34 as do other individual gene products that are differentially expressed in the aggressive phase for which specific antagonistic and agonistic compounds have been developed (eg, to the negative regulatory lysophosphatidic acid receptor Edg-235 ).
How to proceed toward clinical practice? Because the number of genes that comprises the molecular profile is limited, development of diagnostic assays based on custom-made mini-chips or multiplex polymerase chain reaction for use in clinical practice might be feasible.
Our data indicate that on the basis of this FL stratification profile, we can now more reliably stratify FL for low-grade and high-grade disease than by morphologic grading and clinical prognostic parameters. The FL stratification profile provides an important improvement and may be an essential aid to guide the choice of therapy in patients with FL to provide optimal disease control, optimal failure-free survival, and quality of life. The actual impact on patient care should be evaluated in prospective clinical trials.
Prepublished online as Blood First Edition Paper, September 2, 2004; DOI 10.1182/blood-2004-06-2298.
Supported by grants from the Netherlands Cancer Institute.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to acknowledge Koos van der Hoeven, Gert Jan Timmers, Gustaaf van Imhoff, John Raemaekers, Renee Barge, Wilma Smit, Charlotte van Iperen, Harmen van Kamp, Gerhard Woolthuis, Rutger Jan Vermeer, Jose Zijlstra, the pathologists of the Boven IJ Hospital, Amsterdam and Zuiderzee Hospital, Lelystad for providing samples and clinical data; Tony van de Velde and Els Willemse for managing medical records data; Mike Heimerix, Marcel Daemen, and Arenda Schuurman, Niels Bakx for preparation of the microarray slides; and Guus Hart, Ed Schuuring, Petra Nederlof, Bas Kreike, Britta Weigelt, Huib Storm, Ed Roos, Jacques Neefjes, and Erwin van Montfort for helpful suggestions and stimulating discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal